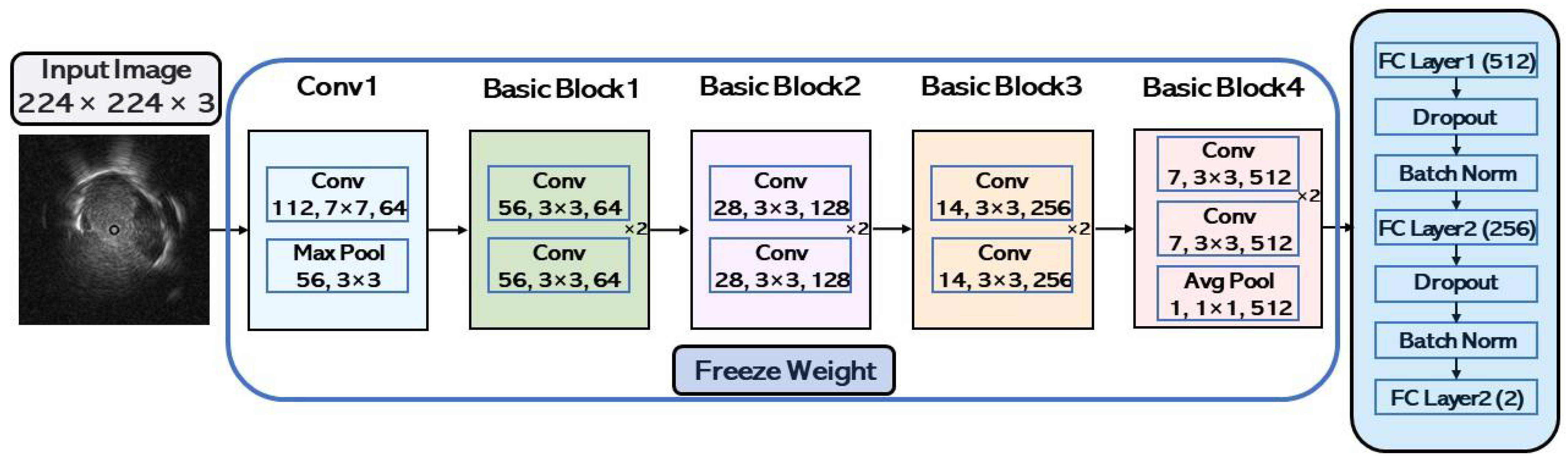
1. Introduction
In accordance with GLOBOCAN 2020 data, which encompasses cancer incidence and mortality estimates compiled by the International Agency for Research on Cancer, the global tally for new cancer cases in 2020 amounted to approximately 19.3 million, with a corresponding death toll of 10.0 million (excluding nonmelanoma skin cancer, the figures stand at 18.1 million new cases and 9.9 million deaths). Among these cases, lung cancer held the second position in terms of incidence, accounting for 11.4% of all reported new cases. However, it claimed the highest number of lives, accounting for 18% of the total deaths [
1]. Lung cancer remains the utmost proponent of cancer death worldwide, with a mere 15% overall five-year survival rate. Most cases are diagnosed at an advanced stage when treatment outcomes are not ideal and newly diagnosed patients develop locally advanced or metastatic diseases [
2]. Thus, early diagnosis is imperative for propelling lung cancer survival rates, successful treatment, and management. Unfortunately, early lung cancer detection methods are currently insufficient.
A thoracic radiograph (TR), commonly known as a chest X-ray (CXR), serves as the primary modality employed in the initial assessment of individuals with suspected lung malignancy. Its widespread accessibility, technical feasibility, minimal risk, and cost-effectiveness contribute to its frequent utilization in this context. The biopsy is another prominent clinical method for lung cancer diagnosis, but taking a biopsy from peripheral pulmonary nodules is notably challenging for physicians. Although computed tomography (CT)-guided percutaneous biopsies are often preferred, pneumothorax, bleeding, and other procedure complications are sometimes problematic. Transbronchial biopsy is customary in Japan, with several steps to ensure a high diagnostic yield: (1) accurately reach the target, (2) confirm the target lesion, (3) collect the biopsy sample from the target, and (4) adequate sampling from the target. In addition, rEBUS technology aids in confirming that the device has reached the target. However, a successful sampling is not always guaranteed because rEBUS is not a real-time guided biopsy procedure. Nonetheless, rEBUS’s image analysis will facilitate additional sampling during the biopsy procedure and treatment recommendations.
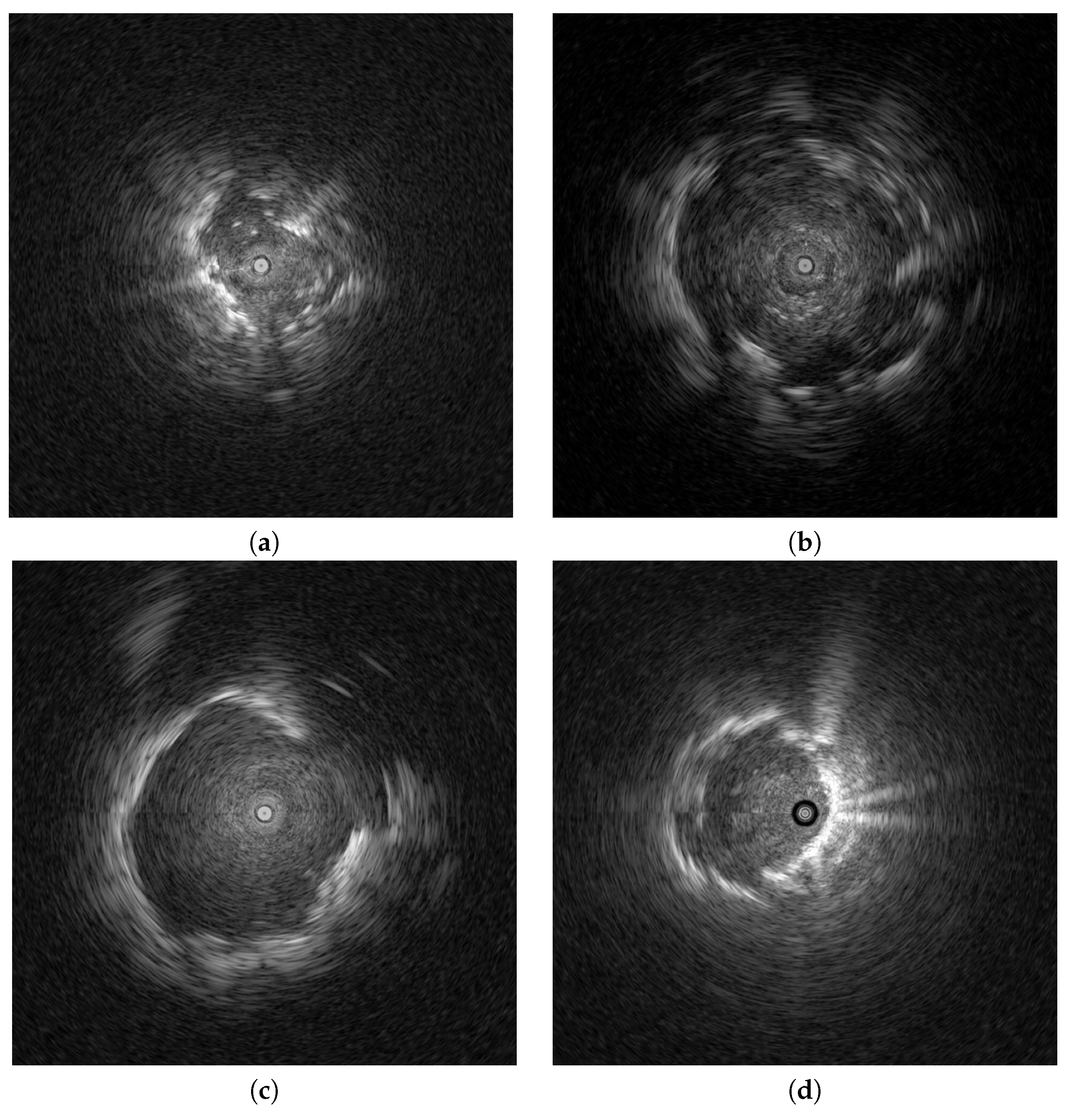
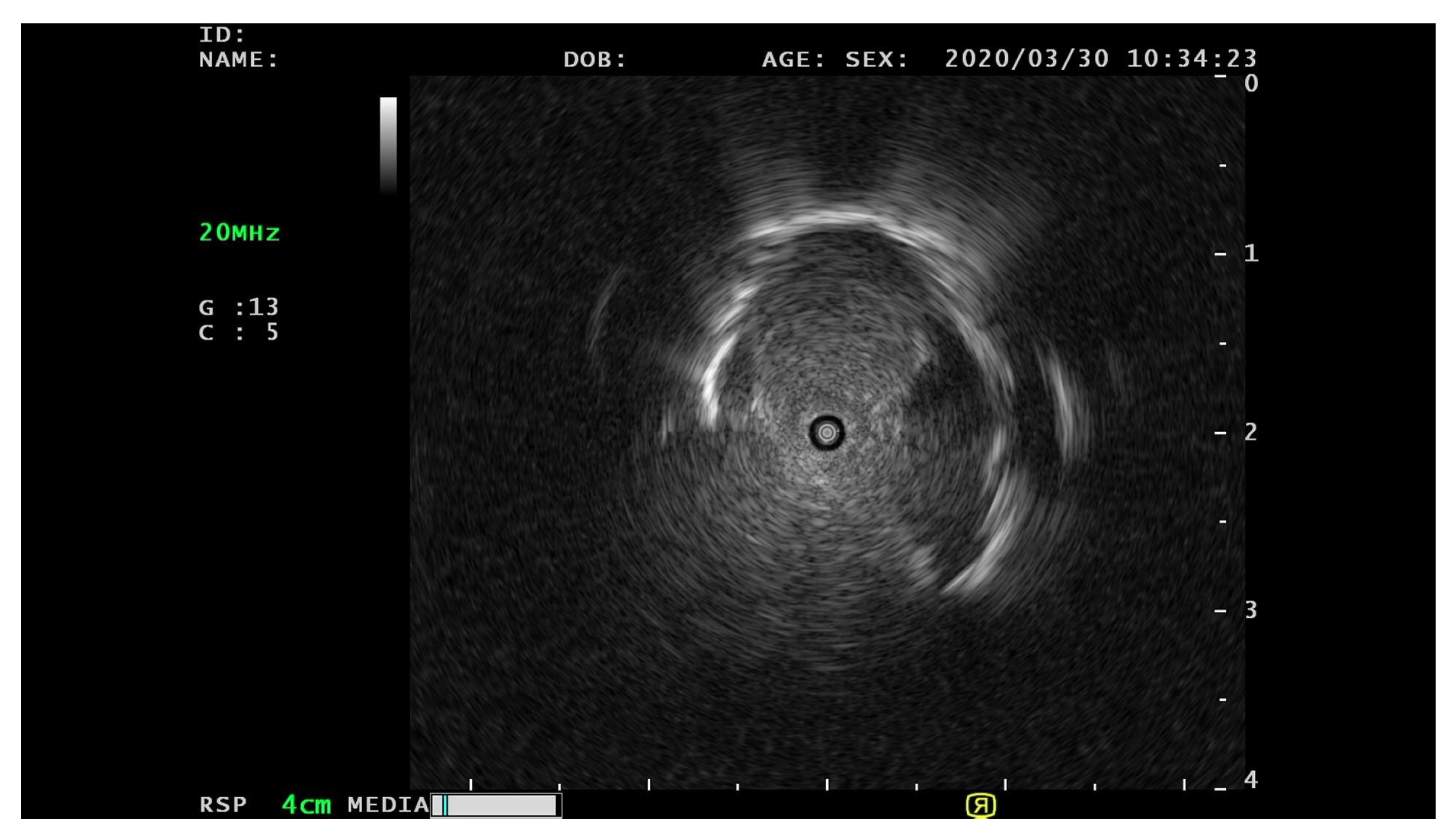
Endobronchial ultrasonography (EBUS) has recently been used to screen for peripheral pulmonary lesions. This radiation-free medical technique uses ultrasound and real-time scanning to diagnose and stage lung diseases, incorporating an endoscope to access the lungs through the airways. It provides high-resolution images for real-time lung tissue and surrounding structure visualizations. EBUS is routinely employed for lung nodule evaluations, lung cancer staging, and various lung disease diagnoses, enabling physicians to make informed decisions regarding treatment options. Clinical studies have attempted to distinguish peripheral pulmonary lesions (PPLs) from EBUS images [
3,
4,
5,
6]; however, benign and malignant PPL distinctions are subjective to physician experience and vex even the most experienced physicians. With current artificial intelligence (AI) advancements, AI technology has been increasingly integrated into the medical field. Many computer-aided diagnoses (CAD) have been presented to assist physicians through medical image processing.
Khomkham et al. [
3] employed two radiomics features, namely the adaptive weighted sum of the upper triangular gray-level co-occurrence matrix (GLCM) and the adaptive weighted sum of the lower triangular GLCM, to visualize the classifications of peripheral pulmonary lesions using EBUS images. The classification of these lesions was performed using the genetic algorithm in conjunction with a support vector machine (SVM) [
7] approach. Within the following year, a three-branch classification framework was proposed for differentiating PPLs. The radiomics feature and patient data are used in Branch 1 to train the random forest classifier [
8]. In Branch 2, convolutional neural network (CNN)-based models are prepared with EBUS images. Lastly, multi-patch EBUS images are selected from the original EBUS images to train the multi-patch-based model in Branch 3. Thus, three models are aggregated using the weighted ensemble strategy. Chen et al. [
4] proposed a CAD system that automatically differentiates benign and malignant PPLs for early lung cancer detection with EBUS images. Notably, this CAD system incorporates the transformer learning technique, and rotation and flipping augmentation methods are also included in the training dataset to avoid overfitting. CNN models extract features, which the SVM then classifies.
Jinsa et al. [
9] also designed a computer-aided classification method to differentiate lung cancer using CT images from an artificial neural network (ANN). Statistical parameters, such as mean, standard deviation, skewness, kurtosis, fifth central moment, and sixth central moment, are used for classification. Alakwaa et al. [
10] introduced a CAD system for lung cancer classification utilizing CT scans and a dataset from the 2017 Kaggle Data Science Bowl. In this two-stage system, a three-dimensional convolutional neural network (3D-CNN) classifies CT scans as lung cancer positive or negative. First, the modified U-Net detects nodule candidates in CT images of segmented lungs. Then, the regions where most nodule candidates are likely located are fed into a 3D-CNN to provide a final classification result. Alternatively, Chaunzwa et al. [
11] employed a radiomics methodology to predict the histology of non-small cell lung cancer tumors using standard-of-care computed tomography (CT) data.
Moreover, a comparable discriminative performance yielded reliable results by implementing machine learning classifiers such as k-nearest neighbor and SVM for CNN-derived quantitative radiomics features. Their best-performing CNNs functioned as effective probabilistic classifiers in a heterogeneous test set, providing qualitatively interpretable prediction visuals and proving that deep learning-based radiomics can identify histological lung cancer phenotypes. However, early-stage benign and malignant nodules’ remarkable similarity hinders accurate detection and classification. Therefore, Nasrullah et al. [
12] developed an automated framework for detecting and classifying lung nodules by utilizing two deep 3D customized mixed-link network (CMixNet) architectures. This framework aims to address the challenges associated with lung nodule analysis. For the nodule detection task, the faster R-CNN algorithm is employed to leverage efficiently-learned features obtained from the CMixNet architecture and a U-Net-like encoder-decoder architecture. On the other hand, the nodule classification task utilizes a gradient boosting machine (GBM) that operates on the learned features extracted from the designed 3D CMixNet structure. To minimize false positives and misdiagnosed results stemming from various error types, the final decision incorporates physiological symptoms and clinical biomarkers.
Chen et al. [
13] conducted a study focusing on the diagnosis of lung cancer using deep attention-based multiple-instance learning (MIL) and radiomics features extracted from CT scan images. Their research approach treated the task of lung cancer diagnosis as a multiple-instance learning problem; radiomics features were selected as the input features source, and deep attention-based MIL was responsible for classification. The attention mechanism improved interpretability by estimating each instance’s value for the final diagnosis set. Although CAD systems have proven remarkable efficacy in lung cancer diagnosis, an ongoing challenge is the need for balanced datasets to obtain satisfactory results with most existing techniques. Recognizing that an imbalanced dataset can significantly impact a machine learning model’s performance and accuracy is crucial. Models tend to favor the predominant class when an uneven class distribution is apparent in a dataset, which decreases sensitivity for minority class detection.
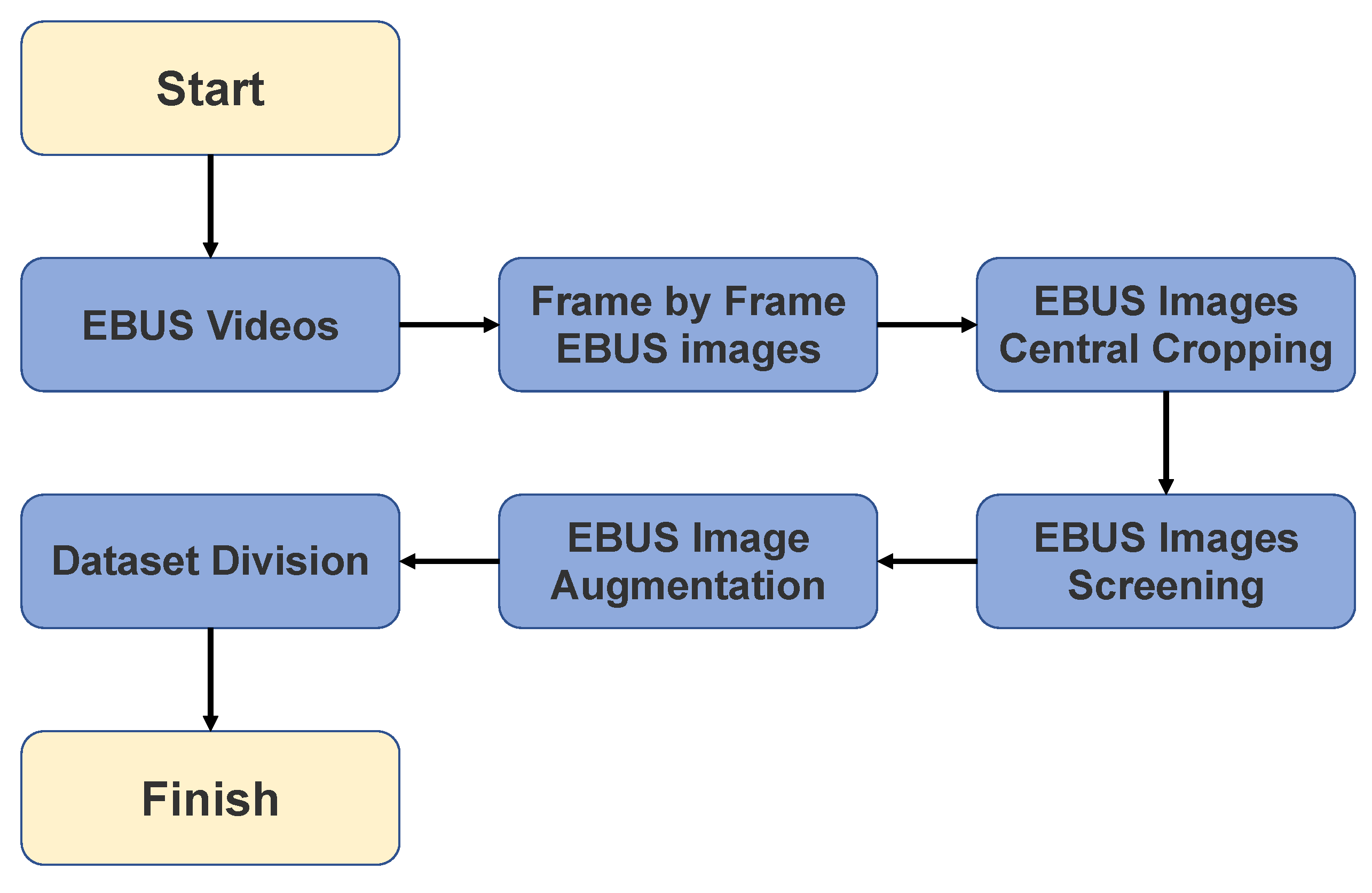
This paper presents a pulmonary lesion classification framework that combines down-sampling and ensemble techniques; the down-sampling technique alleviates the imbalanced dataset, and ensemble learning improves CAD system performance. The ensemble method is pivotal for achieving better results in various machine-learning tasks as it combines multiple models, each trained with distinct initial conditions or algorithms. By aggregating these diverse models’ predictions, the ensemble technique can capture an expansive range of patterns for more accurate predictions. The proposed work has the following contributions:
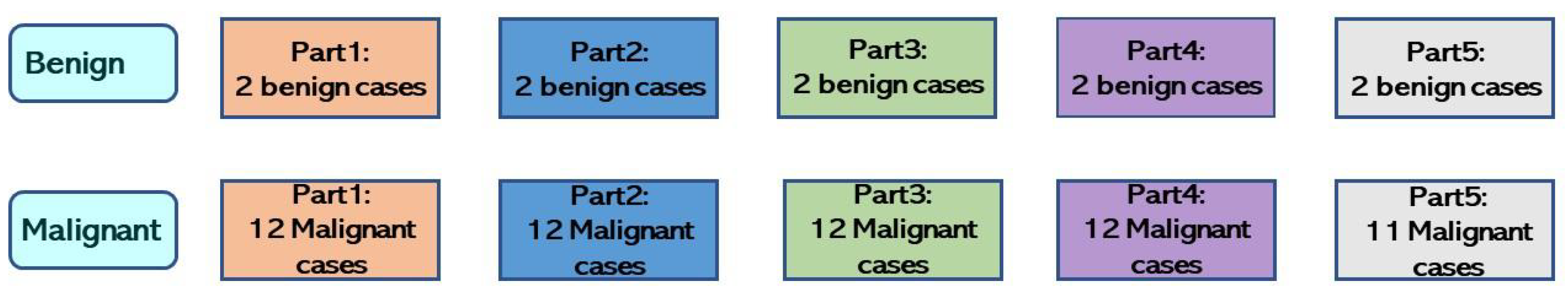
The CAD system implements a down-sampling technique that alleviates repercussions from imbalanced data to improve performance. Every benign case in the training set was used during the model’s training phase, whereas malignant cases were down-sampled and averaged out.
If only malignant cases are down-sampled, the training dataset will not be fully utilized, and data will be wasted; therefore, the CAD system harnesses an ensemble learning technique. All trained models are integrated to combine all benign and malignant dataset features for the final classification.
5. Discussion and Conclusions
5.1. Discussion
This study incorporated bagging ensemble and down-sampling techniques to overcome the hurdles from an imbalanced dataset and achieve better performance. The down-sampling techniques and asymmetric data augmentation used are known to mitigate the imbalanced dataset’s impact on the experimental results. As the number of benign cases was considerably less than malignant cases, four data augmentation methods were implemented for benign cases, and two were used for malignant cases. According to the experimental results, all ensemble results were better than the baseline result. <in addition, we explored various same and different model combinations and integrated two or three models while considering the amount of data available for each model. We employed five-fold cross-validation to evaluate performance, as this assessment determines a model’s generalization ability, revealing that each fold’s results fluctuated. This variation in results across folds can be influenced by training and validation data split compositions, the dataset’s inherent variability, and the models’ sensitivity to different data subsets.
All bagging ensemble experiment result are shown in
Table 5,
Table 6 and
Table 7. Three fusion methods were used in the bagging experiment.
Table 5 displays the bagging experimental results with the majority voting fusion method, and
Table 4 presents the baseline experimental results. Even with data augmentation to alleviate the imbalanced dataset’s impact, the ResNet-34 backbone did not perform well. One explanation may be that the number of benign cases was insufficient; both baseline and bagging ensemble results evidenced that the PPV was better than the NPV. These findings indicate that trained CNN models can more accurately differentiate malignant cases than benign ones. Notably, the number of malignant cases was much larger than the benign cases in this study; thus, the trained CNN models acquired more knowledge regarding malignant cases than benign ones. Additional benign case data should be collected for future investigations.
Table 6 shows the bagging results with the output average fusion method. The output average fusion method yielded the best results within the ensemble experimental results of three fusion methods and aggregating the same CNN models. A ResNet-18 backbone and two-CNN model combination revealed good results: 0.70 accuracy, 0.63 F1-Score, 0.70 AUC, 0.87 PPV, 0.67 NPV, 0.56 sensitivity, and 0.84 specificity.
Table 7 displays the bagging results with the performance weighting fusion method. When aggregating two or three same CNN models, the ResNet-18, ResNet-34, DenseNet-121, and MobileNet-V2 accuracies were remarkably similar. Among all experimental results, a ResNet-18 backbone and the performance weighting fusion method yield the best specificity value, as shown in
Table 7.
Based on the comparison results between the best baseline and bagging ensemble results (
Table 8), the benign and malignant PPL classification improved. This study conducted five-fold cross-validation to evaluate the proposed CAD system’s performance further. The dataset division results are presented in
Table 3. Although the CNN models performed well in some folds, this was not the case for others. This finding reveals that the data distribution of the five-fold dataset was not the same, as using the same CNN model backbone indicated significant differences between results. Another concern involves using pre-trained CNN models for feature extraction. The training results were considerably better than the validation and testing results, potentially due to the CNN models’ depth. In addition, compared with ImageNet images, EBUS is more practical. Deep CNN models are not necessary for feature extraction; therefore, we intend to construct shallow convolutional layers for feature extraction in the future. In addition to using the fully connected layers, we will explore SVM, decision trees, and random forests as classifiers.
Benign or malignant PPLs cannot be determined solely based on EBUS images in clinical practice, as the image features cannot be adequately observed. Thus, a biopsy is often performed to distinguish the PPL type. Despite the developed CAD system’s remarkable performance, there were some limitations. Our study assessed six models and utilized ensemble learning to improve performance, specifically integrating three distinct models. However, numerous excellent deep learning and machine learning algorithms can still be applied to our study. For example, we will explore the fusion of machine learning and deep learning algorithms in our future experiments. We also intend to ensemble a CNN with other state-of-the-art models. Another hurdle was the limited sample size in this study, which impedes our CAD framework’s reliability and performance generalization. External validation is required to address this issue in the future. We will investigate how our system performs when provided data from different hospitals or machine types to validate our proposed model’s reliability and practicality further. This study referenced investigations conducted by other researchers to support our findings and provide a broader context for our research. As such, we also aim to establish a comparison with these references to validate their results using our dataset and further evaluate our model. Lastly, a shallow CNN model should be designed to compare performance with transfer learning.
5.2. Conclusions
This study developed a computer-aided diagnosis system to differentiate between benign and malignant lung cancer cases from EBUS images. Bagging ensemble learning and down-sampling techniques were applied to our CAD system to resolve an imbalanced dataset and enhance results. As there were far more malignant than benign cases, the malignant cases were down-sampled to consider the entire dataset. Malignant cases were equally divided into three, and each was combined with all the benign cases to create three small datasets. Models were trained with each small dataset and aggregated with the bagging ensemble learning method. Thus, the three trained models contained knowledge for reliable benign and malignant classification. In addition, the three fusion methods were majority voting, output average, and performance weighting. The performance weighting fusion method provided the best bagging ensemble result, achieving a 0.7 accuracy, 0.63 F1-Score, 0.75 AUC, 0.84 PPV, 0.68 NPV, 0.56 sensitivity, and 0.85 specificity.
In conclusion, our proposed computer-aided diagnosis system based on deep learning achieved promising results for diagnosing benign and malignant peripheral pulmonary lesions from endobronchial ultrasonography. However, the dataset used in this study was imbalanced. Excluding data augmentation, we will gather additional PPLs data to verify the CAD system’s efficacy in future studies.