An Effective Med-VQA Method Using a Transformer with Weights Fusion of Multiple Fine-Tuned Models
Abstract
:1. Introduction
- Diagnosis and treatment: By offering a quick and precise method for analyzing medical images, Med-VQA can help medical practitioners diagnose and treat medical disorders. Healthcare experts canearn more about the patient’s condition by inquiring about medical imaging (such as X-rays, CT scans, and MRI scans), which can aid in making a diagnosis and selecting the best course of therapy. In addition, it would reduce the doctors’ efforts byetting the patients obtain answers to the most frequent questions about their images.
- Medical education: By giving users a way toearn from medical images, Med-VQA can be used to instruct medical students and healthcare workers. Students canearn how to assess and understand medical images—a crucial ability in the area of medicine—by posing questions about them.
- Patient education: By allowing patients to ask questions about their medical photos, Med-VQA can help them better comprehend their medical issues. Healthcare practitioners can improve patient outcomes by assisting patients in understanding their problems and available treatments by responding to their inquiries regarding their medical photos.
- Research: arge collections of medical photos can be analyzed using Med-VQA to glean insights that can be applied to medical research. Researchers can better comprehend medical issues and create new remedies by posing questions regarding medical imaging and examining the results.
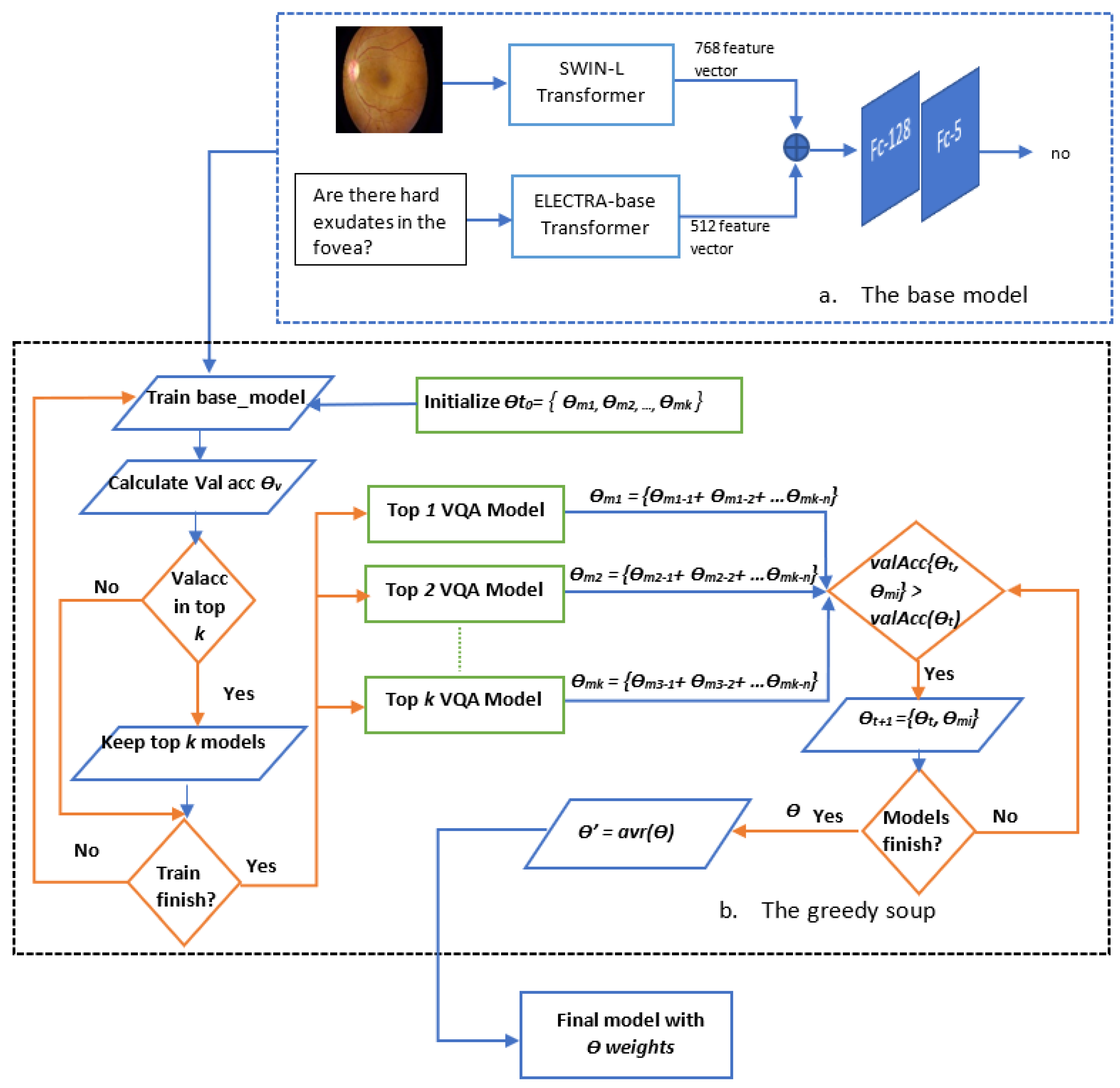
- Since the ELECTRA-base transformer that is pre-trained on aarge corpus of text data using contrastive estimation and outperforms other text transformers, such as BERT and BioBERT, is not utilized yet in medical VQA for the textual feature extraction, the proposed model exploits the ELECTRA-base transformer to extract text features from the question.
- The proposed model aims to solve the issue of thearge feature variation between the question and the image. Therefore, it exploits theast vision transformer, swin-base-patch4-window7-224. The SWIN transformer utilized in this model to extract visual features from the image is used for the first time in medical VQA.
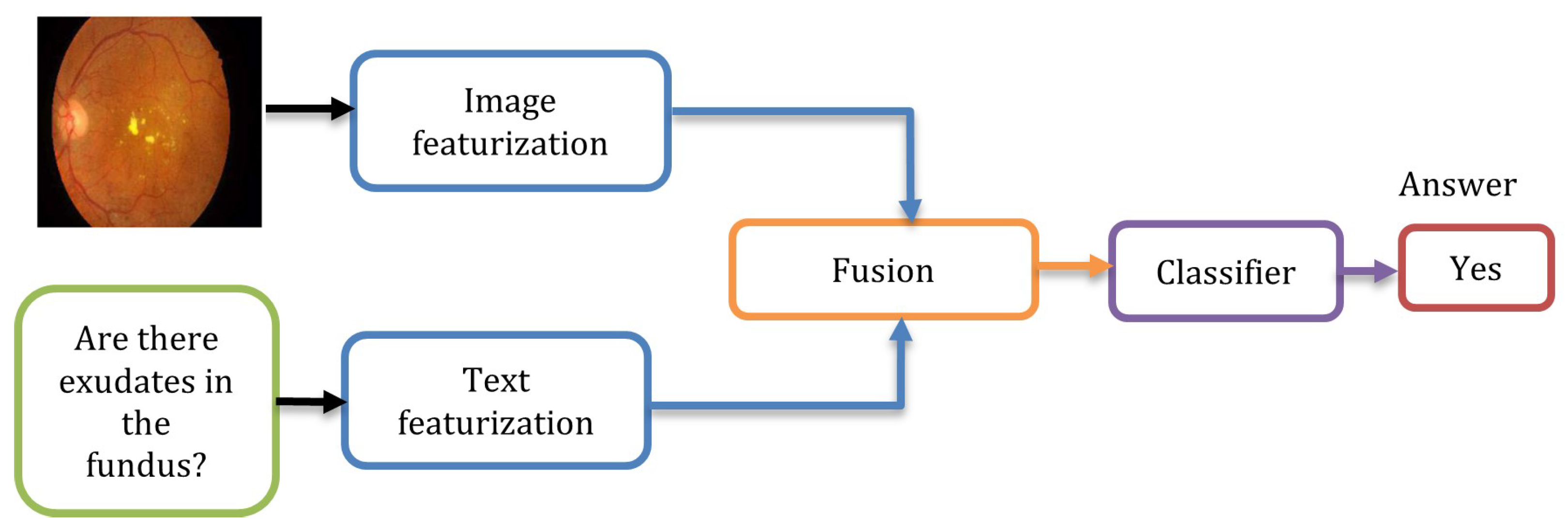
- The extracted visual and textual features are combined and fed into an MLP for classification.
- The proposed model is fine-tuned based on the model parameters that achieved the highest validation accuracy and the greedy soup approach to show the significant impact of the greedy soup method.
- The performance of theast two models are compared with the model by Tascon et al. [5], which we denoted as the SOTA because it is the unique research on this dataset.
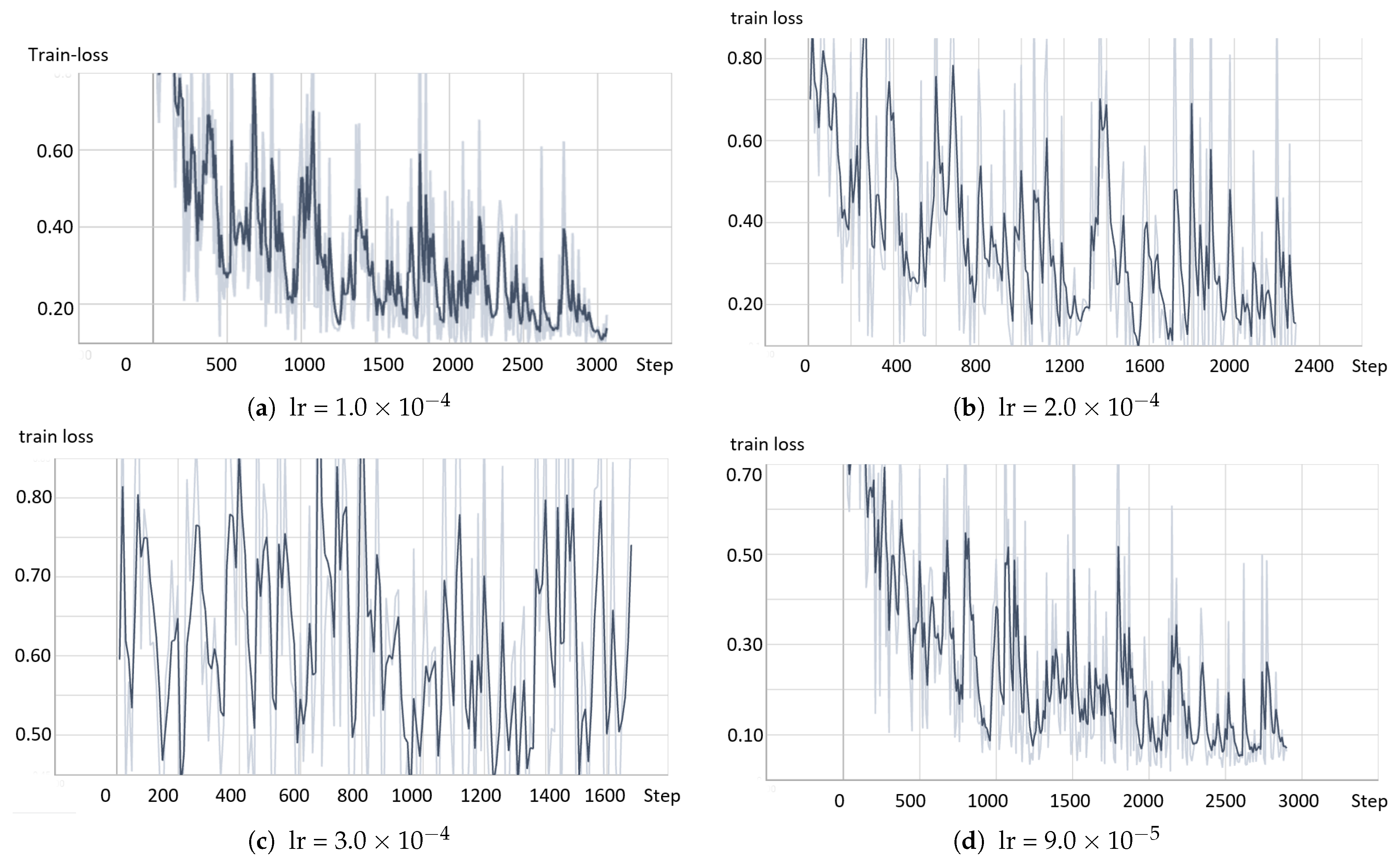
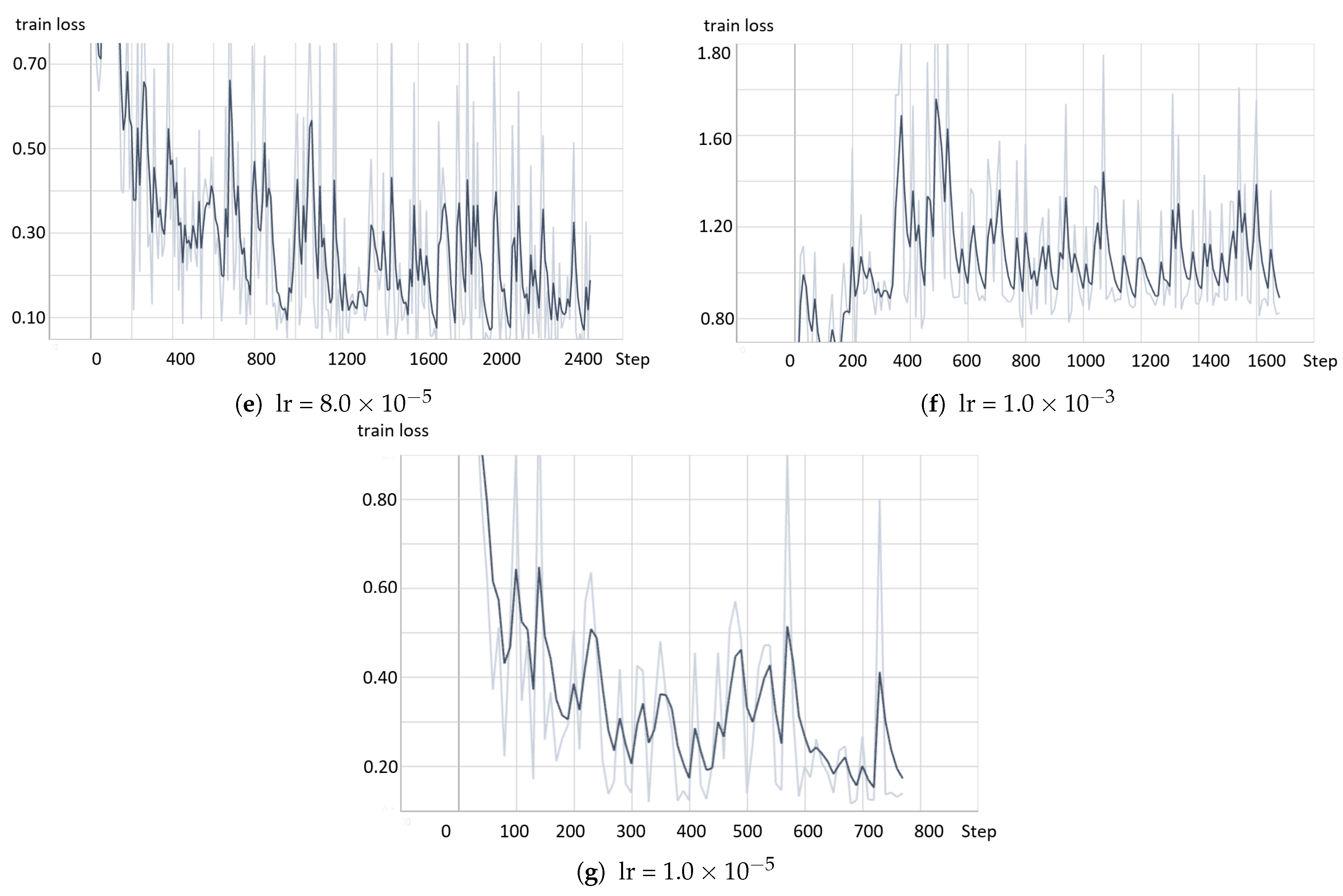
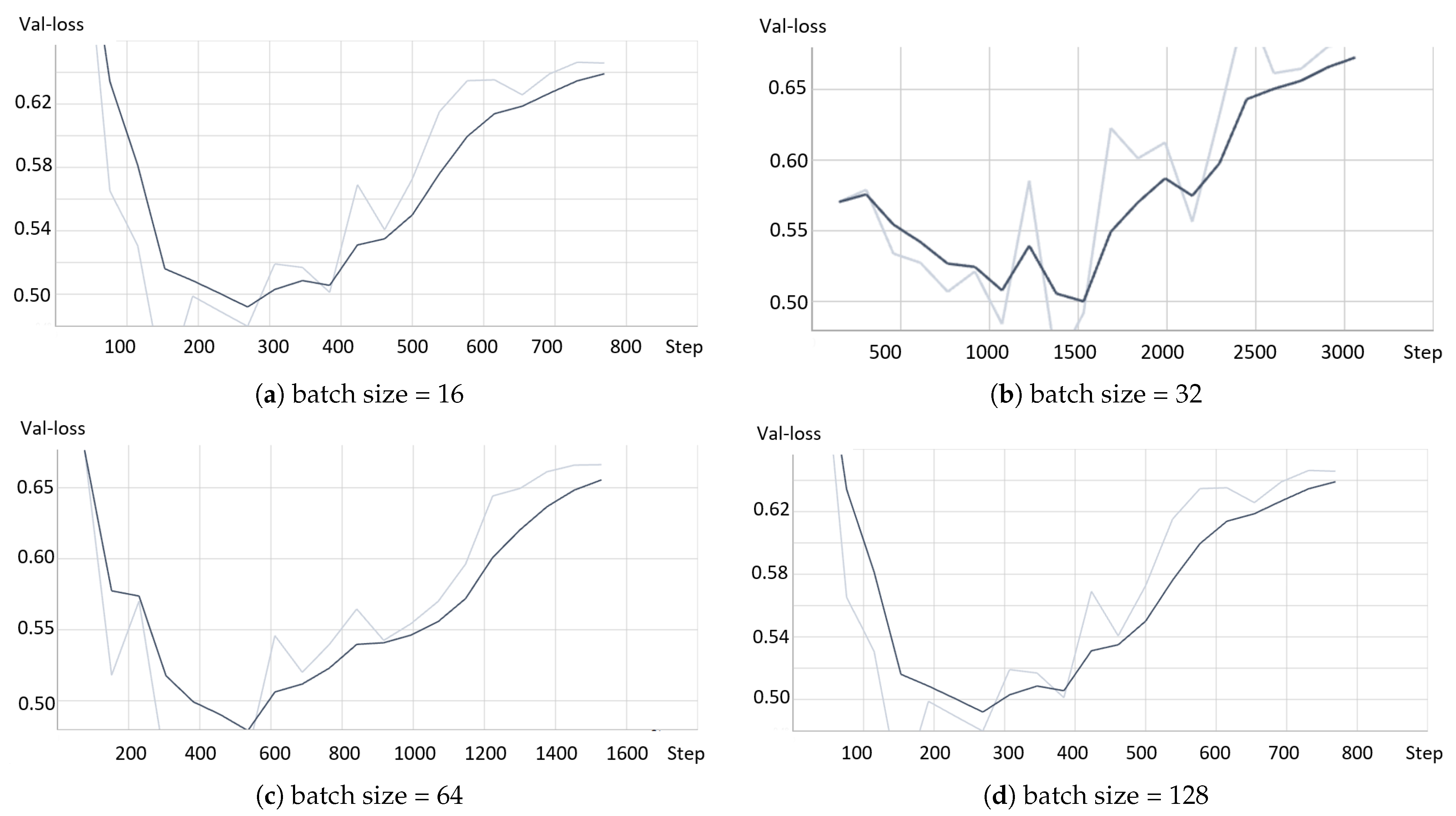
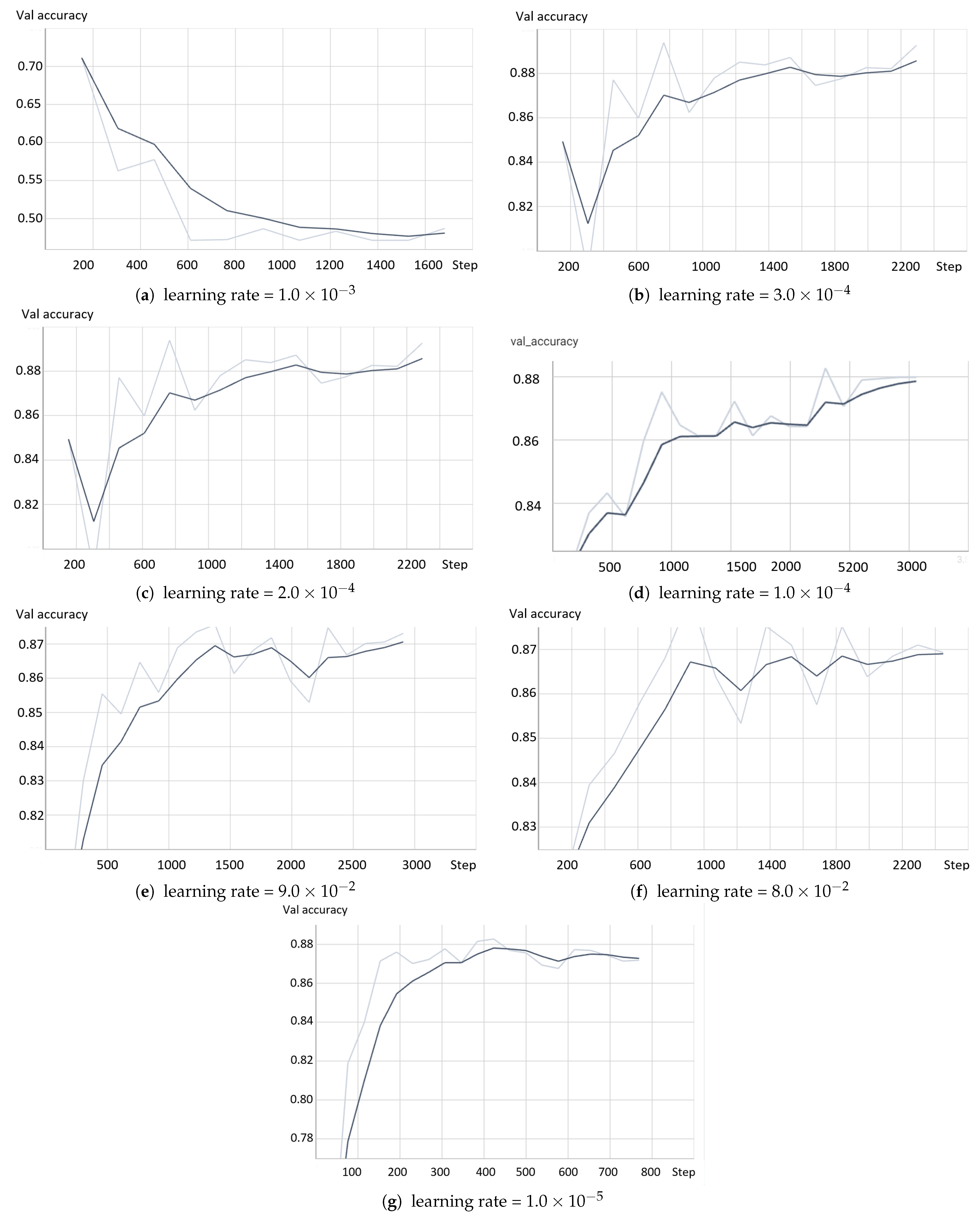
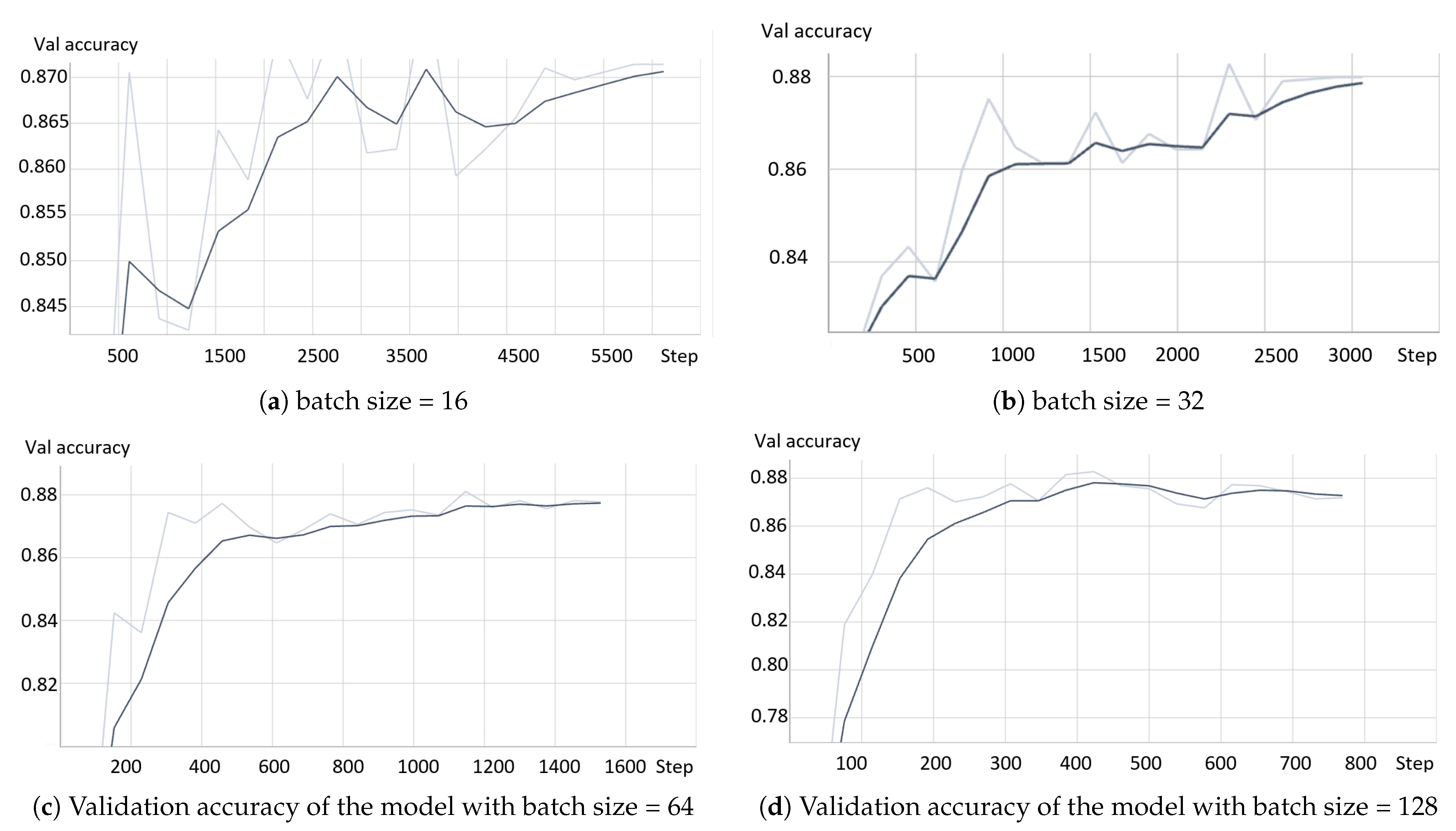
- The model based on the greedy soup fine-tuning technique is optimized to select the bestearning rate and batch size.
2. Related Work
2.1. Vision Featurization
2.2. Text Featurization
2.3. Fusion
3. Methodology
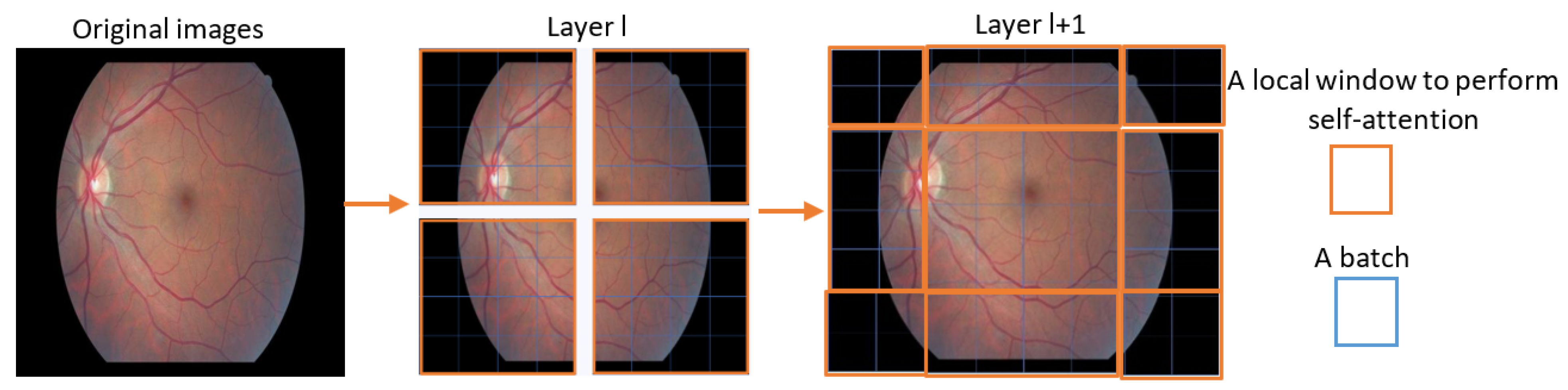
3.1. SWIN Transformer
- PatchEmbedding: The input image is divided into non-overlapping patches, which are flattened into vectors and passed through ainearayer to obtain a set of patch embeddings. The patch embeddings are denoted as X.
- SWIN Transformer Blocks: The SWIN transformer block processes the feature maps of the input patches using a set of shifted windows, producing a set of output feature maps, which are then processed by a SWIN transformerayer that combines the feature maps across the patches and produces a set of feature maps at a higher resolution.where denotes to the patch in row i and column j.
- SWIN Transformerayers: The output feature maps from the SWIN transformer blocks are processed by a SWIN transformerayer that combines the feature maps across the patches and produces a set of feature maps at a higher resolution.where denotes to the patch in row i and column j.
- Global Average Pooling: The output feature maps from the final SWIN transformerayer are passed through a global average poolingayer, which computes the average of each feature map across the spatial dimensions and produces a final feature vector.
3.2. ELECTRA Transformer
- Token Embedding: The input text T is tokenized, and each token is mapped to its corresponding embedding vector. The token embeddings are denoted as X.
- Encoderayers: The ELECTRA transformer uses a stack of encoderayers to process the token embeddings and extract contextualized representations of the input text. Each encoderayer consists of a self-attention mechanism followed by a feedforward network.
- Maskedanguage Modeling: During pre-training, a subset of the input tokens are randomly masked, and the model is trained to predict the original tokens based on the masked tokens. The masked tokens are denoted as M.
- Masked Token Prediction:The output of the encoderayers is used to predict the original tokens based on the masked tokens. The masked token prediction can be expressed as follows:
- –
- First, the output of the encoderayers is passed through ainearayer to obtain a set ofogits for each token position.
- –
- Then, theogits corresponding to the masked token positions are selected and passed through a softmax function to obtain a probability distribution over the vocabulary.
- –
- Finally, the model is trained to maximize theog-likelihood of the original tokens given the masked tokens and the predicted probability distribution.where n and are the number of masked tokens and the index of the original token at position i, respectively.
3.3. MLP Fusion
3.4. SOUPS Fusion Method
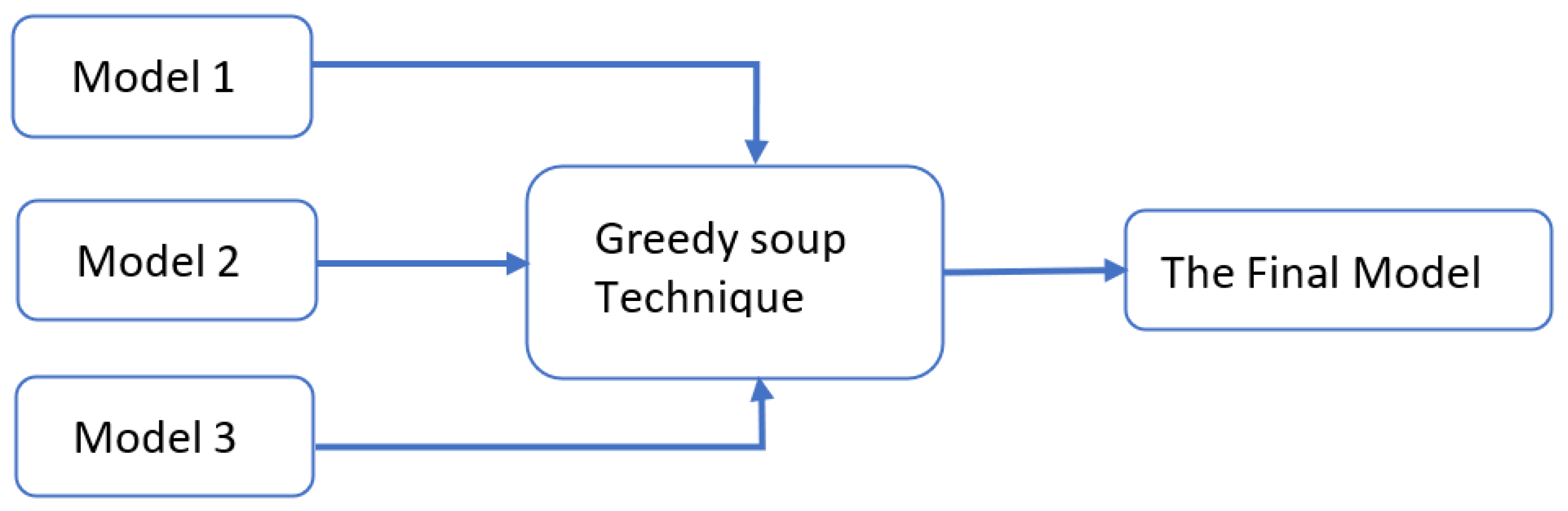
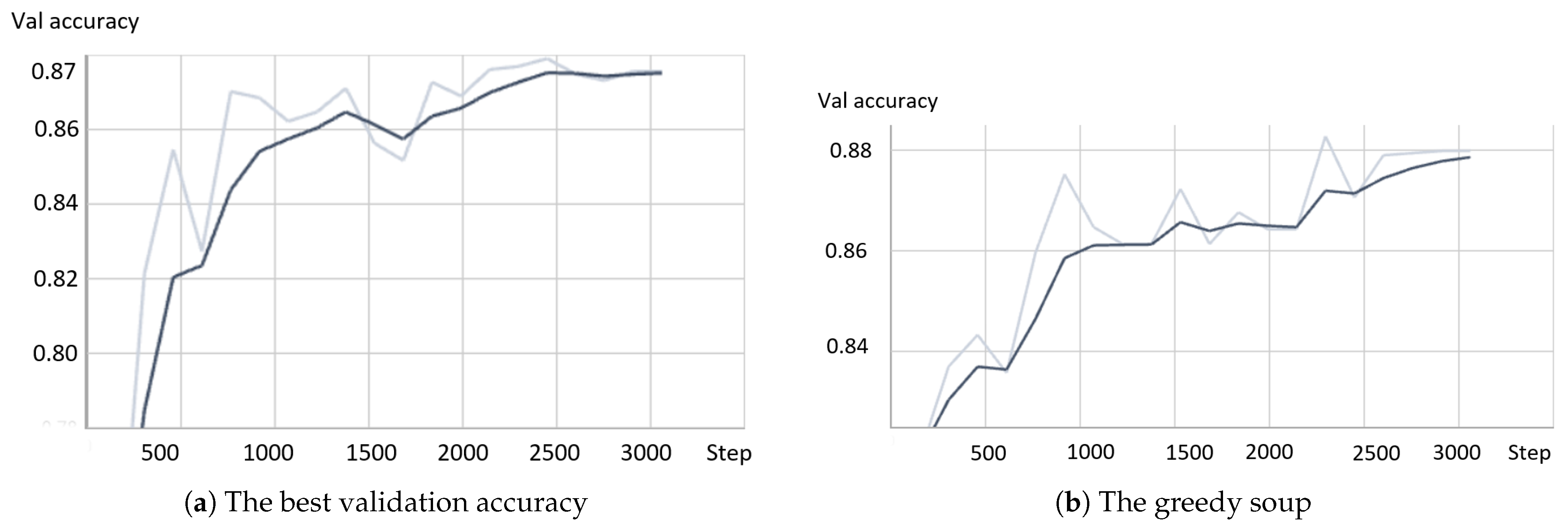
- Greedy soupsThe greedy soups technique is used to fuse the parameters of multiple fine-tuned models, where the model is trained, and validation accuracy is calculated several times during training; the final parameters will be the average of the best k models’ parameters based on the fine-tuned model weights that significantly improve the validation performance by averaging the weights of the new fine-tuned model with theist of previous models weights. The proposed model utilizes this technique to maximize its performance.Let and denote the set of models and their parameters, respectively.et and be soup ingredients or the parameters for the considered models. Each time i the validation is computed, the model is considered if . The models are sorted decreasingly.From the models and their parameters, the model is considered for fusion if, for each time i to k, the is greater than .et denote the considered j models. The final model parameters are the average of the models’ parameters, and they are calculated using the equation:The proposed model is set to be based on the best three models, , where the validation is calculated in the middle and end of each epoch. Algorithm 1 shows the algorithm of greedy soup to fuse the three models with different hyper-parameters (number of steps). Figure 6 presents the overall structure of the greedy soup technique to fuse three models.
- The best valueThe fine-tuning based on the best accuracy value selects the model that achieves the highest score. It is represented using the equation:
| Algorithm 1 Greedy_Soup_Model |
|
4. Experiment
4.1. Environment Setup
4.2. Dataset
4.3. Evaluation Metrics
- Accuracy: Accuracy is a commonly used performance metric for classification problems that measures the proportion of correctly classified samples out of the total number of samples. It is calculated using the equation:where True Positives are the number of positive samples that are correctly identified as positive by the model, True Negatives are the number of negative samples that are correctly identified as negative by the model, False Positives are the number of negative samples that are incorrectly identified as positive by the model, and False Negatives are the number of positive samples that are incorrectly identified as negative by the model.
- Precision: Precision, also known as a positive predictive value, measures the proportion of the predicted true positive samples out of all predicted positive samples. The precision metric’s importance is giving an indication of whether the model has aow false positive rate based on a high precision score. It is calculated using the equation:
- Recall or Sensitivity: Recall, also known as sensitivity or true positive rate, measures the proportion of the predicted positive samples out of the actual positive samples. The recall metric is critical in applications such as medical diagnosis, where it is essential to minimize false negatives. It is calculated using the equation:
- F1-score: The F1 -score is the harmonic mean of precision and recall that measures the model’s accuracy in correctly identifying positive samples. The F1-score is a valuable metric for evaluating classification models, especially when the dataset is imbalanced, and the goal is to ensure that the model performs well on both positive and negative samples. It is calculated using the equation:
- Macro-averaged precision: The macro-averaged precision is calculated by taking the average of the precision scores for each class. The equation for the macro-averaged precision can be expressed as:where n, , and are the total number of classes, the number of true positives for class i, and the number of false positives for class i, respectively.
- Macro-averaged recall: The macro-averaged recall is calculated by taking the average of the recall scores for each class. InaTeX, the equation for the macro-averaged recall can be expressed as:where n, , and are the total number of classes, true positives for class i, and false negatives for class i, respectively.
- Macro-averaged F1-score: The macro-averaged F1-score is calculated by taking the average of the F1-scores for each class. The equation for the macro-averaged F1-score can be expressed as:where n is the total number of classes, and and are the precision and recall values for class i, respectively.
- Weighted-average precision: The weighted-average precision is calculated by taking the weighted average of the precision scores for each class. The equation for the weighted-average precision can be expressed as:where n, , , and are the total number of classes, the number of true positives for class i, the number of false positives for class i, and the weight for class i, respectively.
- Weighted average recall: The weighted average recall is calculated by taking the weighted average of the recall scores for each class. The equation for the weighted-average precision can be expressed as:where n,, , and are the total number of classes, the number of true positives for class i, the number of false negatives for class i, and the weight for class i, respectively.
- Weighted-average F1-score: The weighted-average F1-score is calculated by taking the weighted average of the F1-scores for each class. InaTeX, the equation for the weighted-average F1-score can be expressed as:where n, , , and are the total number of classes, the precision values for class i, recall values for class i, and the weight for class i, respectively.
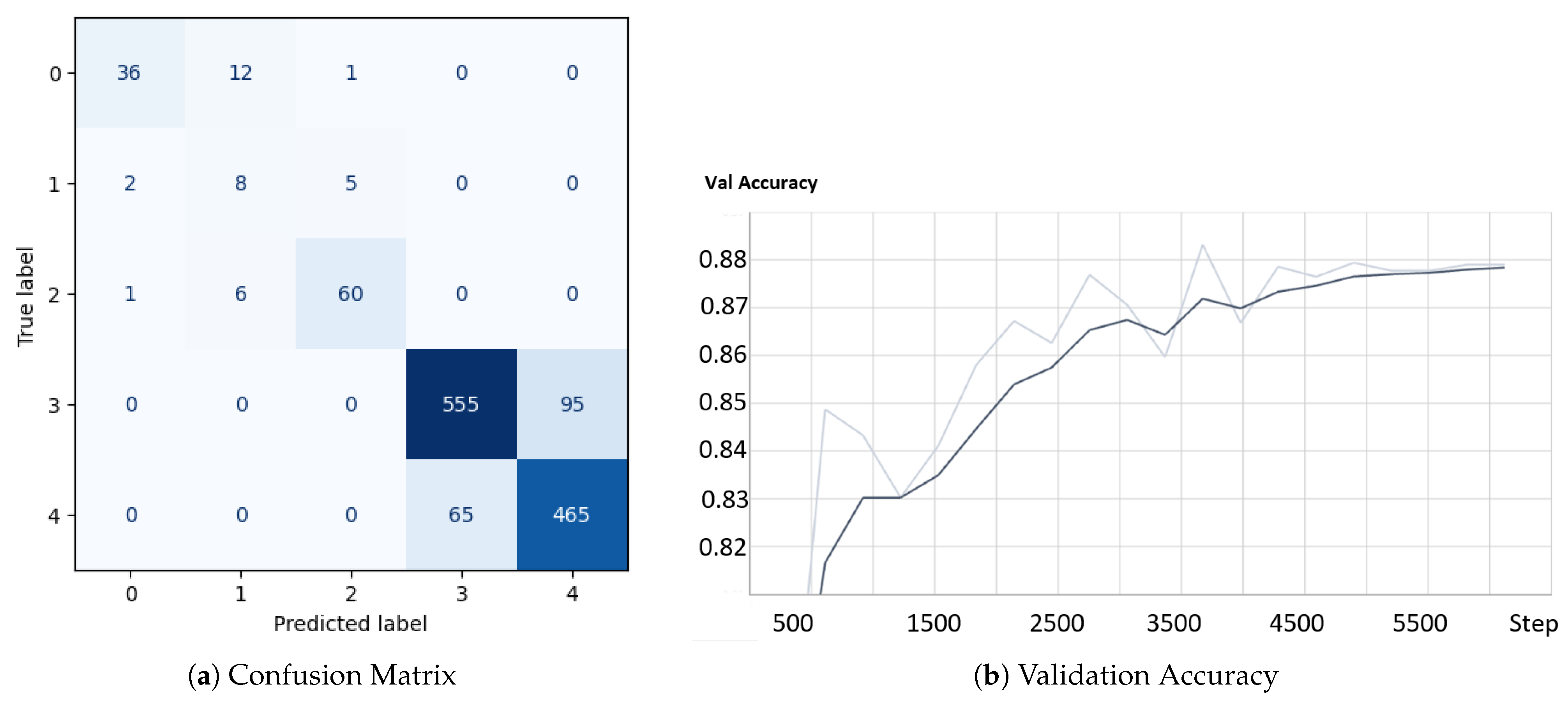
4.4. Results and Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A



References
- Zhu, Y.; Groth, O.; Bernstein, M.; Fei-Fei, L. Visual7w: Grounded question answering in images. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–28 June 2016; pp. 4995–5004. [Google Scholar]
- Abacha, A.B.; Hasan, S.A.; Datla, V.V.; Liu, J.; Demner-Fushman, D.; Müller, H. VQA-Med: Overview of the Medical Visual Question Answering Task at ImageCLEF 2019. In proceeding of Working Notes of CLEF 2019, Lugano, Switzerland, 9–12 September 2019. [Google Scholar]
- Abacha, A.B.; Datla, V.V.; Hasan, S.A.; Demner-Fushman, D.; Müller, H. Overview of the VQA-Med Task at ImageCLEF 2020: Visual Question Answering and Generation in the Medical Domain. In Proceedings of the CLEF 2020—Conference and Labs of the Evaluation Forum, Thessaloniki, Greece, 22–25 September 2020; pp. 1–9. [Google Scholar]
- Liu, B.; Zhan, L.M.; Xu, L.; Ma, L.; Yang, Y.; Wu, X.M. SLAKE: A Semantically-Labeled Knowledge-Enhanced Dataset for Medical Visual Question Answering. In Proceedings of the 2021 IEEE 18th International Symposium on Biomedical Imaging (ISBI), Nice, France, 13–16 April 2021; pp. 1650–1654. [Google Scholar]
- Tascon-Morales, S.; Márquez-Neila, P.; Sznitman, R. Consistency-Preserving Visual Question Answering in Medical Imaging. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2022, Proceedings of the 25th International Conference, Singapore, 18–22 September 2022; Part VIII; Springer: Cham, Switzerland, 2022; pp. 386–395. [Google Scholar]
- Ren, M.; Kiros, R.; Zemel, R. Image question answering: A visual semantic embedding model and a new dataset. Proc. Adv. Neural Inf. Process. Syst. 2015, 1, 5. [Google Scholar]
- Antol, S.; Agrawal, A.; Lu, J.; Mitchell, M.; Batra, D.; Zitnick, C.L.; Parikh, D. VQA: Visual question answering. In Proceedings of the IEEE International Conference on Computer Vision, Santiago, Chile, 7–13 December 2015; pp. 2425–2433. [Google Scholar]
- Malinowski, M.; Rohrbach, M.; Fritz, M. Ask your neurons: A neural-based approach to answering questions about images. In Proceedings of the IEEE International Conference on Computer Vision, Santiago, Chile, 7–13 December 2015; pp. 1–9. [Google Scholar]
- Jiang, A.; Wang, F.; Porikli, F.; Li, Y. Compositional memory for visual question answering. arXiv 2015, arXiv:1511.05676. [Google Scholar]
- Chen, K.; Wang, J.; Chen, L.C.; Gao, H.; Xu, W.; Nevatia, R. ABC-CNN: An Attention Based Convolutional Neural Network for Visual Question Answering. arXiv 2015, arXiv:1511.05960v2. [Google Scholar]
- Ilievski, I.; Yan, S.; Feng, J. A focused dynamic attention model for visual question answering. arXiv 2016, arXiv:1604.01485. [Google Scholar]
- Andreas, J.; Rohrbach, M.; Darrell, T.; Klein, D. Neural module networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–28 June 2016; pp. 39–48. [Google Scholar]
- Song, J.; Zeng, P.; Gao, L.; Shen, H.T. From pixels to objects: Cubic visual attention for visual question answering. arXiv 2022, arXiv:2206.01923. [Google Scholar]
- Andreas, J.; Rohrbach, M.; Darrell, T.; Klein, D. Learning to compose neural networks for question answering. arXiv 2016, arXiv:1601.01705. [Google Scholar]
- Xiong, C.; Merity, S.; Socher, R. Dynamic memory networks for visual and textual question answering. In Proceedings of the International Conference on Machine Learning, PMLR, New York City, NY, USA, 20–22 June 2016; pp. 2397–2406. [Google Scholar]
- Kumar, A.; Irsoy, O.; Ondruska, P.; Iyyer, M.; Bradbury, J.; Gulrajani, I.; Zhong, V.; Paulus, R.; Socher, R. Ask me anything: Dynamic memory networks for natural language processing. In Proceedings of the International Conference on Machine Learning, PMLR, New York City, NY, USA, 20–22 June 2016; pp. 1378–1387. [Google Scholar]
- Noh, H.; Han, B. Training recurrent answering units with joint loss minimization for VQA. arXiv 2016, arXiv:1606.03647. [Google Scholar]
- Gao, L.; Zeng, P.; Song, J.; Li, Y.F.; Liu, W.; Mei, T.; Shen, H.T. Structured two-stream attention network for video question answering. Proc. AAAI Conf. Artif. Intell. 2019, 33, 6391–6398. [Google Scholar] [CrossRef]
- Wang, P.; Wu, Q.; Shen, C.; Hengel, A.v.d.; Dick, A. Explicit knowledge-based reasoning for visual question answering. arXiv 2015, arXiv:1511.02570. [Google Scholar]
- Wang, P.; Wu, Q.; Shen, C.; Dick, A.; Van Den Hengel, A. FVQA: Fact-based visual question answering. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 40, 2413–2427. [Google Scholar] [CrossRef]
- Wu, Q.; Wang, P.; Shen, C.; Dick, A.; Van Den Hengel, A. Ask me anything: Free-form visual question answering based on knowledge from external sources. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–28 June 2016; pp. 4622–4630. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv 2015, arXiv:1409.1556v6. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the CVPR 2016, Las Vegas, NV, USA, 27–28 June 2016; pp. 770–778. [Google Scholar]
- Nguyen, B.D.; Do, T.T.; Nguyen, B.X.; Do, T.; Tjiputra, E.; Tran, Q.D. Overcoming Data Limitation in Medical Visual Question Answering. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Cham, Switzerland, 2019; pp. 522–530. [Google Scholar]
- Do, T.; Nguyen, B.X.; Tjiputra, E.; Tran, M.; Tran, Q.D.; Nguyen, A. Multiple Meta-model Quantifying for Medical Visual Question Answering. arXiv 2021, arXiv:2105.08913. [Google Scholar]
- Hochreiter, S.; Schmidhuber, J. Long short-term memory. Neural Comput. 1997, 9, 1735–1780. [Google Scholar] [CrossRef] [PubMed]
- Schuster, M.; Paliwal, K.K. Bidirectional recurrent neural networks. IEEE Trans. Signal Process. 1997, 45, 2673–2681. [Google Scholar] [CrossRef]
- Devlin, J.; Chang, M.W.; Lee, K.; Toutanova, K. BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding. arXiv 2019, arXiv:1810.04805. [Google Scholar]
- Peng, Y.; Liu, F.; Rosen, M.P. UMass at ImageCLEF Medical Visual Question Answering (Med-VQA) 2018 Task. In Proceedings of the CEUR Workshop, Avignon, France, 10–14 September 2018. [Google Scholar]
- Lu, J.; Yang, J.; Batra, D.; Parikh, D. Hierarchical question-image co-attention for visual question answering. Adv. Neural Inf. Process. Syst. 2016, 29, 289–297. [Google Scholar]
- Shi, Y.; Furlanello, T.; Zha, S.; Anandkumar, A. Question Type Guided Attention in Visual Question Answering. In Proceedings of the ECCV 2018, Munich, Germany, 8–14 September 2018; pp. 151–166. [Google Scholar]
- Li, L.H.; Yatskar, M.; Yin, D.; Hsieh, C.J.; Chang, K.W. VisualBERT: A simple and performant baseline for vision and language. arXiv 2019, arXiv:1908.03557. [Google Scholar]
- Lu, J.; Batra, D.; Parikh, D.; Lee, S. Vilbert: Pretraining task-agnostic visiolinguistic representations for vision-and-language tasks. Adv. Neural Inf. Process. Syst. 2019, 32, 1–11. [Google Scholar]
- Chen, Y.C.; Li, L.; Yu, L.; El Kholy, A.; Ahmed, F.; Gan, Z.; Cheng, Y.; Liu, J. Uniter: Universal image-text representation learning. In Proceedings of the Computer Vision—ECCV 2020, 16th European Conference, Glasgow, UK, 23–28 August 2020; pp. 104–120. [Google Scholar]
- Radford, A.; Kim, J.W.; Hallacy, C.; Ramesh, A.; Goh, G.; Agarwal, S.; Sastry, G.; Askell, A.; Mishkin, P.; Clark, J.; et al. Learning transferable visual models from natural language supervision. In Proceedings of the International Conference on Machine Learning, PMLR, Virtual only, 18–24 July 2021; pp. 8748–8763. [Google Scholar]
- Cong, F.; Xu, S.; Guo, L.; Tian, Y. Caption-Aware Medical VQA via Semantic Focusing and Progressive Cross-Modality Comprehension. In Proceedings of the 30th ACM International Conference on Multimedia, Lisbon, Portugal, 10–14 October 2022; pp. 3569–3577. [Google Scholar]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin transformer: Hierarchical vision transformer using shifted windows. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, BC, Canada, 10–17 October 2021; pp. 10012–10022. [Google Scholar]
- Wortsman, M.; Ilharco, G.; Gadre, S.Y.; Roelofs, R.; Gontijo-Lopes, R.; Morcos, A.S.; Namkoong, H.; Farhadi, A.; Carmon, Y.; Kornblith, S.; et al. Model soups: Averaging weights of multiple fine-tuned models improves accuracy without increasing inference time. In Proceedings of the International Conference on Machine Learning, PMLR, Baltimore, MA, USA, 17–23 July 2022; pp. 23965–23998. [Google Scholar]
- Lowe, D.G. Object recognition from local scale-invariant features. In Proceedings of the Piscataway, NJ, USA of the Seventh IEEE International Conference on Computer Vision, Kerkyra, Greece, 20–27 September 1999; Volume 2, pp. 1150–1157. [Google Scholar]
- Dalal, N.; Triggs, B. Histograms of oriented gradients for human detection. In Proceedings of the 2005 IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR’05), San Diego, CA, USA, 20–25 June 2005; Volume 1, pp. 886–893. [Google Scholar]
- Lienhart, R.; Maydt, J. An extended set of Haar-like features for rapid object detection. In Proceedings of the IEEE International Conference on Image Processing, Rochester, NY, USA, 22–25 September 2002; pp. 900–903. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet Classification with Deep Convolutional Neural Networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Zhang, D.; Cao, R.; Wu, S. Information fusion in visual question answering: A Survey. Inf. Fusion 2019, 52, 268–280. [Google Scholar] [CrossRef]
- Abacha, A.B.; Gayen, S.; Lau, J.J.; Rajaraman, S.; Demner-Fushman, D. NLM at ImageCLEF 2018 Visual Question Answering in the Medical Domain; In Proceedings of Working Notes of CLEF 2018, Avignon, France, 10–14 September 2018.
- Verma, H.; Ramachandran, S. HARENDRAKV at VQA-Med 2020: Sequential VQA with Attention for Medical Visual Question Answering. In Proceedings of the Working Notes of CLEF 2018, Thessaloniki, Greece, 22–25 September 2020. [Google Scholar]
- Bounaama, R.; Abderrahim, M.E.A. Tlemcen University at ImageCLEF 2019 Visual Question Answering Task. In Proceedings of the Working Notes of CLEF 2018, Thessaloniki, Lugano, Switzerland, 9–12 September 2019. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 5–12 June 2015; pp. 1–9. [Google Scholar]
- Fukui, A.; Park, D.H.; Yang, D.; Rohrbach, A.; Darrell, T.; Rohrbach, M. Multimodal Compact Bilinear Pooling for Visual Question Answering and Visual Grounding. In Proceedings of the 2016 Conference on Empirical Methods in Natural Language Processing, Austin, TC, USA, 1–5 November 2016; pp. 457–468. [Google Scholar]
- Kim, J.H.; On, K.W.; Lim, W.; Kim, J.; Ha, J.W.; Zhang, B.T. Hadamard Product for Low-rank Bilinear Pooling. In Proceedings of the 5th International Conference on Learning Representations, ICLR Toulon, France, 24–26 April 2017. [Google Scholar]
- Ben-Younes, H.; Cadene, R.; Cord, M.; Thome, N. MUTAN: Multimodal Tucker Fusion for Visual Question Answering. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 2612–2620. [Google Scholar]
- Huang, J.; Chen, Y.; Li, Y.; Yang, Z.; Gong, X.; Wang, F.L.; Xu, X.; Liu, W. Medical knowledge-based network for Patient-oriented Visual Question Answering. Inf. Process. Manag. 2023, 60, 103241. [Google Scholar] [CrossRef]
- Haridas, H.T.; Fouda, M.M.; Fadlullah, Z.M.; Mahmoud, M.; ElHalawany, B.M.; Guizani, M. MED-GPVS: A Deep Learning-Based Joint Biomedical Image Classification and Visual Question Answering System for Precision e-Health. In Proceedings of the ICC 2022—IEEE International Conference on Communications, Seoul, Republic of Korea, 15–18 August 2022; pp. 3838–3843. [Google Scholar]
- Kovaleva, O.; Shivade, C.; Kashyap, S.; Kanjaria, K.; Wu, J.; Ballah, D.; Coy, A.; Karargyris, A.; Guo, Y.; Beymer, D.B.; et al. Towards Visual Dialog for Radiology. In Proceedings of the 19th SIGBioMed Workshop on Biomedical Language Processing, Online, 9 July 2020; pp. 60–69. [Google Scholar] [CrossRef]
- Liao, Z.; Wu, Q.; Shen, C.; van den Hengel, A.; Verjans, J. AIML at VQA-Med 2020: Knowledge Inference via a Skeleton-based Sentence Mapping Approach for Medical Domain Visual Question Answering. In Proceedings of the Working Notes of CLEF 2020, Thessaloniki, Greece, 22–25 September 2020. [Google Scholar]
- Gong, H.; Huang, R.; Chen, G.; Li, G. SYSU-Hcp at VQA-MED 2021: A data-centric model with efficient training methodology for medical visual question answering. In Proceedings of the Working Notes of CLEF 2021, Bucharest, Romania, 21–24 September 2021; Volume 201. [Google Scholar]
- Wang, H.; Pan, H.; Zhang, K.; He, S.; Chen, C. M2FNet: Multi-granularity Feature Fusion Network for Medical Visual Question Answering. In Proceedings of the PRICAI 2022: Trends in Artificial Intelligence, 19th Pacific Rim International Conference on Artificial Intelligence, PRICAI 2022, Shanghai, China, 10–13 November 2022; Part II. Springer: Cham, Switzerland, 2022; pp. 141–154. [Google Scholar]
- Wang, M.; He, X.; Liu, L.; Qing, L.; Chen, H.; Liu, Y.; Ren, C. Medical visual question answering based on question-type reasoning and semantic space constraint. Artif. Intell. Med. 2022, 131, 102346. [Google Scholar] [CrossRef] [PubMed]
- Manmadhan, S.; Kovoor, B.C. Visual question answering: A state-of-the-art review. Artif. Intell. Rev. 2020, 53, 5705–5745. [Google Scholar] [CrossRef]
- He, X.; Zhang, Y.; Mou, L.; Xing, E.; Xie, P. PathVQA: 30.000+ questions for medical visual question answering. arXiv 2020, arXiv:2003.10286. [Google Scholar]
- Allaouzi, I.; Benamrou, B.; Benamrou, M.; Ahmed, M.B. Deep Neural Networks and Decision Tree Classifier for Visual Question Answering in the Medical Domain. In Proceedings of the Working Notes of CLEF 2018, Avignon, France, 10–14 September 2018. [Google Scholar]
- Zhou, Y.; Kang, X.; Ren, F. Employing Inception-Resnet-v2 and Bi-LSTM for Medical Domain Visual Question Answering. In Proceedings of the Working Notes of CLEF 2018, Avignon, France, 10–14 September 2018. [Google Scholar]
- Talafha, B.; Al-Ayyoub, M. JUST at VQA-Med: A VGG-Seq2Seq Model. In Proceedings of the Working Notes of CLEF 2018, Avignon, France, 10–14 September 2018. [Google Scholar]
- Vu, M.H.; Lofstedt, T.; Nyholm, T.; Sznitman, R. A Question-Centric Model for Visual Question Answering in Medical Imaging. IEEE Trans. Med. Imaging 2020, 39, 2856–2868. [Google Scholar] [CrossRef] [PubMed]
- Kiros, R.; Zhu, Y.; Salakhutdinov, R.; Zemel, R.S.; Torralba, A.; Urtasun, R.; Fidler, S. Skip-Thought Vectors. Adv. Neural Inf. Process. Syst. 2015, 28, 3294–3302. [Google Scholar]
- Yang, Z.; Dai, Z.; Yang, Y.; Carbonell, J.; Salakhutdinov, R.; Le, Q.V. XLNet: Generalized Autoregressive Pretraining for Language Understanding. In Proceedings of the 33rd Advances in Neural Information Processing Systems, Vancouver, BC, Canada, 8–14 December 2019; pp. 5753–5763. [Google Scholar]
- Eslami, S.; de Melo, G.; Meinel, C. Teams at VQA-MED 2021: BBN-orchestra for long-tailed medical visual question answering. In Proceedings of the Working Notes of CLEF 2021, Bucharest, Romania, 21–24 September 2021; pp. 1211–1217. [Google Scholar]
- Schilling, R.; Messina, P.; Parra, D.; Lobel, H. Puc Chile team at VQA-MED 2021: Approaching VQA as a classfication task via fine-tuning a pretrained CNN. In Proceedings of the Working Notes of CLEF 2021, Bucharest, Romania, 21–24 September 2021; pp. 346–351. [Google Scholar]
- Zhou, Y.; Jun, Y.; Chenchao, X.; Jianping, F.; Dacheng, T. Beyond Bilinear: Generalized Multimodal Factorized High-Order Pooling for Visual Question Answering. IEEE Trans. Neural Netw. Learn. Syst. 2018, 29, 5947–5959. [Google Scholar]
- Malinowski, M.; Rohrbach, M.; Fritz, M. Ask Your Neurons: A Deep Learning Approach to Visual Question Answering. Int. J. Comput. Vis. 2017, 125, 110–135. [Google Scholar] [CrossRef]
- Kuniaki, S.; Andrew, S.; Yoshitaka, U.; Tatsuya, H. Dualnet: Domain-invariant network for visual question answering. In Proceedings of the the IEEE International Conference on Multimedia and Expo (ICME) 2017, Hong Kong, 10–14 July 2017; pp. 829–834. [Google Scholar]
- Noh, H.; Seo, P.H.; Han, B. Image Question Answering Using Convolutional Neural Network with Dynamic Parameter Prediction. In Proceedings of the CVPR, Las Vegas, NV, USA, 27–30 June 2016; pp. 30–38. [Google Scholar]
- Kim, J.H.; Lee, S.W.; Kwak, D.; Heo, M.O.; Kim, J.; Ha, J.W.; Zhang, B.T. Multimodal residual learning for visual QA. Adv. Neural Inf. Process. Syst. 2016, 29, 361–369. [Google Scholar]
- Mingrui, L.; Yanming, G.; Hui, W.; Xin, Z. Cross-modal multistep fusion network with co-attention for visual question answering. IEEE Access 2018, 6, 31516–31524. [Google Scholar]
- Bai, Y.; Fu, J.; Zhao, T.; Mei, T. Deep Attention Neural Tensor Network for Visual Question Answering. In Proceedings of the ECCV 2018, Munich, Germany, 8–14 September 2018; pp. 20–35. [Google Scholar]
- Narasimhan, M.; Schwing, A.G. Straight to the Facts: Learning Knowledge Base Retrieval for Factual Visual Question Answering. In Proceedings of the ECCV 2018, Munich, Germany, 8–14 September 2018; pp. 451–468. [Google Scholar]
- Chen, L.; Yan, X.; Xiao, J.; Zhang, H.; Pu, S.; Zhuang, Y. Counterfactual Samples Synthesizing for Robust Visual Question Answering. In Proceedings of the Computer Vision and Pattern Recognition, CVPR 2020, Seattle, WA, USA, 14–19 June 2020; pp. 10800–10809. [Google Scholar]
- Clark, K.; Luong, M.T.; Le, Q.V.; Manning, C.D. Electra: Pre-training text encoders as discriminators rather than generators. arXiv 2020, arXiv:2003.10555. [Google Scholar]
- Rumelhart, D.E.; Hinton, G.E.; McClelland, J.L. A general framework for parallel distributed processing. In Parallel Distributed Processing: Explorations in the Microstructure of Cognition; MIT Press: Cambridge, MA, USA, 1986; Volume 1, pp. 26, 45–76. [Google Scholar]
- Maas, A.L.; Hannun, A.Y.; Ng, A.Y. Rectifier nonlinearities improve neural network acoustic models. In Proceedings of the ICML, Atlanta, GA, USA, 16–21 June 2013; Volume 30, p. 3. [Google Scholar]
- Loshchilov, I.; Hutter, F. Decoupled weight decay regularization. arXiv 2017, arXiv:1711.05101. [Google Scholar]
- Porwal, P.; Pachade, S.; Kamble, R.; Kokare, M.; Deshmukh, G.; Sahasrabuddhe, V.; Meriaudeau, F. Indian diabetic retinopathy image dataset (IDRiD): A database for diabetic retinopathy screening research. Data 2018, 3, 25. [Google Scholar] [CrossRef]
- Decenciere, E.; Cazuguel, G.; Zhang, X.; Thibault, G.; Klein, J.C.; Meyer, F.; Marcotegui, B.; Quellec, G.; Lamard, M.; Danno, R.; et al. TeleOphta: Machine learning and image processing methods for teleophthalmology. IRBM 2013, 34, 196–203. [Google Scholar] [CrossRef]
- Selvaraju, R.R.; Tendulkar, P.; Parikh, D.; Horvitz, E.; Ribeiro, M.T.; Nushi, B.; Kamar, E. Squinting at VQA models: Introspecting vqa models with sub-questions. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 14–19 June 2020; pp. 10003–10011. [Google Scholar]

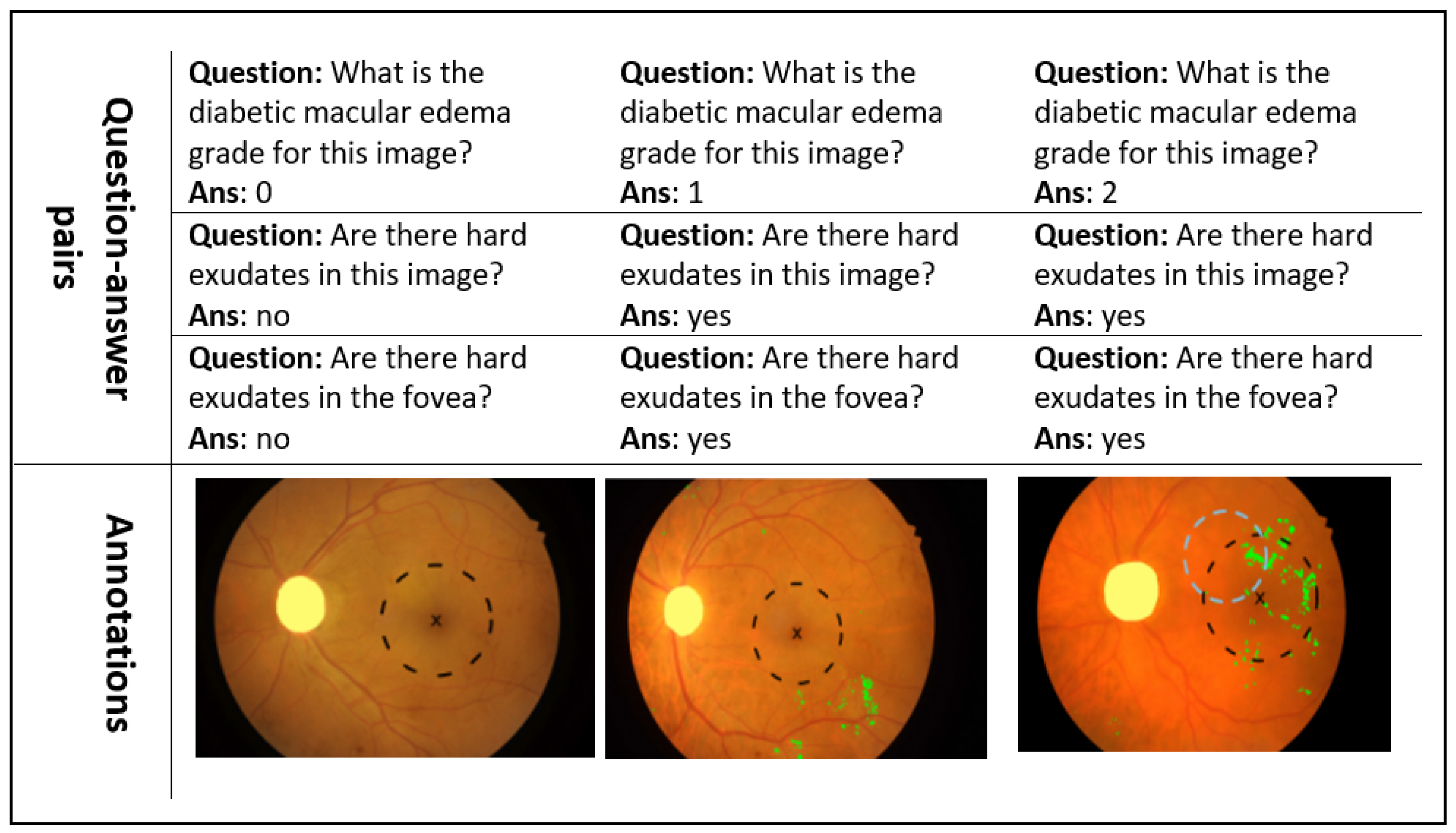



| Yes | No | 0 | 1 | 2 | Total | |
|---|---|---|---|---|---|---|
| Train | 4713 | 4639 | 166 | 41 | 220 | 9779 |
| Validation | 1151 | 1123 | 39 | 8 | 59 | 2380 |
| Test | 530 | 650 | 49 | 15 | 67 | 1311 |
| Total | 6394 | 6412 | 254 | 64 | 346 | 13,470 |
| Answer | Precision | Recall | F1-Score | Instances No. |
|---|---|---|---|---|
| 0 | 0.9512 | 0.7959 | 0.8667 | 49 |
| 1 | 0.4091 | 0.6000 | 0.4865 | 15 |
| 2 | 0.8971 | 0.9104 | 0.9037 | 67 |
| no | 0.9057 | 0.8569 | 0.8806 | 650 |
| yes | 0.8354 | 0.8906 | 0.8621 | 530 |
| accuracy | 0.8680 | 1311 | ||
| macro avg | 0.7997 | 0.8108 | 0.7999 | 1311 |
| weighted avg | 0.8729 | 0.8680 | 0.8693 | 1311 |
| Answer | Precision | Recall | F1-Score | Instances No. |
|---|---|---|---|---|
| 0 | 0.8723 | 0.8367 | 0.8542 | 49 |
| 1 | 0.4286 | 0.4000 | 0.4138 | 15 |
| 2 | 0.8714 | 0.9104 | 0.8905 | 67 |
| no | 0.9012 | 0.8415 | 0.8703 | 650 |
| yes | 0.8202 | 0.8868 | 0.8522 | 530 |
| accuracy | 0.8581 | 1311 | ||
| macro avg | 0.7787 | 0.7751 | 0.7762 | 1311 |
| weighted avg | 0.8604 | 0.8581 | 0.8582 | 1311 |
| Answer | Precision | Recall | F1-Score | Instances No. |
|---|---|---|---|---|
| 0 | 0.9048 | 0.7755 | 0.8352 | 49 |
| 1 | 0.3500 | 0.4667 | 0.4000 | 15 |
| 2 | 0.8841 | 0.9104 | 0.8971 | 67 |
| no | 0.8942 | 0.8585 | 0.8760 | 650 |
| yes | 0.8345 | 0.8755 | 0.8545 | 530 |
| accuracy | 0.8604 | 1311 | ||
| macro avg | 0.7735 | 0.7773 | 0.7725 | 1311 |
| weighted avg | 0.8637 | 0.8604 | 0.8614 | 1311 |
| Answer | Precision | Recall | F1-Score | Instances No. |
|---|---|---|---|---|
| 0 | 0.8936 | 0.8571 | 0.8750 | 49 |
| 1 | 0.4615 | 0.4000 | 0.4286 | 15 |
| 2 | 0.8732 | 0.9254 | 0.8986 | 67 |
| no | 0.8738 | 0.8523 | 0.8629 | 650 |
| yes | 0.8242 | 0.8491 | 0.8364 | 530 |
| accuracy | 0.8497 | 1311 | ||
| macro avg | 0.7853 | 0.7768 | 0.7803 | 1311 |
| weighted avg | 0.8497 | 0.8497 | 0.8495 | 1311 |
| Answer | Precision | Recall | F1-Score | Instances No. |
|---|---|---|---|---|
| 0 | 0.6622 | 1.0000 | 0.7967 | 49 |
| 1 | 0.0000 | 0.0000 | 0.0000 | 15 |
| 2 | 0.9636 | 0.7910 | 0.8689 | 67 |
| no | 0.7727 | 0.5231 | 0.6239 | 650 |
| yes | 0.5822 | 0.8151 | 0.6792 | 530 |
| accuracy | 0.6667 | 1311 | ||
| macro avg | 0.5961 | 0.6258 | 0.5937 | 1311 |
| weighted avg | 0.6925 | 0.6667 | 0.6581 | 1311 |
| Answer | Precision | Recall | F1-Score | Instances No. |
|---|---|---|---|---|
| 0 | 0.7119 | 0.8571 | 0.7778 | 49 |
| 1 | 0.0000 | 0.0000 | 0.0000 | 15 |
| 2 | 0.8194 | 0.8806 | 0.8489 | 67 |
| no | 0.7634 | 0.4815 | 0.5906 | 650 |
| yes | 0.5623 | 0.8170 | 0.6662 | 530 |
| accuracy | 0.6461 | 1311 | ||
| macro avg | 0.5714 | 0.6073 | 0.5767 | 1311 |
| weighted avg | 0.6743 | 0.6461 | 0.6346 | 1311 |
| Answer | Precision | Recall | F1-Score | Instances No. |
|---|---|---|---|---|
| 0 | 0.8000 | 0.8980 | 0.8462 | 49 |
| 1 | 0.8000 | 0.2667 | 0.4000 | 15 |
| 2 | 0.8592 | 0.9104 | 0.8841 | 67 |
| no | 0.8819 | 0.8615 | 0.8716 | 650 |
| yes | 0.8349 | 0.8585 | 0.8465 | 530 |
| accuracy | 0.8574 | 1311 | ||
| macro avg | 0.8352 | 0.7590 | 0.7697 | 1311 |
| weighted avg | 0.8577 | 0.8574 | 0.8557 | 1311 |
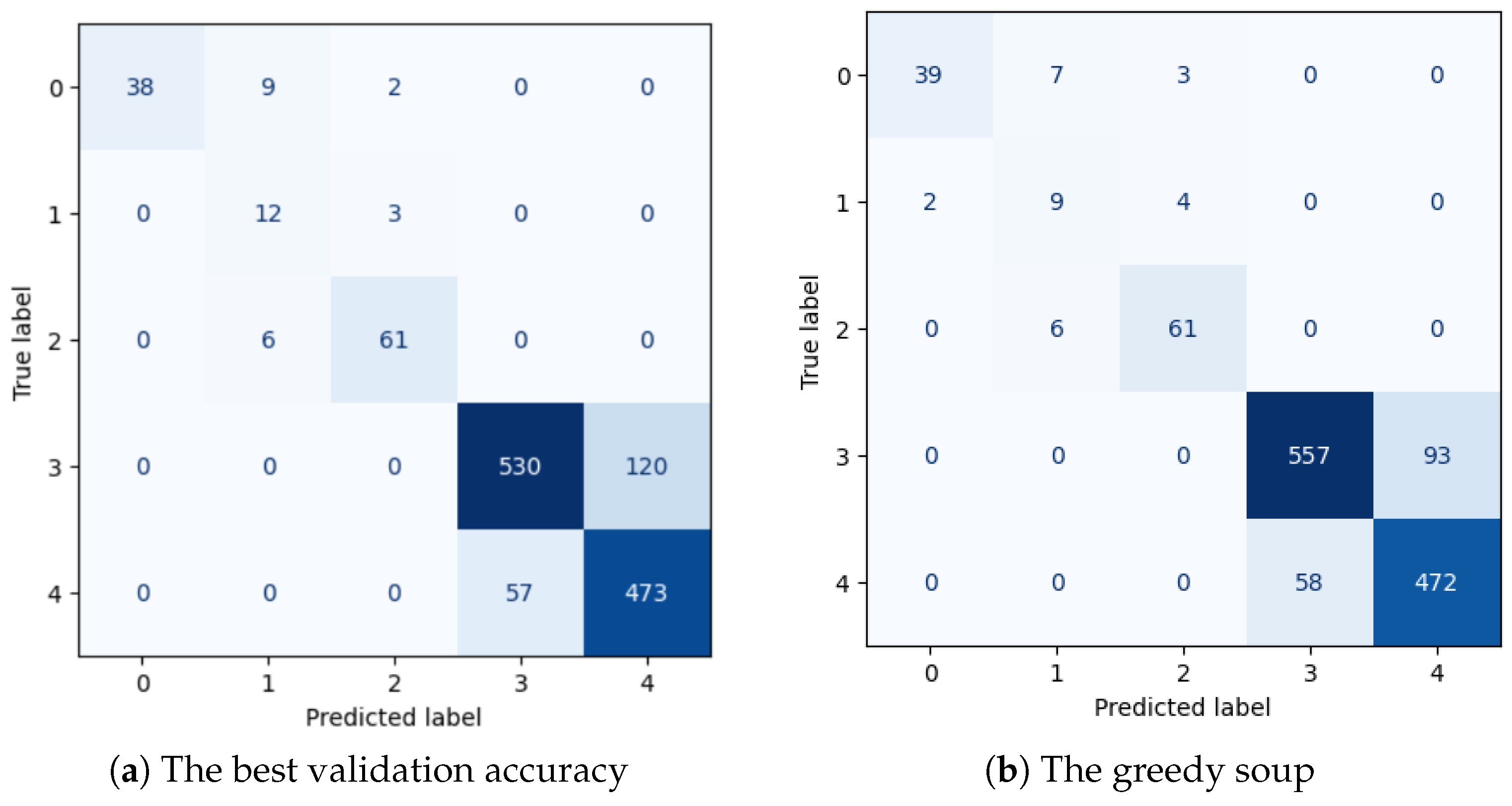
| Answer | Beat-Value-Based Model | Greedy-Soup-Based Model | Samples No. | ||||
|---|---|---|---|---|---|---|---|
| Precision | Recall | F1-Score | Precision | Recall | F1-Score | ||
| 0 | 1.000 | 0.7755 | 0.8736 | 0.9512 | 0.7959 | 0.8667 | 49 |
| 1 | 0.4444 | 0.8000 | 0.5714 | 0.4091 | 0.6000 | 0.4865 | 15 |
| 2 | 0.9242 | 0.9104 | 0.9173 | 0.8971 | 0.9104 | 0.9037 | 67 |
| no | 0.9029 | 0.8154 | 0.8569 | 0.9057 | 0.8569 | 0.8806 | 650 |
| yes | 0.7976 | 0.8925 | 0.8424 | 0.8354 | 0.8906 | 0.8621 | 530 |
| Accuracy | 0.8497 | 0.8680 | 1311 | ||||
| macro avg | 0.8138 | 0.8388 | 0.8123 | 0.7997 | 0.8108 | 0.7999 | 1311 |
| weighted avg | 0.8598 | 0.8497 | 0.8515 | 0.8729 | 0.8680 | 0.8693 | 1311 |
| Answer | Precision | Recall | F1-Score | Samples No. |
|---|---|---|---|---|
| 0 | 0.8913 | 0.8367 | 0.8632 | 49 |
| 1 | 0.4000 | 0.5333 | 0.4571 | 15 |
| 2 | 0.9077 | 0.8806 | 0.8939 | 67 |
| no | 0.8703 | 0.9185 | 0.8937 | 650 |
| yes | 0.8927 | 0.8321 | 0.8613 | 530 |
| accuracy | 0.8741 | 1311 | ||
| macro avg | 0.7924 | 0.8002 | 0.7939 | 1311 |
| weighted avg | 0.8767 | 0.8741 | 0.8745 | 1311 |
| Answer | Precision | Recall | F1-Score | Samples No. |
|---|---|---|---|---|
| 0 | 0.9750 | 0.7959 | 0.8764 | 49 |
| 1 | 0.4706 | 0.5333 | 0.5000 | 15 |
| 2 | 0.8514 | 0.9403 | 0.8936 | 67 |
| no | 0.9011 | 0.7846 | 0.8388 | 650 |
| yes | 0.7720 | 0.8943 | 0.8287 | 530 |
| accuracy | 0.8345 | 1311 | ||
| macro avg | 0.7940 | 0.7897 | 0.7875 | 1311 |
| weighted avg | 0.8442 | 0.8345 | 0.8350 | 1311 |
| Answer | Precision | Recall | F1-Score | Samples No. |
|---|---|---|---|---|
| 0 | 0.9730 | 0.7347 | 0.8372 | 49 |
| 1 | 0.3684 | 0.4667 | 0.4118 | 15 |
| 2 | 0.8400 | 0.9403 | 0.8873 | 67 |
| no | 0.9089 | 0.7985 | 0.8501 | 650 |
| yes | 0.7849 | 0.9019 | 0.8393 | 530 |
| accuracy | 0.8413 | 1311 | ||
| macro avg | 0.7750 | 0.7684 | 0.7652 | 1311 |
| weighted avg | 0.8515 | 0.8413 | 0.8422 | 1311 |
| Answer | Precision | Recall | F1-Score | Samples No. |
|---|---|---|---|---|
| 0 | 0.9231 | 0.7347 | 0.8182 | 49 |
| 1 | 0.3077 | 0.5333 | 0.3902 | 15 |
| 2 | 0.9091 | 0.8955 | 0.9023 | 67 |
| no | 0.8952 | 0.8538 | 0.8740 | 650 |
| yes | 0.8304 | 0.8774 | 0.8532 | 530 |
| accuracy | 0.8574 | 1311 | ||
| macro avg | 0.7731 | 0.7790 | 0.7676 | 1311 |
| weighted avg | 0.8640 | 0.8574 | 0.8594 | 1311 |
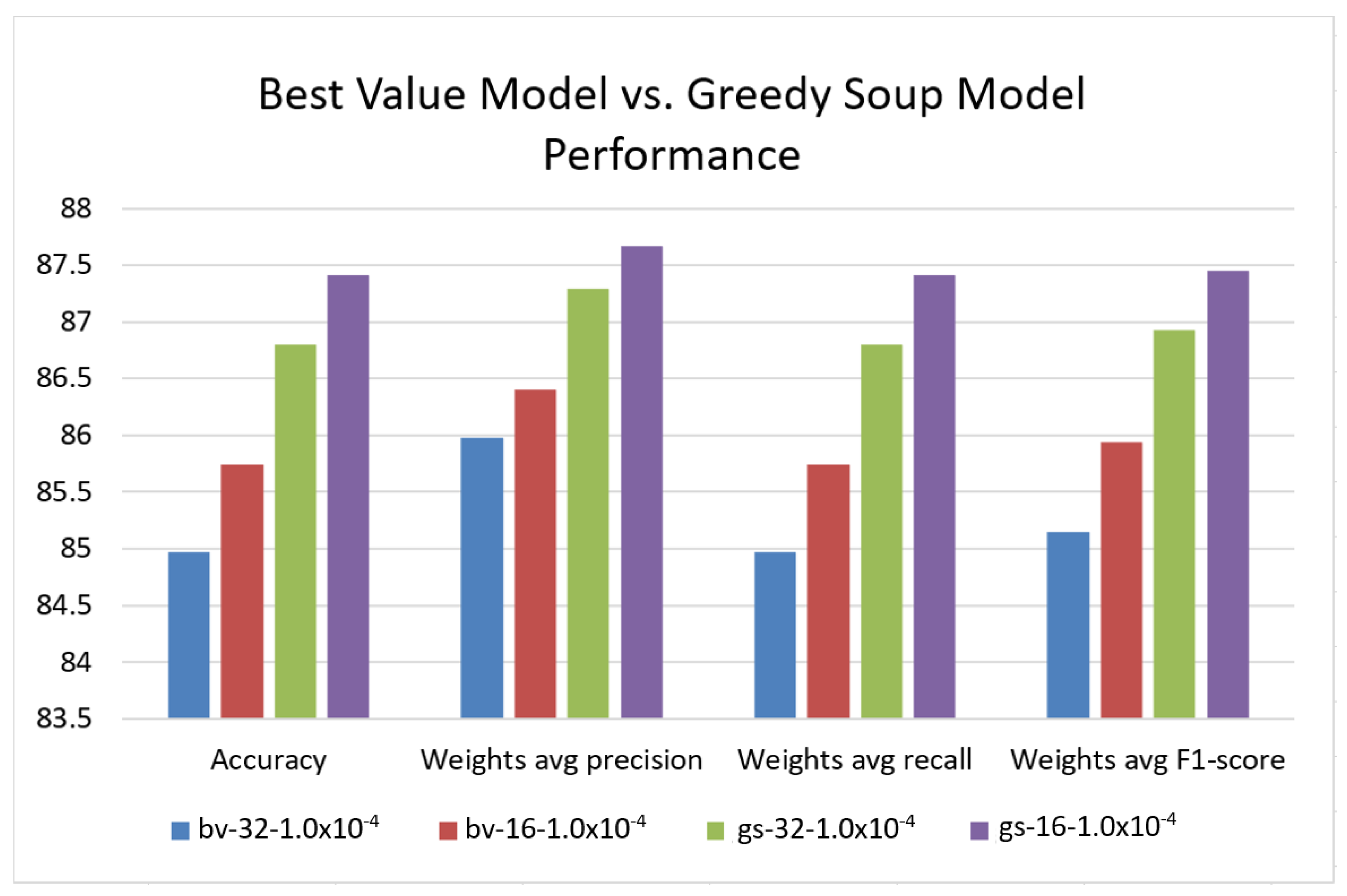
| Model | Accuracy | Weights Avg Precision | Weights Avg Recall | Weights Avg F1-Score |
|---|---|---|---|---|
| bv-32- | 84.97 | 85.98 | 84.97 | 85.15 |
| bv-16- | 85.74 | 86.40 | 85.74 | 85.94 |
| gs-32- | 86.80 | 87.29 | 86.80 | 86.93 |
| gs-16- | 87.41 | 87.67 | 87.41 | 87.45 |
| Model | Accuracy | Avg. Macro Precision | Avg. Macro Recall | Avg. Macro F1-Score | Weighted Avg. Precision | Weighted Avg. Recall | Weighted Avg. F1-Score |
|---|---|---|---|---|---|---|---|
| bv-32- | 0.8497 | 0.8138 | 0.8388 | 0.8123 | 0.8598 | 0.8497 | 0.8515 |
| bv-16- | 0.8574 | 0.7731 | 0.7790 | 0.7676 | 0.8640 | 0.8574 | 0.8594 |
| gs-32- | 0.868 | 0.7997 | 0.8108 | 0.7999 | 0.8729 | 0.8680 | 0.8693 |
| gs-32- | 0.8581 | 0.7787 | 0.7751 | 0.7762 | 0.8604 | 0.8581 | 0.8582 |
| gs-32- | 0.8604 | 0.7735 | 0.7773 | 0.7725 | 0.8637 | 0.8604 | 0.8614 |
| gs-32- | 0.8497 | 0.7853 | 0.7768 | 0.7803 | 0.8497 | 0.8497 | 0.8495 |
| gs-32- | 0.6667 | 0.5961 | 0.6258 | 0.5937 | 0.6925 | 0.6667 | 0.6581 |
| gs-32- | 0.6461 | 0.5714 | 0.6073 | 0.5767 | 0.6743 | 0.6461 | 0.6346 |
| gs-32- | 0.8574 | 0.8352 | 0.7590 | 0.7697 | 0.8577 | 0.8574 | 0.8557 |
| gs-16- | 0.8741 | 0.7924 | 0.8002 | 0.7939 | 0.8767 | 0.8741 | 0.8745 |
| gs-64- | 0.8345 | 0.7940 | 0.7897 | 0.7875 | 0.8442 | 0.8345 | 0.8350 |
| gs-128- | 0.8413 | 0.7750 | 0.7684 | 0.7652 | 0.8515 | 0.8413 | 0.8422 |
| Model | Overall | Grade | Whole | Macula | Region |
|---|---|---|---|---|---|
| SOTA [5] | 83.49 | 80.69 | 84.96 | 87.18 | 83.16 |
| bv-32- | 84.97 | 84.73 | 90.84 | 85.29 | 83.22 |
| bv-16- | 85.74 | 79.39 | 90.84 | 83.21 | 86.27 |
| gs-128- | 84.13 | 80.92 | 90.08 | 88.55 | 83.12 |
| gs-32- | 85.74 | 83.21 | 90.08 | 87.79 | 85.19 |
| gs-32- | 86.8 | 83.21 | 92.37 | 90.84 | 85.95 |
| gs-16- | 87.41 | 82.44 | 88.55 | 87.02 | 88.02 |
| gs-32- | 84.97 | 83.97 | 87.02 | 89.31 | 84.20 |
| gs-32- | 85.81 | 82.44 | 89.31 | 88.55 | 85.40 |
| gs-32- | 86.04 | 80.92 | 89.31 | 89.32 | 85.84 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al-Hadhrami, S.; Menai, M.E.B.; Al-Ahmadi, S.; Alnafessah, A. An Effective Med-VQA Method Using a Transformer with Weights Fusion of Multiple Fine-Tuned Models. Appl. Sci. 2023, 13, 9735. https://doi.org/10.3390/app13179735
Al-Hadhrami S, Menai MEB, Al-Ahmadi S, Alnafessah A. An Effective Med-VQA Method Using a Transformer with Weights Fusion of Multiple Fine-Tuned Models. Applied Sciences. 2023; 13(17):9735. https://doi.org/10.3390/app13179735
Chicago/Turabian StyleAl-Hadhrami, Suheer, Mohamed El Bachir Menai, Saad Al-Ahmadi, and Ahmad Alnafessah. 2023. "An Effective Med-VQA Method Using a Transformer with Weights Fusion of Multiple Fine-Tuned Models" Applied Sciences 13, no. 17: 9735. https://doi.org/10.3390/app13179735
APA StyleAl-Hadhrami, S., Menai, M. E. B., Al-Ahmadi, S., & Alnafessah, A. (2023). An Effective Med-VQA Method Using a Transformer with Weights Fusion of Multiple Fine-Tuned Models. Applied Sciences, 13(17), 9735. https://doi.org/10.3390/app13179735






