1. Introduction
The multiclass classification problem is one of the most common and important tasks in modern machines and deep learning applications in various fields [
1,
2,
3,
4,
5]. One such area is medicine, where the solution to such problems allows for improving the systems of diagnostics, rehabilitation, and the life quality support of patients. Currently, there are numerous works devoted to using neural networks for solving classification problems in medicine.
For example, the classification of cancer on pathological images of cervical tissues using deep learning algorithms was considered by Pan Huang, et al. in [
6]. The authors used ResNet50 [
7], DenseNet12, Inception_v3 [
8], and VGGNet19 [
9] for an almost balanced dataset consisting of 468 RGB images, including 150 images of the norm, 85 low-grade squamous intraepithelial lesions images, and 104 images of high-grade squamous intraepithelial lesions. Swaraj and Verma [
10] solved the COVID-19 classification problem on chest X-ray images. The dataset consisted of 15,371 posterior-to-anterior chest X-ray images. The authors used LeNet [
11], VGG-16 [
12], ResNet-50, AlexNet [
13], Xception [
14] and Inception_v3 architectures as deep learning models for training. As a result, LeNet showed an accuracy of 74.75% on a validation dataset, AlexNet—70.04%, ResNet-50—75.71%, VGG-16—87.34%, Inception_v3—84.29%, Xception—67.76%. State of art algorithms are also used for solving actual medical problems. For example, works [
15,
16] provide an overview of the application of transformers models for solving various medical problems related to the analysis of medical images. The types of problems that can be solved by transformer models, their architecture, and metrics were considered. The paper [
17] states the approach of applying transformers to solving the problem of classifying medical images of the stage of chronic venous disease for self-diagnosis of a patient by using the image of the patient’s legs.
Deep learning algorithms are used to solve classification problems in other areas of science and technology too. Gangsar and Tiwari [
18] studied vibration and current monitoring for effective fault prediction in an induction motor by using multiclass support vector machine (MSVM) algorithms. Ceyhan et al. [
19] shared the results of experiments aimed at classifying wheat varieties using deep learning methods, which made it possible to create an original approach for such classification, which can be used in industry, including on equipment with low memory/small computing power.
Despite the widespread use of deep learning methods, their training faces several challenges. Therefore, one of the critical problems in multiclass classification tasks is the imbalance of the dataset. This is especially important when using contemporary pre-trained neural networks which were mentioned before, where the last layers of the neural network are retrained. With an unbalanced dataset, the error of relating an object to the correct class increases, which cannot always be eliminated by changing the training parameters of a neural network.
Note that most of the data obtained in solving practical problems are not balanced. This is due to the nature of such problems. For example, most medical datasets are imbalanced because the distribution of patients according to the stages of the disease is uneven. Moreover, the distribution of the disease stages of those who sought medical help is also uneven. The number of correct banking transactions will always be significantly greater than the number of fraudulent transactions. There are more distinct types of civilian vehicles than special ones. The distribution of images of different animals that we can use for classification is also not uniform. There will be more images of domestic animals than images of rare or wild animals.
There are a number of works devoted to various techniques for increasing the metrics of training models on an unbalanced dataset. A feasible way to improve the quality of the classification is to change the number of objects in the dataset by data augmentation, data mining, etc. It is commonly said that data augmentation can increase the values of training metrics.
The authors of article [
20] propose the modified VGG16 model for the classification of pneumonia X-ray images. The presented IVGG13 model trained on an augmented dataset produced good metrics with the F1-measure compared with the best convolutional neural networks for medical image recognition. At a time when COVID-19 was the most common in the world, Jain et al. [
21] presented an approach to form a classifier neural network for COVID-19 diagnoses by X-ray images. This approach contained 4 phases including data augmentation. As shown in [
22], the strategy of training used with data augmentation techniques that avoid overfitting problems on ConvNets shows the best results.
Song et al. [
23] used BLogitBoost-J, LogitBoost [
24], and BABoost [
25] methods for multiclass classification on imbalanced datasets of Thyroid and Glass from the UCI data repository. Both datasets contained only numerical (non-images) samples. The thyroid dataset had 3 classes and the sample quantity for each class was 168:368:6666. The glass dataset consisted of 6 types. The ratio of the sample quantity was 70:76:17:13:9:29. LogitBoost algorithm, extended from Adaboost, is an adaptive algorithm that can acquire higher prediction precision. Because it is based on conditional Bernoulli likelihood without prior probability in consideration, the LogitBoost algorithm will lead to a high minority class prediction error. Song et al. [
25] propose an improved BLogitBoost, based on a stratified normalization method, to deduce the badness of LogitBoost in the case of two-class unbalanced data. The authors proposed a new algorithm named BLogitboost-J. The BABoost, in turn, was also an improved AdaBoost algorithm for unbalanced classification data. The difference is that in each round of boosting, the BABoost algorithm assigns more weights to the misclassified examples in the minority class by a user-specified parameter.
Bhadra et al. in [
26] classified three different environmental chemical stimuli, using fifteen statistical features, extracted from the plant electrical signals. They used the imbalanced dataset consisting of 37,834 data blocks for three chemical stimuli. The ratio was 628:1488:35718. The authors applied Monte Carlo under-sampling of major classes and they used eight algorithms (AdaBoost, Decision tree, Gaussian Naive Bayes, k-nearest Neighbors, Multilayer perceptron classifier, Quadratic discriminant analysis, Random Forest, and Support vector machine) to compare their performance.
Nurrahman et al. [
27] solved multiclass classification anemia and iron deficiency using the XGBoost method. The data set consisted of 11,327 samples into 4 categories. The sample ratio was as follows: iron deficiency anemia—9.40%; iron deficiency—14.95%; Anemia—11.93%; Normal—63.72%. The data preprocessing included MissForest imputation (a method for filling gaps in data), feature selection using the Boruta method, and feature generation using SMOTE. Then they applied the XGBoost classification algorithm.
Meisen and Kraus [
28] compare several approaches to deal with the problem of unbalanced sidescan sonar datasets for classification tasks. They used the imbalanced dataset consisting of 1290 images for 3 classes. The ratio was 30:630:630. This data was labeled, normalized, and prepared for use by neural networks. In this paper, they have introduced a new method of transfer learning for GANs on sidescan sonar data named TransfGAN which uses ray-traced images for pre-training. On their sidescan sonar image dataset augmentation with synthetic images from TransfGAN increases the balanced accuracy by more than 10% while also achieving a 3% higher macro F1 score.
Bhowan et al. in [
29] compared the effectiveness of two genetic programming classification strategies. The first uses the standard (zero) class-threshold, while the second uses the “best” class-threshold determined dynamically on a solution-by-solution basis during evolution. The authors conducted experiments for five benchmark binary classification problems on datasets from the UCI Repository of Machine Learning Databases, and the Intelligent Systems Lab at the University of Amsterdam. One of the five datasets used for the experiment contained 267 records derived from cardiac Single Photon Emission Computed Tomography (Spect) images. There are 55 “abnormal” records (20.6%) and 212 “normal” records (79.4%), an imbalance ratio of approximately 1:4. Study results suggest that there is no overall difference between the two strategies and that both strategies can evolve good solutions in binary classification when used in combination with an effective fitness function.
At the same time, data mining usage can also have a positive impact on the performance of neural networks. In ref. [
30] various approaches to medical data mining were considered, as well as the learning outcomes after applying each of the approaches. It also showed that all considered mining methods increase the accuracy of the neural networks. Bellazzi and Zupan [
31] stated that many data mining methods can be successfully applied to a variety of practical problems in clinical medicine and are an excellent means of increasing network performance for medical data processing tasks.
So, most of the work either uses different data-filling techniques and different boosting algorithms. One more approach is to use implicit ways to replicate the smaller class until you have as many samples as in the larger one, for example, class weight for some neural network models. However, this way cannot be considered as acceptable for computer vision tasks.
For binary classification problems, cost-sensitive learning can be used. However, this approach is too difficult to generalize for multiclass classifications.
Various ensemble methods can also be used to solve problems of multiclass image classification. However, the result of their application strongly depends on the problem being solved and the dataset used.
The dataset imbalance affects the training results of each neural network (NN) differently, as does data preprocessing similar to data augmentation and data mining. It is also the subject of interest to study which of the methods or their combinations for reducing the imbalance of a dataset is most preferable for a particular neural network.
The aim of the work was to compare the results of using contemporary neural networks for multiclass classification on a highly imbalanced image dataset and to formulate an approach for normalizing such datasets to improve the quality of a deep neural network (DNN) model training.
To achieve this goal, we studied the sensitivity to a dataset imbalance of the following contemporary neural networks: Xception, ViT-384 [
32], ViT-224, VGG19, ResNet34 [
33], ResNet50, ResNet101 [
34], Inception_v3, DenseNet201 [
35], DenseNet161 [
36], and DeIT [
37]. Different imbalance reduction techniques and their ensembles were used to determine this sensitivity. Based on the analysis of the obtained learning metrics, recommendations were formulated on the use of the ensemble method for dataset normalization.
The remainder of this paper is organized as follows. The data processing methods, data mining ways, as well as trained neural networks are described in
Section 2. The performance evaluation and research results are stated in
Section 3. The validation of the suggested approach is in
Section 4. Finally,
Section 5 discusses the study’s findings, results, and the main conclusion of the work.
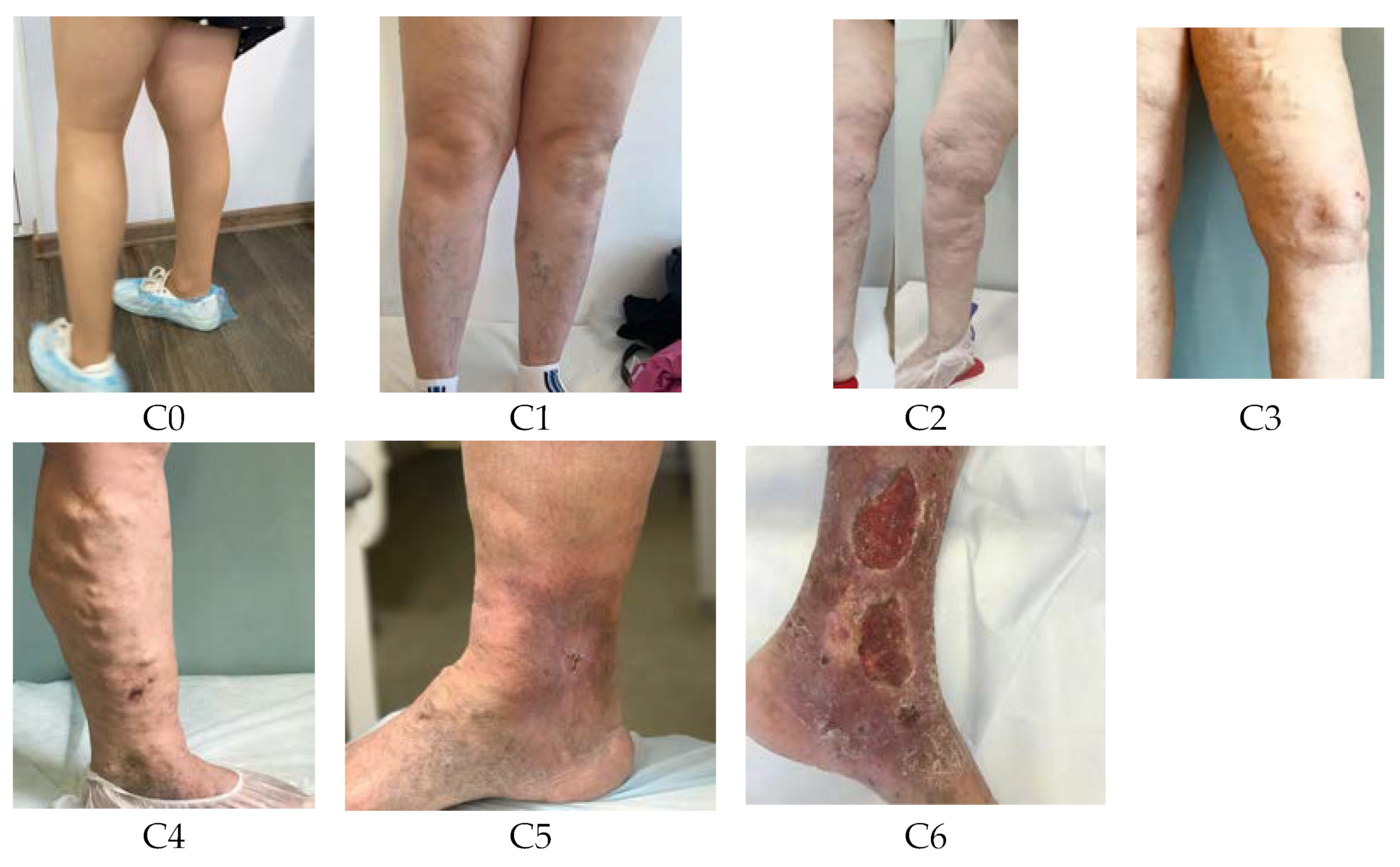
3. Results
As can be seen in
Table 5,
Table 6,
Table 7,
Table 8,
Table 9,
Table 10,
Table 11,
Table 12,
Table 13,
Table 14 and
Table 15, training on an unbalanced dataset leads to the fact that the model can poorly see the small classes, or not see at all. ResNet models are extremely sensitive to class imbalance. So, the ResNet34 model does not see the C5 class at all when training on the initial dataset (TPR = 0.00), but TPR ≈ 0.46 after training on small datasets (small_ds_1, small_ds_2, small_ds_3 and aug_ds). For ResNet50, we can see an increase of TPR for class C5 from 0.29 (initial ds) to ≈0.58(small ds) and 0.60 (aug_ds). For the C6 class, the TPR increases from 0.56 (initial ds) to 0.77 (aug_ds). TPR for class C5 for ResNet101 increases from 0.15 (initial ds) to 0.67(aug_ds).
For VIT models we can improve TPR by 0.5 for small classes if we will use a balanced dataset. For example, VIT-base-patch16-224: TPR = 0.12(initial ds) and TPR = 0.61 (aug_ds). DeIT models are more stabilized for the training on an unbalanced dataset. For them, the improvement in metrics is no more than 0.2.
DenseNet proved to be overly sensitive to training on an unbalanced dataset. For them, training on a balanced dataset is preferable, then class recognition metrics will be significantly improved (by 0.6 or more).
A similar conclusion can be drawn for Inception_v3, Xception, and VGG. The training of these models on a balanced dataset allows improved metrics by 0.2 for some classes.
It should be noted that there is no deterioration in the recognition of classes that contain a large number of objects in the initial data set. It can also be seen from the obtained results that the metrics on the “small_ds_1”, “small_ds_2”, and “small_ds_3” datasets are different and depend on those objects that remained in these datasets after the initial dataset was cut off.
The best and the most stable metrics of the neural networks under consideration were obtained on the “aug_ds” dataset, which was formed from the initial dataset, when the initial dataset was cropped by the number of images in C6 class + 15%. Then the missing images in all classes were augmented.
Thus, it seems appropriate for unbalanced datasets to form balanced datasets on their basis according to the described methodology, and train neural network models using the resulting balanced dataset.
5. Discussion
The approach presented in the paper for the construction of a balanced dataset based on the initial unbalanced dataset allows a significant increase in the metrics for the classification of the smallest classes. For some models, this increase is incredibly significant–from zero metrics, when the model does not see this class at all, to TPR values more than 0.6. At the same time, there is always a slight deterioration (by a few percent) in metrics for large classes. The proposed approach was tested on various unbalanced datasets that contained color and grayscale images. The metrics for small classes for all datasets were significantly improved.
In fact, the proposed approach is an ensemble approach for preprocessing unbalanced datasets. It includes two steps.
The first step is to reduce all classes except the smallest one. However, unlike other approaches, we suggest reducing not to the size of the smallest class, but to the size of the smallest class + 15% of it. The second step is a random augmentation of the smallest class. Thus, on the one hand, we balance the dataset, while retaining more information than with conventional downsampling. However, the dataset is not too clogged with artificially generated information, as with conventional upsampling.
At the same time, it is of interest to what percentage we can augment the smallest class without losing the quality of training the model. This may be the subject of further study. It is also the subject of interest to what extent the proposed approach can improve the models’ metrics for datasets containing non-image samples.
The suggested approach can help to solve the problem of small class recognition in various important computer vision problems where unbalanced datasets are widespread, for example, medical tasks of computer vision, and tasks of recognizing non-standard or critical situations in many fields of science and technology.