FPN-SE-ResNet Model for Accurate Diagnosis of Kidney Tumors Using CT Images
Abstract
:1. Introduction
1.1. Diagnosis
1.2. Computer-Aided Diagnosis
1.3. Medical Imaging
1.4. Convolutional Neural Network
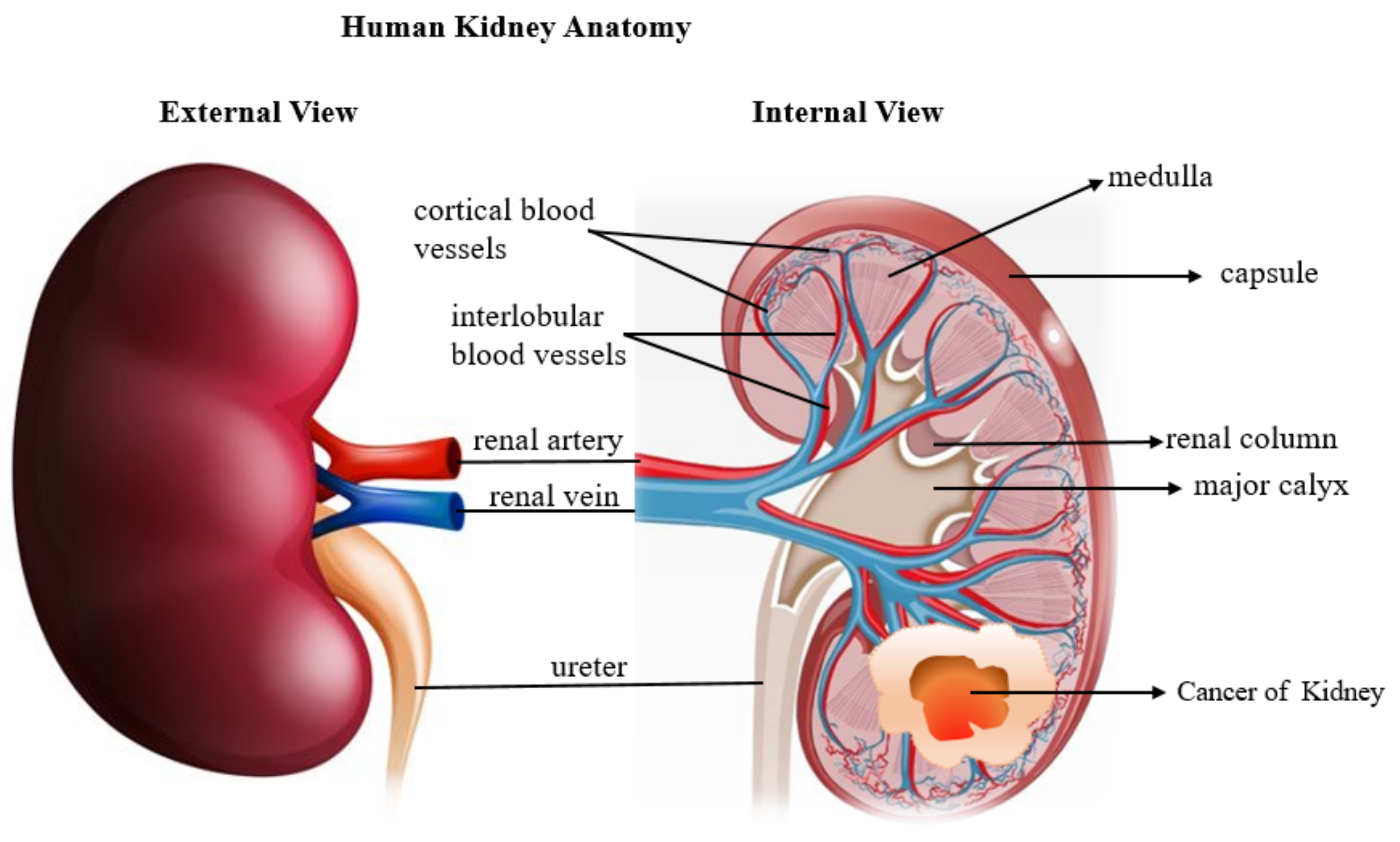
1.5. Kidney Cancer
1.6. Current Methods
1.7. Problem Description
1.8. Paper Structure
2. Materials and Methods
2.1. Dataset
2.2. Preprocessing
2.3. Feature Pyramid Network
2.4. SE-ResNets
2.5. SE-ResNet + FPN Network
2.6. Evaluation Metrics
2.7. Loss Function
2.8. Proposed Solution
2.8.1. Backbone Architectures
2.8.2. Methodology
2.9. Implementation
2.10. Model Setup
3. Results and Discussion
3.1. Experimental Results
3.2. Discussion
3.2.1. Result Analysis
3.2.2. Model Accuracy and Model Loss
3.2.3. Prediction on Validation Images
3.2.4. Prediction on Testing Images
3.2.5. Computational Efficiency and Training Time
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhou, X.; Menche, J.; Barabási, A.L.; Sharma, A. Human Symptoms-Disease Network. Nat. Commun. 2014, 5, 4212. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, J. Diagnostic Reasoning in Action. IEEE Trans. Syst. Man Cybern. 1993, 23, 981–992. [Google Scholar] [CrossRef]
- Clark, L.A.; Cuthbert, B.; Lewis-Fernández, R.; Narrow, W.E.; Reed, G.M. Three Approaches to Understanding and Classifying Mental Disorder: ICD-11, DSM-5, and the National Institute of Mental Health’s Research Domain Criteria (RDoC). Psychol. Sci. Public Interes. 2017, 18, 72–145. [Google Scholar] [CrossRef]
- Dalby, W. Section of Otology. Br. Med. J. 1895, 2, 1289–1294. [Google Scholar] [CrossRef] [PubMed]
- Scheuermann, R.H.; Ceusters, W.; Smith, B. Toward an Ontological Treatment of Disease and Diagnosis Department of Pathology and Division of Biomedical Informatics, University of Texas. AMIA Summit Transl. Bioinform. 2009, 2009, 116–120. [Google Scholar]
- Croft, P.; Altman, D.G.; Deeks, J.J.; Dunn, K.M.; Hay, A.D.; Hemingway, H.; LeResche, L.; Peat, G.; Perel, P.; Petersen, S.E.; et al. The Science of Clinical Practice: Disease Diagnosis or Patient Prognosis? Evidence about “What Is Likely to Happen” Should Shape Clinical Practice. BMC Med. 2015, 13, 1–8. [Google Scholar] [CrossRef]
- Torres, A.; Nieto, J.J. Fuzzy Logic in Medicine and Bioinformatics. J. Biomed. Biotechnol. 2006, 2006, 1–7. [Google Scholar] [CrossRef]
- Alam, R.; Cheraghi-Sohi, S.; Panagioti, M.; Esmail, A.; Campbell, S.; Panagopoulou, E. Managing Diagnostic Uncertainty in Primary Care: A Systematic Critical Review. BMC Fam. Pract. 2017, 18, 79. [Google Scholar] [CrossRef] [PubMed]
- Malmir, B.; Amini, M.; Chang, S.I. A Medical Decision Support System for Disease Diagnosis under Uncertainty. Expert Syst. Appl. 2017, 88, 95–108. [Google Scholar] [CrossRef]
- Nilashi, M.; Ibrahim, O.B.; Ahmadi, H.; Shahmoradi, L. An Analytical Method for Diseases Prediction Using Machine Learning Techniques. Comput. Chem. Eng. 2017, 106, 212–223. [Google Scholar] [CrossRef]
- Nilashi, M.; Ibrahim, O.; Ahmadi, H.; Shahmoradi, L.; Farahmand, M. A Hybrid Intelligent System for the Prediction of Parkinson’s Disease Progression Using Machine Learning Techniques. Biocybern. Biomed. Eng. 2018, 38, 1–15. [Google Scholar] [CrossRef]
- Nilashi, M.; Ibrahim, O.; Dalvi, M.; Ahmadi, H.; Shahmoradi, L. Accuracy Improvement for Diabetes Disease Classification: A Case on a Public Medical Dataset. Fuzzy Inf. Eng. 2017, 9, 345–357. [Google Scholar] [CrossRef]
- Abdelrahman, A.; Viriri, S. Kidney Tumor Semantic Segmentation Using Deep Learning: A Survey of State-of-the-Art. J. Imaging 2022, 8, 55. [Google Scholar] [CrossRef] [PubMed]
- Salih, O.; Duffy, K.J. Optimization Convolutional Neural Network for Automatic Skin Lesion Diagnosis Using a Genetic Algorithm. Appl. Sci. 2023, 13, 3248. [Google Scholar] [CrossRef]
- Gur, D.; Sumkin, J.H.; Rockette, H.E.; Ganott, M.; Hakim, C.; Hardesty, L.; Poller, W.R.; Shah, R.; Wallace, L. Changes in Breast Cancer Detection and Mammography Recall Rates after the Introduction of a Computer-Aided Detection System. J. Natl. Cancer Inst. 2004, 96, 185–190. [Google Scholar] [CrossRef]
- Destounis, S.V.; DiNitto, P.; Logan-Young, W.; Bonaccio, E.; Zuley, M.L.; Willison, K.M. Can Computer-Aided Detection with Double Reading of Screening Mammograms Help Decrease the False-Negative Rate? Initial Experience. Radiology 2004, 232, 578–584. [Google Scholar] [CrossRef]
- Doi, K. Computer-Aided Diagnosis in Medical Imaging: Historical Review, Current Status and Future Potential. Comput. Med. Imaging Graph. 2007, 31, 198–211. [Google Scholar] [CrossRef]
- Salih, O.; Viriri, S. Skin Lesion Segmentation Using Stochastic Region-Merging and Pixel-Based Markov Random Field. Symmetry 2020, 12, 1224. [Google Scholar] [CrossRef]
- Li, Q.; Li, F.; Suzuki, K.; Shiraishi, J.; Abe, H.; Engelmann, R.; Nie, Y.; MacMahon, H.; Doi, K. Computer-Aided Diagnosis in Thoracic CT. Semin. Ultrasound CT MRI 2005, 26, 357–363. [Google Scholar] [CrossRef]
- Liu, Y.Y.; Huang, Z.H.; Huang, K.W. Deep Learning Model for Computer-Aided Diagnosis of Urolithiasis Detection from Kidney–Ureter–Bladder Images. Bioengineering 2022, 9, 811. [Google Scholar] [CrossRef]
- Panayides, A.S.; Amini, A.; Filipovic, N.D.; Sharma, A.; Tsaftaris, S.A.; Young, A.; Foran, D.; Do, N.; Golemati, S.; Kurc, T.; et al. AI in Medical Imaging Informatics: Current Challenges and Future Directions. IEEE J. Biomed. Heal. Inf. 2020, 24, 1837–1857. [Google Scholar] [CrossRef]
- Lim, E.J.; Castellani, D.; So, W.Z.; Fong, K.Y.; Li, J.Q.; Tiong, H.Y.; Gadzhiev, N.; Heng, C.T.; Teoh, J.Y.C.; Naik, N.; et al. Radiomics in Urolithiasis: Systematic Review of Current Applications, Limitations, and Future Directions. J. Clin. Med. 2022, 11, 5151. [Google Scholar] [CrossRef] [PubMed]
- Vishwanath, V.; Jafarieh, S.; Rembielak, A. The Role of Imaging in Head and Neck Cancer: An Overview of Different Imaging Modalities in Primary Diagnosis and Staging of the Disease. J. Contemp. Brachyther. 2020, 12, 512–518. [Google Scholar] [CrossRef]
- Bi, W.L.; Hosny, A.; Schabath, M.B.; Giger, M.L.; Birkbak, N.J.; Mehrtash, A.; Allison, T.; Arnaout, O.; Abbosh, C.; Dunn, I.F.; et al. Artificial Intelligence in Cancer Imaging: Clinical Challenges and Applications. CA Cancer J. Clin. 2019, 69, 127–157. [Google Scholar] [CrossRef] [PubMed]
- Salmanpour, M.R.; Shamsaei, M.; Saberi, A.; Setayeshi, S.; Klyuzhin, I.S.; Sossi, V.; Rahmim, A. Optimized Machine Learning Methods for Prediction of Cognitive Outcome in Parkinson’s Disease. Comput. Biol. Med. 2019, 111, 103347. [Google Scholar] [CrossRef] [PubMed]
- LeCun, Y.; Bottou, L.; Bengio, Y.; Haffner, P. Gradient-Based Learning Applied to Document Recognition. Proc. IEEE 1998, 86, 2278–2323. [Google Scholar] [CrossRef]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.A.W.M.; van Ginneken, B.; Sánchez, C.I. A Survey on Deep Learning in Medical Image Analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef]
- Sarvamangala, D.R.; Kulkarni, R.V. Convolutional Neural Networks in Medical Image Understanding: A Survey. Evol. Intell. 2022, 15, 1–22. [Google Scholar] [CrossRef]
- Chan, H.P.; Hadjiiski, L.M.; Samala, R.K. Computer-Aided Diagnosis in the Era of Deep Learning. Med. Phys. 2020, 47, e218–e227. [Google Scholar] [CrossRef]
- Li, Z.; Liu, F.; Yang, W.; Peng, S.; Zhou, J. A Survey of Convolutional Neural Networks: Analysis, Applications, and Prospects. IEEE Trans. Neural Netw. Learn. Syst. 2022, 33, 6999–7019. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Yang, W.; Gao, Y.; Shen, D. Does Manual Delineation Only Provide the Side Information in CT Prostate Segmentation? Lect. Notes Comput. Sci. 2017, 10435, 692–700. [Google Scholar] [CrossRef]
- He, B.; Xiao, D.; Hu, Q.; Jia, F. Automatic Magnetic Resonance Image Prostate Segmentation Based on Adaptive Feature Learning Probability Boosting Tree Initialization and CNN-ASM Refinement. IEEE Access 2017, 6, 2005–2015. [Google Scholar] [CrossRef]
- Mortazi, A.; Karim, R.; Rhode, K.; Burt, J.; Bagci, U. CardiacNET: Segmentation of Left Atrium and Proximal Pulmonary Veins from MRI Using Multi-View CNN. Lect. Notes Comput. Sci. 2017, 10434, 377–385. [Google Scholar] [CrossRef]
- Patravali, J.; Jain, S.; Chilamkurthy, S. 2D-3D Fully Convolutional Neural Networks for Cardiac MR Segmentation. Lect. Notes Comput. Sci. 2018, 10663, 130–139. [Google Scholar] [CrossRef]
- Moeskops, P.; Viergever, M.A.; Mendrik, A.M.; De Vries, L.S.; Benders, M.J.N.L.; Isgum, I. Automatic Segmentation of MR Brain Images with a Convolutional Neural Network. IEEE Trans. Med. Imaging 2016, 35, 1252–1261. [Google Scholar] [CrossRef]
- Wang, G.; Li, W.; Zuluaga, M.A.; Pratt, R.; Patel, P.A.; Aertsen, M.; Doel, T.; David, A.L.; Deprest, J.; Ourselin, S.; et al. Interactive Medical Image Segmentation Using Deep Learning with Image-Specific Fine Tuning. IEEE Trans. Med. Imaging 2018, 37, 1562–1573. [Google Scholar] [CrossRef]
- Salmanpour, M.R.; Hosseinzadeh, M.; Masoud, S. Computer Methods and Programs in Biomedicine Fusion-Based Tensor Radiomics Using Reproducible Features: Application to Survival Prediction in Head and Neck Cancer. Comput. Methods Programs Biomed. 2023, 240, 107714. [Google Scholar] [CrossRef]
- Salmanpour, M.R.; Rezaeijo, S.M.; Hosseinzadeh, M.; Rahmim, A. Deep versus Handcrafted Tensor Radiomics Features: Prediction of Survival in Head and Neck Cancer Using Machine Learning and Fusion Techniques. Diagnostics 2023, 13, 1696. [Google Scholar] [CrossRef]
- Jahangirimehr, A.; Abdolahi Shahvali, E.; Rezaeijo, S.M.; Khalighi, A.; Honarmandpour, A.; Honarmandpour, F.; Labibzadeh, M.; Bahmanyari, N.; Heydarheydari, S. Machine Learning Approach for Automated Predicting of COVID-19 Severity Based on Clinical and Paraclinical Characteristics: Serum Levels of Zinc, Calcium, and Vitamin D. Clin. Nutr. ESPEN 2022, 51, 404–411. [Google Scholar] [CrossRef]
- Lee, K.; Zung, J.; Li, P.; Jain, V.; Seung, H.S. Superhuman Accuracy on the SNEMI3D Connectomics Challenge. arXiv 2017, arXiv:1706.00120. [Google Scholar]
- Isensee, F.; Petersen, J.; Klein, A.; Zimmerer, D.; Jaeger, P.F.; Kohl, S.; Wasserthal, J.; Koehler, G.; Norajitra, T.; Wirkert, S.; et al. NnU-Net: Self-Adapting Framework for U-Net-Based Medical Image Segmentation. Inform. Aktuell 2019, 22. [Google Scholar] [CrossRef]
- Vu, M.H.; Grimbergen, G.; Nyholm, T.; Löfstedt, T. Evaluation of Multislice Inputs to Convolutional Neural Networks for Medical Image Segmentation. Med. Phys. 2020, 47, 6216–6231. [Google Scholar] [CrossRef] [PubMed]
- Motzer, R.J.; Bander, N.H.; Nanus, D.M. Renal-Cell Carcinoma. N. Engl. J. Med. 1996, 335, 865–875. [Google Scholar] [CrossRef]
- Liu, J.; Yildirim, O.; Akin, O.; Tian, Y. AI-Driven Robust Kidney and Renal Mass Segmentation and Classification on 3D CT Images. Bioengineering 2023, 10, 116. [Google Scholar] [CrossRef]
- Shehata, M.; Abouelkheir, R.T.; Gayhart, M.; Van Bogaert, E.; Abou El-Ghar, M.; Dwyer, A.C.; Ouseph, R.; Yousaf, J.; Ghazal, M.; Contractor, S.; et al. Role of AI and Radiomic Markers in Early Diagnosis of Renal Cancer and Clinical Outcome Prediction: A Brief Review. Cancers 2023, 15, 2835. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer Statistics, 2015. CA. Cancer J. Clin. 2015, 65, 5–29. [Google Scholar] [CrossRef]
- Muglia, V.F.; Prando, A. Renal Cell Carcinoma: Histological Classification and Correlation with Imaging Findings. Radiol. Bras. 2015, 48, 166–174. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, P.A.; Moch, H.; Cubilla, A.L.; Ulbright, T.M.; Reuter, V.E. The 2016 WHO Classification of Tumours of the Urinary System and Male Genital Organs—Part B: Prostate and Bladder Tumours. Eur. Urol. 2016, 70, 106–119. [Google Scholar] [CrossRef]
- Rendon, R.A. Active Surveillance as the Preferred Management Option for Small Renal Masses. J. Can. Urol. Assoc. 2010, 4, 136–138. [Google Scholar] [CrossRef]
- Mindrup, S.R.; Pierre, J.S.; Dahmoush, L.; Konety, B.R. The Prevalence of Renal Cell Carcinoma Diagnosed at Autopsy. BJU Int. 2005, 95, 31–33. [Google Scholar] [CrossRef]
- Nadimi-Shahraki, M.H.; Taghian, S.; Mirjalili, S.; Abualigah, L. Binary Aquila Optimizer for Selecting Effective Features from Medical Data: A COVID-19 Case Study. Mathematics 2022, 10, 1929. [Google Scholar] [CrossRef]
- Rezaeijo, S.M.; Nesheli, S.J.; Serj, M.F.; Birgani, M.J.T. Segmentation of the Prostate, Its Zones, Anterior Fibromuscular Stroma, and Urethra on the MRIs and Multimodality Image Fusion Using U-Net Model. Quant. Imaging Med. Surg. 2022, 12, 4786–4804. [Google Scholar] [CrossRef]
- Ekinci, S.; Izci, D.; Eker, E.; Abualigah, L. An Effective Control Design Approach Based on Novel Enhanced Aquila Optimizer for Automatic Voltage Regulator; Springer: Berlin/Heidelberg, Germany, 2023; Volume 56. [Google Scholar]
- Tsuneki, M. Deep Learning Models in Medical Image Analysis. J. Oral Biosci. 2022, 64, 312–320. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In BT—Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015; Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F., Eds.; Springer International Publishing: Berlin/Heidelberg, Germany, 2015. [Google Scholar]
- Yang, G.; Li, G.; Pan, T.; Kong, Y. Automatic Segmentation of Kidney and Renal Tumor in CT Images Based on 3D Fully Convolutional Neural Network with Pyramid Pooling Module. In Proceedings of the 2018 24th International Conference on Pattern Recognition (ICPR), Beijing, China, 20–24 August 2018; pp. 3790–3795. [Google Scholar] [CrossRef]
- Diniz, J.O.B.; Ferreira, J.L.; Diniz, P.H.B.; Silva, A.C.; de Paiva, A.C. Esophagus Segmentation from Planning CT Images Using an Atlas-Based Deep Learning Approach. Comput. Methods Programs Biomed. 2020, 197, 105685. [Google Scholar] [CrossRef] [PubMed]
- Ducros, N.; Mur, A.L.; Peyrin, F. A Completion Network for Reconstruction from Compressed Acquisition. In Proceedings of the IEEE 17th International Symposium on Biomedical Imaging (ISBI), Iowa City, IA, USA, 3–7 April 2020; pp. 619–623. [Google Scholar] [CrossRef]
- Türk, F.; Lüy, M.; Barışçı, N. Kidney and Renal Tumor Segmentation Using a Hybrid V-Net-Based Model. Mathematics 2020, 8, 1772. [Google Scholar] [CrossRef]
- Lin, D.T.; Lei, C.C.; Hung, S.W. Computer-Aided Kidney Segmentation on Abdominal CT Images. IEEE Trans. Inf. Technol. Biomed. 2006, 10, 59–65. [Google Scholar] [CrossRef]
- D’Arco, A.; Ferrara, M.A.; Indolfi, M.; Tufano, V.; Sirleto, L. Implementation of Stimulated Raman Scattering Microscopy for Single Cell Analysis. Nonlinear Opt. Appl. X 2017, 10228, 102280S. [Google Scholar] [CrossRef]
- Khalifa, F.; Gimel’farb, G.; Abo El-Ghar, M.; Sokhadze, G.; Manning, S.; McClure, P.; Ouseph, R.; El-Baz, A. A New Deformable Model-Based Segmentation Approach for Accurate Extraction of the Kidney from Abdominal CT Images. In Proceedings of the 18th IEEE International Conference on Image Processing, Brussels, Belgium, 11–14 September 2011; pp. 3393–3396. [Google Scholar] [CrossRef]
- Yang, G.; Gu, J.; Chen, Y.; Liu, W.; Tang, L.; Shu, H.; Toumoulin, C. Automatic Kidney Segmentation in CT Images Based on Multi-Atlas Image Registration. In Proceedings of the 36th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Chicago, IL, USA, 26–30 August 2014; pp. 5538–5541. [Google Scholar] [CrossRef]
- Cuingnet, R.; Prevost, R.; Lesage, D.; Cohen, L.D.; Mory, B.; Ardon, R. Automatic Detection and Segmentation of Kidneys in 3D CT Images Using Random Forests. Lect. Notes Comput. Sci. 2012, 7512, 66–74. [Google Scholar] [CrossRef]
- Jin, C.; Shi, F.; Xiang, D.; Jiang, X.; Zhang, B.; Wang, X.; Zhu, W.; Gao, E.; Chen, X. 3D Fast Automatic Segmentation of Kidney Based on Modified AAM and Random Forest. IEEE Trans. Med. Imaging 2016, 35, 1395–1407. [Google Scholar] [CrossRef]
- Hsiao, C.H.; Lin, P.C.; Chung, L.A.; Lin, F.Y.S.; Yang, F.J.; Yang, S.Y.; Wu, C.H.; Huang, Y.; Sun, T.L. A Deep Learning-Based Precision and Automatic Kidney Segmentation System Using Efficient Feature Pyramid Networks in Computed Tomography Images. Comput. Methods Programs Biomed. 2022, 221, 106854. [Google Scholar] [CrossRef] [PubMed]
- Heller, N.; Sathianathen, N.; Kalapara, A.; Walczak, E.; Moore, K.; Kaluzniak, H.; Rosenberg, J.; Blake, P.; Rengel, Z.; Oestreich, M.; et al. The KiTS19 Challenge Data: 300 Kidney Tumor Cases with Clinical Context, CT Semantic Segmentations, and Surgical Outcomes. arXiv 2019, arXiv:1904.00445. [Google Scholar]
- Aresta, G.; Araújo, T.; Kwok, S.; Chennamsetty, S.S.; Safwan, M.; Alex, V.; Marami, B.; Prastawa, M.; Chan, M.; Donovan, M.; et al. BACH: Grand Challenge on Breast Cancer Histology Images. Med. Image Anal. 2019, 56, 122–139. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.; Ge, W.; Yu, Y. GraphFPN: Graph Feature Pyramid Network for Object Detection. In Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV), Montreal, QC, Canada, 10–17 October 2021; pp. 2743–2752. [Google Scholar] [CrossRef]
- Lin, T.-Y.; Dollár, P.; Girshick, R.; He, K.; Hariharan, B.; Belongie, S. Feature Pyramid Networks for Object Detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 2117–2125. [Google Scholar]
- Liu, S.; Qi, L.; Qin, H.; Shi, J.; Jia, J. PANet: Path Aggregation Network for Instance Segmentation. arXiv 2019, arXiv:1803.01534v3. [Google Scholar]
- Zhang, Z.; Zhang, X.; Peng, C.; Xue, X.; Sun, J. ExFuse: Enhancing Feature Fusion for Semantic Segmentation. Lect. Notes Comput. Sci. 2018, 11214, 273–288. [Google Scholar] [CrossRef]
- Lin, D.; Shen, D.; Shen, S.; Ji, Y.; Lischinski, D.; Cohen-Or, D.; Huang, H. Zigzagnet: Fusing Top-down and Bottom-up Context for Object Segmentation. Proc. IEEE Comput. Soc. Conf. Comput. Vis. Pattern Recognit. 2019, 2019, 7482–7491. [Google Scholar] [CrossRef]
- Toshev, A.; Szegedy, C. Deeppose: Human Pose Estimation via Deep Neural Networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Columbus, OH, USA, 23–28 June 2014; pp. 1653–1660. [Google Scholar]
- Wu, G.; Ji, X.; Yang, G.; Jia, Y.; Cao, C. Signal-to-Image: Rolling Bearing Fault Diagnosis Using ResNet Family Deep-Learning Models. Processes 2023, 11, 1527. [Google Scholar] [CrossRef]
- Task, C. Se-ResNet with GAN Based Data Augmentation Applied to Acoustic Scene Classificaiton. Detect. Classif. Acoust. Scenes Events 2018, 2018, 10063424. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet Classification with Deep Convolutional Neural Networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. In Proceedings of the 3rd International Conference on Learning Representations—ICLR 2015, San Diego, CA, USA, 7–9 May 2015; pp. 1–14. [Google Scholar]
- Lecun, Y.; Bengio, Y.; Hinton, G. Deep Learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Taha, A.A.; Hanbury, A. Metrics for Evaluating 3D Medical Image Segmentation: Analysis, Selection, and Tool. BMC Med. Imaging 2015, 15, 29. [Google Scholar] [CrossRef]
- Popovic, A.; de la Fuente, M.; Engelhardt, M.; Radermacher, K. Statistical Validation Metric for Accuracy Assessment in Medical Image Segmentation. Int. J. Comput. Assist. Radiol. Surg. 2007, 2, 169–181. [Google Scholar] [CrossRef]
- Lin, T.-Y.; Goyal, P.; Girshick, R.; He, K.; Dollár, P. Focal Loss for Dense Object Detection. In Proceedings of the IEEE International Conference on Computer Vision, Honolulu, HI, USA, 21–26 July 2017; pp. 2980–2988. [Google Scholar]
- Kodym, O.; Španěl, M.; Herout, A. Segmentation of Head and Neck Organs at Risk Using CNN with Batch Dice Loss. Lect. Notes Comput. Sci. 2019, 11269, 105–114. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, S.; Li, C.; Wang, J. Rethinking the Dice Loss for Deep Learning Lesion Segmentation in Medical Images. J. Shanghai Jiaotong Univ. 2021, 26, 93–102. [Google Scholar] [CrossRef]
- Milletari, F.; Navab, N.; Ahmadi, S.A. V-Net: Fully Convolutional Neural Networks for Volumetric Medical Image Segmentation. In Proceedings of the 4th International Conference on 3D Vision (3DV), Stanford, CA, USA, 25–28 October 2016; pp. 565–571. [Google Scholar] [CrossRef]
- Elharrouss, O.; Akbari, Y.; Almaadeed, N.; Al-Maadeed, S. Backbones-Review: Feature Extraction Networks for Deep Learning and Deep Reinforcement Learning Approaches. arXiv 2022, arXiv:2206.08016. [Google Scholar]
- Liang, S.; Gu, Y. SRENet: A Spatiotemporal Relationship-Enhanced 2D-CNN-Based Framework for Staging and Segmentation of Kidney Cancer Using CT Images. Appl. Intell. 2022, 53, 17061–17073. [Google Scholar] [CrossRef]
- Sun, P.; Mo, Z.; Hu, F.; Liu, F.; Mo, T.; Zhang, Y.; Chen, Z. Kidney Tumor Segmentation Based on FR2PAttU-Net Model. Front. Oncol. 2022, 12, 853281. [Google Scholar] [CrossRef] [PubMed]
- Abdelrahman, A.; Viriri, S. EfficientNet Family U-Net Models for Deep Learning Semantic Segmentation of Kidney Tumors on CT Images. Front. Comput. Sci. 2023, 5, 1235622. [Google Scholar]









| Properties | Values |
|---|---|
| Number of all Image | 7899 |
| Number of Training Images | 5841 |
| Number of Validation Images | 1027 |
| Number of Testing Images | 1031 |
| Image format | Png |
| Modality | CT |
| Hyperparameter | Settings |
|---|---|
| Activation | `softmax’ |
| Optimizer | Adam |
| Loss function | Focal Loss, Dice Loss |
| Learning rate | 0.0001 |
| Batch size | 32 |
| Epochs | 50 |
| Metrics | IoU, F1-score |
| Input images size | 128 × 128 |
| IoU Scores | Obtained Values |
|---|---|
| Background IoU Score | 0.999 |
| Kidney IoU Score | 0.980 |
| Tumor IoU Score | 0.984 |
| Mean IoU Score | 0.988 |
| SE-ResNet | Background | Kidney | Tumor | Mean |
|---|---|---|---|---|
| Seresnet18 | 0.999 | 0.972 | 0.971 | 0.981 |
| Seresnet34 | 0.999 | 0.972 | 0.971 | 0.981 |
| Seresnet50 | 0.999 | 0.980 | 0.984 | 0.988 |
| Seresnet101 | 0.999 | 0.976 | 0.982 | 0.986 |
| Seresnet152 | 0.999 | 0.979 | 0.984 | 0.987 |
| SE-ResNet | Loss | IoU Score | F1-Score |
|---|---|---|---|
| Seresnet18 | 0.753 | 0.979 | 0.989 |
| Seresnet34 | 0.753 | 0.979 | 0.989 |
| Seresnet50 | 0.752 | 0.987 | 0.993 |
| Seresnet101 | 0.752 | 0.985 | 0.992 |
| Seresnet152 | 0.752 | 0.986 | 0.993 |
| Reference | Architecture | Kidney Dice | Tumor Dice | Kidney IoU Scores | Tumor IoU Scores |
|---|---|---|---|---|---|
| [87] | nnU-Net | 0.969 | 0.919 | - | - |
| [87] | Hybrid V-Net | 0.962 | 0.913 | - | - |
| [87] | U-Net3+ | 0.959 | 0.909 | - | - |
| [87] | SRE Net 2D-CNN | 0.979 | 0.925 | - | - |
| [88] | FR2PAttU-Net | 0.948 | 0.911 | - | - |
| [89] | U-Net EfficientNet-B0 | 0.984 | 0.980 | 0.969 | 0.960 |
| [89] | U-Net EfficientNet-B1 | 0.981 | 0.980 | 0.963 | 0.961 |
| [89] | U-Net EfficientNet-B2 | 0.983 | 0.982 | 0.966 | 0.965 |
| [89] | U-Net EfficientNet-B3 | 0.985 | 0.980 | 0.970 | 0.960 |
| [89] | U-Net EfficientNet-B4 | 0.987 | 0.984 | 0.974 | 0.968 |
| [89] | U-Net EfficientNet-B5 | 0.986 | 0.983 | 0.972 | 0.966 |
| [89] | U-Net EfficientNet-B6 | 0.985 | 0.981 | 0.971 | 0.963 |
| [89] | U-Net EfficientNet-B7 | 0.988 | 0.981 | 0.977 | 0.962 |
| Proposed | FPN- Seresnet18 | 0.986 | 0.985 | 0.972 | 0.971 |
| Proposed | FPN- Seresnet34 | 0.986 | 0.985 | 0.972 | 0.971 |
| Proposed | FPN- Seresnet50 | 0.990 | 0.992 | 0.980 | 0.984 |
| Proposed | FPN- Seresnet101 | 0.988 | 0.991 | 0.976 | 0.982 |
| Proposed | FPN- Seresnet152 | 0.989 | 0.992 | 0.979 | 0.984 |
| Proposed | FPN- Resnet50 | 0.987 | 0.988 | 0.974 | 0.977 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abdelrahman, A.; Viriri, S. FPN-SE-ResNet Model for Accurate Diagnosis of Kidney Tumors Using CT Images. Appl. Sci. 2023, 13, 9802. https://doi.org/10.3390/app13179802
Abdelrahman A, Viriri S. FPN-SE-ResNet Model for Accurate Diagnosis of Kidney Tumors Using CT Images. Applied Sciences. 2023; 13(17):9802. https://doi.org/10.3390/app13179802
Chicago/Turabian StyleAbdelrahman, Abubaker, and Serestina Viriri. 2023. "FPN-SE-ResNet Model for Accurate Diagnosis of Kidney Tumors Using CT Images" Applied Sciences 13, no. 17: 9802. https://doi.org/10.3390/app13179802






