Assessing Glycosphingolipid Profiles in Human Health and Disease Using Non-Imaging MALDI Mass Spectrometry
Abstract
1. Introduction
2. Structural Characteristics, Classification, and Nomenclature of GSLs
3. MALDI Non-Imaging MS Analysis of GSLs
3.1. Fragmentation Map of GSLs
3.2. The Influence of Matrices Used in MALDI Non-Imaging MS Analysis of GSLs
- DHB (2,5-dihydroxybenzoic acid)
- DHA (2,6-dihydroxyacetophenone)
- CHCA (α-cyano-4-hydroxycinnamic acid)
- 5-methoxysalicylic acid (MSA)
- ATT (6-azo-2-thiothymine)
- THAP (trihydroxyacetophenone)
3.3. Instrumentation in MALDI Non-Imaging MS Analysis of GSLs
3.4. MALDI Non-Imaging MS Analysis of GSLs in the Healthy State
3.4.1. GSLs in Human Milk
3.4.2. GSLs from Human Brain Tissues
3.4.3. GSLs Extracted from Human Blood Cells
3.5. MALDI Non-Imaging MS Analysis of GSLs in Pathology
3.5.1. GSLs in Storage Diseases
- 1.
- GM1 Gangliosidosis
- 2.
- Tay–Sachs disease
- 3.
- Spielmeyer–Vogt type amaurotic idiocy (Niemann–Pick disease type C)
- 4.
- Sudanophilic leukodistrophy
- 5.
- Gaucher disease
- 6.
- Farber disease
- 7.
- Fabry disease
3.5.2. GSLs in Cancer
- 1.
- Colon tumors
- 2.
- Brain tumors
3.5.3. GSLs in Neurodegenerative Diseases
3.6. MALDI Non-Imaging MS Analysis of GSLs in Various Cell Cultures
3.6.1. GSLs in Breast Cancer Cell Lines
3.6.2. GSLs in Melanoma and Neuroblastoma Cell Lines
3.6.3. GSLs in Hepatic Cancer Cell Lines
3.6.4. GSLs in Other Cell Lines
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AD | Alzheimer’s disease |
| APCI | Atmospheric pressure chemical ionization |
| CE | Capillary electrophoresis |
| Cer | Ceramide |
| CI | Chemical ionization |
| CID | Collision-induced dissociation |
| EI | Electron impact ionization |
| ESI | Electrospray ionization |
| ETD | Electron transfer dissociation |
| FTICR-MS | Fourier transform ion cyclotron resonance mass spectrometry |
| Fuc | Fucose |
| FUT | Fucosyl transferase |
| Gal | Galactose |
| GalCer | Galactosiylceramide |
| GC | Gas chromatography |
| GalNAc | N-acetyl galactosamine |
| GalT | Galactosyl transferase |
| Glc | Glucose |
| GlcCer | Glucosylceramide |
| GlcNAc | N-acetyl-glucosamine |
| GSLs | Glycosphingolipids |
| Hex | Hexose |
| HPLC | High-performance liquid chromatography |
| IT | Ion trap |
| KDN | Deaminoneuraminic acid |
| LacCer | Lactosylceramide |
| LC | Liquid chromatography |
| Man | Manose |
| MALDI | Matrix-assisted laser desorption ionization |
| MS | Mass spectrometry |
| MS/MS | Tandem mass spectrometry |
| MSn | Multistage fragmentation mass spectrometry |
| Neu5Ac | Acetyl-neuraminic acid |
| Neu5Gc | N-glicolyl-neuraminic acid |
| O-Ac | O-acetyl |
| P | Parkinson’s disease |
| QIT | Quadrupole ion trap |
| QTOF MS | Quadrupole time-of-flight mass spectrometry |
| SA | Sialic acid |
| sLeA | Sialyl LewisA |
| sLeX | Sialyl LewisX |
| ST | Sialyl transferase |
| S/NC | Signal-to-noise ratio |
| TLC | Thin-layer chromatography |
| TOF MS | Time-of-flight mass spectrometry |
Appendix A
| Sample (Organ, Tissue, Fluid) | Type | Instrumentation/Operation Mode | Matrices | GSLs | Comments | Reference |
|---|---|---|---|---|---|---|
| Milk | Human milk; bovine milk | IonSpec Pro MALDI FTICR MS (IonSpec, Irvine, CA, USA) equipped with a 7.0 T superconducting magnet. A pulsed Nd: YAG laser (355 nm) was used as the external MALDI source. Tandem MS analysis was achieved via CID and IRMPD. Both positive and negative ion modes. | DHB | GM3 GD3 | The study presents a rapid means to characterize and analyze milk GSLs, using MALDI FTCIR MS coupled with CID and IRMPD MS/MS. It compares the distribution of abundances of GSLs in human and bovine milk samples. | [122] |
| Milk | Human milk; bovine milk (GM3 and GD3 standards) | Voyager-DE-STR system (PerSeptive Biosystems) MALDI-TOF MS equipped with a nitrogen laser emitting at 337 nm. Positive linear ion mode, with delayed extraction. | DHB | GM3 GD3 | The aim of the study is to compare the difference between human and bovine milk gangliosides, the latter being used to produce milk formula. The amount of long-chain fatty acids was higher in bovine milk and tricosanoic acid dominated in bovine milk gangliosides, compared to human milk. | [118] |
| Human brain | Normal human brain tissue samples alongside ganglioside standards | MALDI-FTMS (IonSpec, Irvine, CA, USA) with a Cryomagnetics (OakRidge, TN, USA) 7 Tesla active-shielded superconducting electromagnet. Both positive and negative ion modes. | ATT | GM1, GD1a, GT1b, GQ1b, GP1b1c | This study demonstrates that transiently elevating pressure in an MALDI-FTMS source into the 1–10 mbar range during ionization decreases the metastable fragmentation of gangliosides. This allows the detection of the molecular ion species without loss of the highly liable sialic acid residues. | [120] |
| Human brain | Normal vs. diseased brain tissue samples (AD), CSF, and serum | MALDI-TOF MS and on a Bruker Daltonics mass spectrometer equipped with a reflector; additional fragmentation analysis was conducted using MALDI-TOF/TOF analysis (Bruker Daltonics, Bremen, Germany). | DHB | GM1, GM2, GM3, GD1-a, GD1b-lactone | The study demonstrates that using glycoblotting-based glycome analysis can be used for human brain tissue post mortem. The Aβ glycosylation (which can become aberrant or absent in tissues from AD brain) leads to elevation of the N-glycans with proximal fucose, bisecting-GlcNAc, tri- and tetra-antennary types, and small gangliosides GM1 and GM2—in serum. There were found smaller amounts of GD1a/GD1b-lactone in AD brain tissues and a significantly lesser amount of gangliosides in the frontal cortex sample, compared to the control sample. | [134] |
| Human brain | Human brain and hippocampus tissue samples from patients with AD, Parkinson’s disease, and control samples. | TLC-Blot-MALDI-TOF MS system (QSTAR XL, Applied Biosystems/MDS Sciex, Foster City, CA, USA). AXIMA-QIT-TOF mass spectrometer (Shimadzu, Kyoto, Japan) in negative ion mode. Ionization was performed with a 337 nm pulsed N2 laser. Precursor and fragment ions obtained using CID using Ar were ejected from the ion trap and analyzed by a reflectron TOF detector. | DHB | GM1, GD1a, GD1b, GT1b | The study uses a new approach by using the lipids transferred from a TLC plate to a PVDF membrane and direct mass spectrometric analysis. The authors found that GD1b and GT1b levels were lower in AD, compared with PD or control patients. Furthermore, there was a significant reduction in d:20:1/C18:0 ceramide-containing gangliosides in AD samples. | [158] |
| Human brain | Normal human brain tissue vs. several diseased human brain tissues | DE MALDI-TOF MS on a Voyager Elite XL, (PerSeptive Biosystem, Framingham, MA, USA), pulsed with a 337 nm nitrogen laser. Both positive and negative ion modes (reflector mode). | CHCA DHB | GD1a, GD1b, GD2, GalNAc-GD1a | The study aims to determine if using DE MALDI-TOF MS is suitable for determining individual molecular species of gangliosides in the GM and GD series from various human brain tissue samples, as well as some animal tissue samples (caprine erythrocytes, porcine spinal cord, and brain) by comparing the spectra results with known masses of gangliosides. | [123] |
| Human blood | Human leukocytes | MALDI-TOF MS on a TofSpec-E (Micromass, Manchester, UK). Negative ion mode (reflector mode). | ATT | The study tested binding of human and avian influenza viruses to gangliosides isolated from different human tissues/fluids (only leukocytes are detailed). | [183] | |
| Human blood | Human platelets | DE MALDI-TOF MS on a Voyager Elite XL, (PerSeptive Biosystem, Framingham, MA, USA) pulsed with a 337 nm nitrogen laser. Both positive and negative ion modes (reflector mode). | CHCA DHB | sphingomyelin, glucosylceramide, lactosylceramide, globotrialosylceramide, globotetraosylceramide, and GM2 | The article refers to methods of extracting and analyzing sphingolipids from a single sample. The sample preparation steps are described in detail. | [162] |
| Human blood | Human erythrocytes | MALDI-TOF MS on a Bruker Daltonik (Bruker Daltonics, Bremen, Germany), with pulsed nitrogen laser at 337 nm wavelength, coupled with a nanoHPLC. Both positive and negative ion modes (reflector mode). | DHB ATT/DAC 2,4,6trihydroxyacetophenone/DAC | LacCer Gb3Cer Gb4Cer | This study establishes a protocol for screening neutral GSL mixtures (GSLs from human erythrocytes and monosialogangliosides from murine MDAY-D2 cell line) by coupling nanoHPLC/MALDI MS. | [127] |
| Sample (Organ, Tissue, Fluid) | Type | Instrumentation/Operation Mode | Matrices | GSLs | Comments | Reference |
|---|---|---|---|---|---|---|
| Human brain | Sudanophilic leukodystrophy disease; Spielmeyer–Vogt type of juvenile amaurotic idiocy; GM1 gangliosidosis; Gaucher disease; nuclear jaundice; Schilder’s disease; schizophrenia; brain edema. | DE MALDI-TOF MS (Voyager Elite XL Biospectrometry Workstation, PerSeptive Biosystem; Framingham, MA, USA, reflector mode) nitrogen laser at 337 nm wavelength was used for ionization. Spectra were acquired in negative ion mode. | αCHCA DHB | GM1 | This comprehensive study uses mass spectrometry to determine the long-chain base composition of glycosphingolipids, including gangliosides. The main focus is the changes that occur with age in the long-chain base of GM1. For this purpose, brain tissue samples from patients with various diseases were studied, alongside brain tissue samples from Eker rats, with and without renal carcinoma. | [142] |
| Human brain | Normal vs. diseased brain tissue samples (AD), CSF, and serum. | MALDI-TOF MS on a Bruker Daltonics mass spectrometer equipped with a reflector; additional fragmentation analysis was conducted using MALDI-TOF/TOF analysis (Bruker Daltonics, Bremen, Germany). | DHB | GM1, GM2, GM3, GD1-a, GD1b-lactone. | The study demonstrates that using glycoblotting-based glycome analysis can be used for human brain tissue post mortem. The Aβ glycosylation (which can become aberrant or absent in tissues from AD brain) leads to elevation of the N-glycans with proximal fucose, bisecting-GlcNAc, tri- and tetra-antennary types, and small gangliosides GM1 and GM2—in serum. There were found smaller amounts of GD1a/GD1b-lactone in AD brain tissues, and also a significantly lesser amount of gangliosides in the frontal cortex sample, compared to the control sample. | [134] |
| Human brain | Human brain and hippocampus tissue samples from patients with AD, Parkinson’s disease, and control samples. | TLC-Blot-MALDI-TOF MS system (QSTAR XL, Applied Biosystems/MDS Sciex, Foster City, CA, USA) AXIMA-QIT-TOF mass spectrometer (Shimadzu, Kyoto, Japan) in negative ion mode. Ionization was performed with a 337 nm pulsed N2 laser. Precursor and fragment ions obtained using CID using Ar were ejected from the ion trap and analyzed by a reflectron TOF detector. | DHB | GM1, GD1a, GD1b, GT1b. | The study uses a new approach by using the lipids transferred from a TLC plate to a PVDF membrane and direct mass spectrometric analysis. The authors found that GD1b and GT1b levels were lower in AD, compared with PD or control patients. Furthermore, there was a significant reduction in d:20:1/C18:0 ceramide-containing gangliosides in AD samples. | [158] |
| Human brain | GM1 gangliosidosis brain. Brain tissue sample from Tay–Sachs disease. Brain tumor tissue. GD1b, GD2, GD2a and GalNAc-GD1a extracted from human brain. | DE MALDI-TOF MS on a Voyager Elite XL, PerSeptive Biosystem, pulsed with a 337 nm nitrogen laser (reflector mode). Spectra were acquired in both positive and negative ion modes. | CHCA DHB | GM1, GM2, GM3, GD3, GD2, GD1a, GD1b, GalNAc-GD1a, mixture of various gangliosides. | The study aims to determine if using DE MALDI-TOF MS is suitable for determining individual molecular species of gangliosides in the GM and GD series, from various human brain tissue samples, as well as some animal tissue samples (caprine erythrocytes, porcine spinal cord, and brain) by comparing the spectra results with known masses of gangliosides. | [123] |
| Human brain; Human liver; Human spleen | Cerebrum, cerebellum, and liver from GM1 gangliosidosis patient. Liver from Farber disease patient. Liver and spleen from Gaucher disease type 2 patient. Liver from Niemann–Pick disease type C; liver from Potter syndrome patient. | DE MALDI-TOF MS on a Voyager Elite XL, (PerSeptive Biosystem, Framingham, MA, USA) reflector mode), pulsed with a 337 nm nitrogen laser. Spectra were acquired in both positive negative ion modes. | CHCA DHB | GM1, ceramide, sphingomyelin, monohexosylceramides. | The study demonstrates that DE MALDI-TOF MS can allow rapid screening for sphingolipidosis, with minimal tissue. The main focus of the study is sphingolipids (sphingomyelin, monohexosylceramide, and ceramide), but ganglioside GM1 is also studied. | [137] |
| Human brain | Human brain tumor tissue, brain tissue from a patient with sudanophilic leukodystrophy. | DE MALDI-TOF MS on a Voyager Elite XL (PerSeptive Biosystem, Framingham, MA, USA) reflector mode, pulsed with a 337 nm nitrogen laser. Spectra were acquired in both positive and negative ion modes. | CHCA DHB | GM1, GM3. | The article refers to methods of extracting and analyzing sphingolipids from a single sample. The sample preparation steps are described in detail. For gala-series glycosphingoplipids, monkey brain tissue is used, for globo-series glycosphingolipids, porcine erythrocytes are used, and for gangliosides, the before-mentioned human brain tissues. Lyso compound of GM3 was isolated from the human brain tumor tissue, with ion peaks at m/z 871.78 (lysoGM3 d18:1) and 899.79 (lysoGM3 d20:1). Intact GM1 from a patient with sudanophilic leukodystrophy was analyzed directly, with GM1 identified at m/z 1545.42 (d18:1C18:0) and m/z 1573.46 (d20:1C18:0). | [162] |
| Human brain | Human brain tissue from a patient with Tay–Sachs disease. | QIT-TOF-MS (Shimadzu, Kyoto, Japan), equipped with an AP-MALDI (Mass Technologies, Burtonsville, MD, USA) Nitrogen laser pulsed at 337 nm. Spectra were acquired in negative ion mode. | CHCA | GM2 | This article presents protocols for mass spectrometry used in analyzing gangliosides. Regarding MALDI MS, the gangliosides used were GM2 from a patient with Tay–Sachs disease, as well as GM1, GD2, GD1a, GD1b, and GT1b purified from bovine brain. | [150] |
| Human brain | Human brain tissue from Tay–Sachs disease (GM2), purified ganglioside from human brain (GT1a), alongside gangliosides from animal tissues. | QIT-TOF-MS (Shimadzu, Kyoto, Japan), equipped with an AP-MALDI (Mass Technologies, Burtonsville, MD, USA) Nitrogen laser pulsed at 337 nm. Spectra were acquired in negative ion mode. | CHCA | GM1, GM2, GD2, GD1a, GD1b, GT1a. | The study presents the advantages of AP-MALDI in characterizing the structures of gangliosides. | [140] |
| Human brain | Secondary brain tumor tissue (metastasis), originating from a non-small-cell lung carcinoma. | Bruker ultrafleXtreme MALDI-TOF, (Bruker Daltonics, Bremen, Germany). nitrogen laser pulsed at 355 nm wavelength spectra were acquired in negative ion mode. | 2,5 DHB | Gluco- and galacto-cerebrosides, lactosylceramides, hexosylceramides, lacto-, neolacto-, globo, and isoglobo-GSLs, GA2, GM4, GM3, GM2, GM1, GD1, GD2, GD3, GT3. | The study investigates the glycolipid profile of a brain metastasis sample (with NSCLC-type origin) using MALDI-TOF. The sample presented mainly GD2 and GD3 and some unusual de-amino-neuraminic acid and N-Glycolyl-neuraminic acid. | [216] |
| Human colon | Human colon tissue with diagnosed colorectal cancer from 13 patients. | MALDI-TOF MS ultrafleXtreme (Bruker Daltonics, Bremen, Germany). Spectra were acquired in the negative ion reflector mode. | DHB | (iso)globo GSLs, (neo)lacto GSLs, GM1, GD2, GD1, GD3. | The study uses MALDI-TOF (/TOF) MS for identifying labeled AA glycans, enzymatically released from GSLs to reveal significant differences between normal human colon tissue and colorectal cancer tissue. The main changes found are increased fucosylation, decreased acetylation, decreased sulfation, and reduced expression of globo-type glycans, as well as disialyl gangliosides. | [135] |
| Sample (Organ, Tissue, Fluid) | Type | Instrumentation/Operation Mode | Matrices | GSLs | Comments | Reference |
|---|---|---|---|---|---|---|
| Cell line culture | Normal human liver cell line (HL-7702) hepatic cancer cell lines (MHCC97L, MHCC97H, HCCLM3) | ultrafleXtreme MALDI-TOF/TOF MS (Bruker Daltonics, Bremen, Germany). Spectra were acquired in positive ion and reflectron mode, and the intense ions from the mass spectra were subsequently selected and subjected to MS/MS. | 2,5 DHB | GSL-glycans | By using lectin microarrays to analyze GSL glycans, there is no need to separate certain GSLs by traditional methods such as HPLC due to the nature of high-throughput omics-level screening of lectin microarrays. Up- and/or downregulations of corresponding glycopatterns such as α1 − 2 fucosylation and α2 − 3 sialylation, and changes of certain glycostructures were observed. | [136] |
| hESC (human embryonic stem cells) | Two undifferentiated hESC lines, 16-d differentiated embryoid body (EB) outgrowth cells | ABI 4700 MALDI-TOF/TOF Proteomics Analyzer (Applied Biosystems) for MS, CID MALDI MS/MS was performed on a Q/TOF Ultima MALDI (Waters Micromass, Manchester, UK). Both operated in reflectron positive ion mode. | 2,5 DHB A-CHCA | Globo-series GSLs, lacto-series GSLs, ganglio-series GSLs. | The study aims to provide insight into the stage-specific transition and mechanism for alterations of GSL core structures during hESC differentiation. | [288] |
| Cell line cultures | Breast cancer cells (Hs 578T, MDA-MB-231, MCF-7, SUM159PT); melanoma cell line (SK-MEL-28); neuroblastoma cell line (LAN-1) | MALDI-QIT-TOF Shimadzu AXIMA Resonance MS (Manchester, UK). Spectra were acquired in negative ion mode. | THAP | Ganglio-series GSLs (GM1, GM2, GM3, GD2, GD3, GT2, GT30), and O-acetylated gangliosides (O-Ac-GM1, O-Ac-GD3, O-Ac-GD2, O-Ac-GT2, O-Ac-GT3). | The aim of the study is to use structural approaches in order to comprehend ganglioside diversity in melanoma, neuroblastoma, and breast cancer cells, focusing on O-acetylated species, that are usually lost under alkaline conditions and require specific analytical procedures. | [265] |
| Cell line cultures | Jurkat cells (line of human T lymphocytes) | MALDI-TOF-MS (Bruker microflex, Bruker Daltonics, Bremen, Germany). Negative ion mode. | THAP | GM3, GM2, GM1, GD1a. | This study demonstrates that the use of metabolic oligosaccharide engineering enables a mammalian cell line to produce photoactivatable gangliosides. The results show promise for using photoreaction to capture and characterize ganglioside-mediated interactions. | [287] |
| Breast cancer cell lines | MCF7 MDA-MB231 MDA-MB468 HMLE-Twist-ER | MALDI-TOF/TOF 5800 system (Sciex; Vaughan, Canada). Spectra were acquired in positive ion mode. | 2,5 DHB | GD2, GD3. | This study demonstrates that GD3s alone can sustain CSC (cancer stem cells) properties and also promote malignant cancer properties. | [257] |
| Breast cancer cell lines | MCF-t, MDA-MB-231, Hs578T | 4800 MALDI-TOF/TOF Proteomics Analyzer (Applied Biosystems). MS, operated in the positive reflectron mode. | 2,5 DHB | LacCer Ganglio-series: GM1a, GM1b, GM1a/b, GM3, GM2, GD1 (GD1a, GD1b, GD1c), GD3, GT3, lacto-series: GL7 globo-series GSLs (Gb3, Gb4), GA2. | This study analyzes the effect of TNF on ganglioside biosynthesis and expression in breast cancer cells from different molecular subtypes. | [269] |
| Cell culture | Parkin-mutant human skin primary fibroblasts vs. normal control fibroblasts (NHDF) | Bruker Microflex LRF MS and Bruker Daltonics Ultraflex Extreme MALDI-TOF/TOF (Bruker Daltonics, Bremen, Germany) for MS/MS analyses. Spectra were acquired in positive or negative reflector mode. | 9-AA | GM3, GM2. | This study demonstrates that the proportions of phospholipids and glycosphingolipids are altered in the lipid profiles of Parkin-mutant fibroblasts. | [286] |
References
- Schnaar, R.L.; Sandhoff, R.; Tiemeyer, M.; Kinoshita, T. Glycosphingolipids. In Essentials of Glycobiology, 4th ed.; Varki, A., Cummings, R.D., Esko, J.D., Eds.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2022; Chapter 11. [Google Scholar]
- Hakomori, S.I. Structure and Function of Sphingoglycolipids in Transmembrane Signalling and Cell-Cell Interactions. Biochem. Soc. Trans. 1993, 21, 583–595. [Google Scholar] [CrossRef] [PubMed]
- Hakomori, S.I.; Igarashi, Y. Functional Role of Glycosphingolipids in Cell Recognition and Signaling. J. Biochem. 1995, 118, 1091–1103. [Google Scholar] [CrossRef] [PubMed]
- Kojima, N.; Hakomori, S.I. Cell Adhesion, Spreading, and Motility of G(M3)-Expressing Cells Based on Glycolipid-Glycolipid Interaction. J. Biol. Chem. 1991, 266, 17552–17558. [Google Scholar] [CrossRef]
- Hakomori, S. Structure and Function of Glycosphingolipids and Sphingolipids: Recollections and Future Trends. Biochim. Biophys. Acta-Gen. Subj. 2008, 1780, 325–346. [Google Scholar] [CrossRef]
- Smart, E.J.; Graf, G.A.; Mcniven, M.A.; Sessa, W.C.; Engelman, J.A.; Scherer, P.E.; Okamoto, T.; Lisanti, M.P. Caveolins, Liquid-Ordered Domains, and Signal Transduction overview: Caveolae and caveola-related domains are liquid-ordered microdomains. Mol. Cell. Biol. 1999, 19, 7289–7304. [Google Scholar] [CrossRef] [PubMed]
- Brown, D.A.; Rose, J.K. Sorting of GPI-Anchored Proteins to Glycolipid-Enriched Membrane Subdomains during Transport to the Apical Cell Surface. Cell 1992, 68, 533–544. [Google Scholar] [CrossRef]
- Liu, P.; Ying, Y.; Ko, Y.G.; Anderson, R.G.W. Localization of Platelet-Derived Growth Factor-Stimulated Phosphorylation Cascade to Caveolae. J. Biol. Chem. 1996, 271, 10299–10303. [Google Scholar] [CrossRef]
- Mineo, C.; James, G.L.; Smart, E.J.; Anderson, R.G.W. Localization of Epidermal Growth Factor-Stimulated Ras/Raf-1 Interaction to Caveolae Membrane. J. Biol. Chem. 1996, 271, 11930–11935. [Google Scholar] [CrossRef]
- Chun, M.; Liyanage, U.K.; Lisanti, M.P.; Lodish, H.F. Signal Transduction of a G Protein-Coupled Receptor in Caveolae: Colocalization of Endothelin and Its Receptor with Caveolin. Proc. Natl. Acad. Sci. USA 1994, 91, 11728–11732. [Google Scholar] [CrossRef] [PubMed]
- Gorodinsky, A.; Harris, D.A. Glycolipid-Anchored Proteins in Neuroblastoma Cells Form Detergent- Resistant Complexes without Caveolin. J. Cell Biol. 1995, 129, 619–627. [Google Scholar] [CrossRef]
- Bickel, P.E.; Scherer, P.E.; Schnitzer, J.E.; Oh, P.; Lisanti, M.P.; Lodish, H.F. Flotillin and Epidermal Surface Antigen Define a New Family of Caveolae- Associated Integral Membrane Proteins. J. Biol. Chem. 1997, 272, 13793–13802. [Google Scholar] [CrossRef]
- Hakomori, S.I. The Glycosynapse. Proc. Natl. Acad. Sci. USA 2002, 99, 225–232. [Google Scholar] [CrossRef]
- Hakomori, S. Glycosynapses: Microdomains Controlling Carbohydrate-Dependent Cell Adhesion and Signaling. An. Acad. Bras. Cienc. 2004, 76, 553–572. [Google Scholar] [CrossRef]
- Handa, K.; Hakomori, S.I. Carbohydrate to Carbohydrate Interaction in Development Process and Cancer Progression. Glycoconj. J. 2012, 29, 627–637. [Google Scholar] [CrossRef] [PubMed]
- Santacroce, P.V.; Basu, A. Probing Specificity in Carbohydrate-Carbohydrate Interactions with Micelles and Langmuir Monolayers. Angew. Chemie-Int. Ed. 2003, 42, 95–98. [Google Scholar] [CrossRef]
- Garcia-Ruiz, C.; Morales, A.; Fernández-Checa, J.C. Glycosphingolipids and Cell Death: One Aim, Many Ways. Apoptosis 2015, 20, 607–620. [Google Scholar] [CrossRef] [PubMed]
- Iwabuchi, K.; Nakayama, H.; Oizumi, A.; Suga, Y.; Ogawa, H.; Takamori, K. Role of Ceramide from Glycosphingolipids and Its Metabolites in Immunological and Inflammatory Responses in Humans. Mediat. Inflamm. 2015, 2015, 120748. [Google Scholar] [CrossRef] [PubMed]
- Lingwood, C.A. Glycosphingolipid Functions. Cold Spring Harb. Perspect. Biol. 2011, 3, a004788. [Google Scholar] [CrossRef]
- Ravindran, M.S.; Tanner, L.B.; Wenk, M.R. Sialic Acid Linkage in Glycosphingolipids Is a Molecular Correlate for Trafficking and Delivery of Extracellular Cargo. Traffic 2013, 14, 1182–1191. [Google Scholar] [CrossRef]
- Hakomori, S. Traveling for the Glycosphingolipid Path. Glycoconj. J. 2000, 17, 627–647. [Google Scholar] [CrossRef]
- Frey, S.L.; Lee, K.Y.C. Number of Sialic Acid Residues in Ganglioside Headgroup Affects Interactions with Neighboring Lipids. Biophys. J. 2013, 105, 1421–1431. [Google Scholar] [CrossRef] [PubMed]
- Ohmi, Y.; Ohkawa, Y.; Yamauchi, Y.; Tajima, O.; Furukawa, K.; Furukawa, K. Essential Roles of Gangliosides in the Formation and Maintenance of Membrane Microdomains in Brain Tissues. Neurochem. Res. 2012, 37, 1185–1191. [Google Scholar] [CrossRef]
- Walkley, S.U.; Zervas, M.; Wiseman, S. Gangliosides as Modulators of Dendritogenesis in Normal and Storage Disease-Affected Pyramidal Neurons. Cereb. Cortex 2000, 10, 1028–1037. [Google Scholar] [CrossRef] [PubMed]
- Schachner, M.; Bartsch, U. Multiple Functions of the Myelin-Associated Glycoprotein MAG (Siglec-4a) in Formation and Maintenance of Myelin. Glia 2000, 29, 154–165. [Google Scholar] [CrossRef]
- Vyas, A.A.; Schnaar, R.L. Brain Gangliosides: Functional Ligands for Myelin Stability and the Control of Nerve Regeneration. Biochimie 2001, 83, 677–682. [Google Scholar] [CrossRef]
- Hilbush, B.S.; Levine, J.M. Modulation of a Ca2+ Signaling Pathway by G(M1) Ganglioside in PC12 Cells. J. Biol. Chem. 1992, 267, 24789–24795. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.H.; Goldenring, J.R.; DeLorenzo, R.J.; Yu, R.K. Gangliosides Inhibit Phospholipid-sensitive Ca2+-dependent Kinase Phosphorylation of Rat Myelin Basic Proteins. J. Neurosci. Res. 1986, 15, 159–166. [Google Scholar] [CrossRef]
- Takahashi, T.; Suzuki, T. Role of Sulfatide in Normal and Pathological Cells and Tissues. J. Lipid Res. 2012, 53, 1437–1450. [Google Scholar] [CrossRef]
- Porubsky, S.; Speak, A.O.; Salio, M.; Jennemann, R.; Bonrouhi, M.; Zafarulla, R.; Singh, Y.; Dyson, J.; Luckow, B.; Lehuen, A.; et al. Globosides but Not Isoglobosides Can Impact the Development of Invariant NKT Cells and Their Interaction with Dendritic Cells. J. Immunol. 2012, 189, 3007–3017. [Google Scholar] [CrossRef]
- Nakayama, H.; Nagafuku, M.; Suzuki, A.; Iwabuchi, K.; Inokuchi, J.I. The Regulatory Roles of Glycosphingolipid-Enriched Lipid Rafts in Immune Systems. FEBS Lett. 2018, 592, 3921–3942. [Google Scholar] [CrossRef]
- Zhang, T.; De Waard, A.A.; Wuhrer, M.; Spaapen, R.M. The Role of Glycosphingolipids in Immune Cell Functions. Front. Immunol. 2019, 10, 90. [Google Scholar] [CrossRef]
- Haslam, S.M.; Julien, S.; Burchell, J.M.; Monk, C.R.; Ceroni, A.; Garden, O.A.; Dell, A. Characterizing the Glycome of the Mammalian Immune System. Immunol. Cell Biol. 2008, 86, 564–573. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, H.; Ogawa, H.; Takamori, K.; Iwabuchi, K. GSL-Enriched Membrane Microdomains in Innate Immune Responses. Arch. Immunol. Ther. Exp. 2013, 61, 217–228. [Google Scholar] [CrossRef] [PubMed]
- Yu, R.K.; Ariga, T. The role of glycosphingolipids in neurological disorders. Mechanisms of immune action. Ann. N. Y. Acad. Sci. 1998, 845, 285–306. [Google Scholar] [CrossRef]
- Dodge, J.C.; Treleaven, C.M.; Pacheco, J.; Cooper, S.; Bao, C.; Abraham, M.; Cromwell, M.; Sardi, S.P.; Chuang, W.L.; Sidman, R.L.; et al. Glycosphingolipids Are Modulators of Disease Pathogenesis in Amyotrophic Lateral Sclerosis. Proc. Natl. Acad. Sci. USA 2015, 112, 8100–8105. [Google Scholar] [CrossRef] [PubMed]
- Schnaar, R.L. Gangliosides of the Vertebrate Nervous System. J. Mol. Biol. 2016, 428, 3325–3336. [Google Scholar] [CrossRef]
- Han, X. Lipid Alterations in the Earliest Clinically Recognizable Stage of Alzheimers Disease: Implication of the Role of Lipids in the Pathogenesis of Alzheimers Disease. Curr. Alzheimer Res. 2005, 2, 65–77. [Google Scholar] [CrossRef]
- Wallom, K.L.; Fernández-Suárez, M.E.; Priestman, D.A.; Vruchte, D.; Huebecker, M.; Hallett, P.J.; Isacson, O.; Platt, F.M. Glycosphingolipid Metabolism and Its Role in Ageing and Parkinson’s Disease. Glycoconj. J. 2022, 39, 39–53. [Google Scholar] [CrossRef] [PubMed]
- Zöller, I.; Meixner, M.; Hartmann, D.; Büssow, H.; Meyer, R.; Gieselmann, V.; Eckhardt, M. Absence of 2-Hydroxylated Sphingolipids Is Compatible with Normal Neural Development but Causes Late-Onset Axon and Myelin Sheath Degeneration. J. Neurosci. 2008, 28, 9741–9754. [Google Scholar] [CrossRef]
- Sasaki, N.; Toyoda, M.; Ishiwata, T. Gangliosides as Signaling Regulators in Cancer. Int. J. Mol. Sci. 2021, 22, 5076. [Google Scholar] [CrossRef]
- Hakomori, S.I.; Zhang, Y. Glycosphingolipid Antigens and Cancer Therapy. Chem. Biol. 1997, 4, 97–104. [Google Scholar] [CrossRef]
- Park, D.D.; Xu, G.; Wong, M.; Phoomak, C.; Liu, M.; Haigh, N.E.; Wongkham, S.; Yang, P.; Maverakis, E.; Lebrilla, C.B. Membrane Glycomics Reveal Heterogeneity and Quantitative Distribution of Cell Surface Sialylation. Chem. Sci. 2018, 9, 6271–6285. [Google Scholar] [CrossRef]
- Bellis, S.L.; Reis, C.A.; Varki, A.; Kannagi, R.; Stanley, P. Glycosylation Changes in Cancer. In Essentials of Glycobiology, 4th ed.; Varki, A., Cummings, R.D., Esko, J.D., Eds.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2022; Chapter 47. [Google Scholar]
- Li, B.; Qin, Y.; Yu, X.; Xu, X.; Yu, W. Lipid Raft Involvement in Signal Transduction in Cancer Cell Survival, Cell Death and Metastasis. Cell Prolif. 2022, 55, e13167. [Google Scholar] [CrossRef] [PubMed]
- Mirkin, B.L.; Clark, S.H.; Zhang, C. Inhibition of Human Neuroblastoma Cell Proliferation and EGF Receptor Phosphorylation by Gangliosides GM1, GM3, GD1A and GT1B. Cell Prolif. 2002, 35, 105–115. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, K.; Ohmi, Y.; Ohkawa, Y.; Bhuiyan, R.H.; Zhang, P.; Tajima, O.; Hashimoto, N.; Hamamura, K.; Furukawa, K. New Era of Research on Cancer-Associated Glycosphingolipids. Cancer Sci. 2019, 110, 1544–1551. [Google Scholar] [CrossRef]
- Kovbasnjuk, O.; Mourtazina, R.; Baibakov, B.; Wang, T.; Elowsky, C.; Choti, M.A.; Kane, A.; Donowitz, M. The Glycosphingolipid Globotriaosylceramide in the Metastatic Transformation of Colon Cancer. Proc. Natl. Acad. Sci. USA 2005, 102, 19087–19092. [Google Scholar] [CrossRef] [PubMed]
- Hamamura, K.; Furukawa, K.; Hayashi, T.; Hattori, T.; Nakano, J.; Nakashima, H.; Okuda, T.; Mizutani, H.; Hattori, H.; Ueda, M.; et al. Ganglioside GD3 Promotes Cell Growth and Invasion through P130Cas and Paxillin in Malignant Melanoma Cells. Proc. Natl. Acad. Sci. USA 2005, 102, 11041–11046. [Google Scholar] [CrossRef]
- Yoshida, S.; Fukumoto, S.; Kawaguchi, H.; Sato, S.; Ueda, R.; Furukawa, K. Ganglioside GD2 in Small Cell Lung Cancer Cell Lines: Enhancement of Cell Proliferation and Mediation of Apoptosis. Cancer Res. 2001, 61, 4244–4252. [Google Scholar]
- Wang, L.; Wang, Y.; Sato, T.; Yamagata, S.; Yamagata, T. Ganglioside GD1a Suppresses TNFα Expression via Pkn1 at the Transcriptional Level in Mouse Osteosarcoma-Derived FBJ Cells. Biochem. Biophys. Res. Commun. 2008, 371, 230–235. [Google Scholar] [CrossRef] [PubMed]
- Ohkawa, Y.; Miyazaki, S.; Miyata, M.; Hamamura, K.; Furukawa, K.; Furukawa, K. Essential Roles of Integrin-Mediated Signaling for the Enhancement of Malignant Properties of Melanomas Based on the Expression of GD3. Biochem. Biophys. Res. Commun. 2008, 373, 14–19. [Google Scholar] [CrossRef] [PubMed]
- Shibuya, H.; Hamamura, K.; Hotta, H.; Matsumoto, Y.; Nishida, Y.; Hattori, H.; Furukawa, K.; Ueda, M.; Furukawa, K. Enhancement of Malignant Properties of Human Osteosarcoma Cells with Disialyl Gangliosides GD2/GD3. Cancer Sci. 2012, 103, 1656–1664. [Google Scholar] [CrossRef]
- Hamamura, K.; Tsuji, M.; Hotta, H.; Ohkawa, Y.; Takahashi, M.; Shibuya, H.; Nakashima, H.; Yamauchi, Y.; Hashimoto, N.; Hattori, H.; et al. Functional Activation of Src Family Kinase Yes Protein Is Essential for the Enhanced Malignant Properties of Human Melanoma Cells Expressing Ganglioside GD3. J. Biol. Chem. 2011, 286, 18526–18537. [Google Scholar] [CrossRef] [PubMed]
- Hyuga, S.; Yamagata, S.; Tai, T.; Yamagata, T. Inhibition of Highly Metastatic FBJ-LL Cell Migration by Ganglioside GD1a Highly Expressed in Poorly Metastatic FBJ-S1 Cells. Biochem. Biophys. Res. Commun. 1997, 231, 340–343. [Google Scholar] [CrossRef]
- Cao, T.; Zhang, T.; Wang, L.; Zhang, L.; Adachi, T.; Sato, T.; Yamagata, S.; Yamagata, T. Ganglioside GD1a Suppression of NOS2 Expression via ERK1 Pathway in Mouse Osteosarcoma FBJ Cells. J. Cell. Biochem. 2010, 110, 1165–1174. [Google Scholar] [CrossRef]
- Furukawa, K.; Hamamura, K.; Ohkawa, Y.; Ohmi, Y.; Furukawa, K. Disialyl Gangliosides Enhance Tumor Phenotypes with Differential Modalities. Glycoconj. J. 2012, 29, 579–584. [Google Scholar] [CrossRef] [PubMed]
- Mitsuzuka, K.; Handa, K.; Satoh, M.; Arai, Y.; Hakomori, S. A Specific Microdomain (“glycosynapse 3”) Controls Phenotypic Conversion and Reversion of Bladder Cancer Cells through GM3-Mediated Interaction of A3β1 Integrin with CD9. J. Biol. Chem. 2005, 280, 35545–35553. [Google Scholar] [CrossRef] [PubMed]
- Lewis, A.L.; Szymanski, C.M.; Schnaar, R.L.; Aebi, M. Bacterial and Viral Infections. In Essentials of Glycobiology, 4th ed.; Varki, A., Cummings, R.D., Esko, J.D., Eds.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2022; Chapter 42. [Google Scholar]
- Sackstein, R.; Hoffmeister, K.M.; Stowell, S.R.; Kinoshita, T.; Varki, A.; Freeze, H.H. Glycans in Acquired Human Diseases. In Essentials of Glycobiology, 4th ed.; Varki, A., Cummings, R.D., Esko, J.D., Eds.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2022; Chapter 46. [Google Scholar]
- Freeze, H.H.; Steet, R.; Suzuki, T.; Kinoshita, T.; Schnaar, R.L. Genetic Disorders of Glycan Degradation. In Essentials of Glycobiology, 4th ed.; Varki, A., Cummings, R.D., Esko, J.D., Eds.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2022; Chapter 44. [Google Scholar]
- Breiden, B.; Sandhoff, K. Lysosomal Glycosphingolipid Storage Diseases. Annu. Rev. Biochem. 2019, 88, 461–485. [Google Scholar] [CrossRef]
- Rustam, Y.H.; Reid, G.E. Analytical Challenges and Recent Advances in Mass Spectrometry Based Lipidomics. Anal. Chem. 2018, 90, 374–397. [Google Scholar] [CrossRef]
- Farwanah, H.; Kolter, T. Lipidomics of Glycosphingolipids. Metabolites 2012, 2, 134–164. [Google Scholar] [CrossRef] [PubMed]
- Merrill, A.H.; Sullards, M.C.; Allegood, J.C.; Kelly, S.; Wang, E. Sphingolipidomics: High-Throughput, Structure-Specific, and Quantitative Analysis of Sphingolipids by Liquid Chromatography Tandem Mass Spectrometry. Methods 2005, 36, 207–224. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, B.; Schiller, J. MALDI-TOF MS, Analysis of Lipids from Cells, Tissues and Body Fluids. In Lipids in Health and Disease. Subcellular Biochemistry; Quinn, P.J., Wang, X., Eds.; Springer: Berlin/Heidelberg, Germany, 2008; Chapter 21; p. 49. [Google Scholar]
- Barrientos, R.C.; Zhang, Q. Recent Advances in the Mass Spectrometric Analysis of Glycosphingolipidome—A Review. Anal. Chim. Acta 2020, 1132, 134–155. [Google Scholar] [CrossRef]
- Leopold, J.; Popkova, Y.; Engel, K.M.; Schiller, J. Recent Developments of Useful MALDI Matrices for the Mass Spectrometric Characterization of Lipids. Biomolecules 2018, 8, 173. [Google Scholar] [CrossRef] [PubMed]
- Levery, S.B. Studies of the Primary and Secondary Structure of Glycosphingolipids. Ph.D. Thesis, University of Washington, Seattle, WA, USA, 1993. [Google Scholar]
- Guo, Z. The Structural Diversity of Natural Glycosphingolipids (GSLs). J. Carbohydr. Chem. 2022, 41, 63–154. [Google Scholar] [CrossRef]
- Van Eijk, M.; Ferra, M.J.; Boot, R.G.; Aerts, J.M.F.G. Lyso-Glycosphingolipids: Presence and Consequences. Essays Biochem. 2020, 64, 565–578. [Google Scholar]
- Pruett, S.T.; Bushnev, A.; Hagedorn, K.; Adiga, M.; Haynes, C.A.; Sullards, M.C.; Liotta, D.C.; Merrill, A.H. Biodiversity of Sphingoid Bases (“sphingosines”) and Related Amino Alcohols. J. Lipid Res. 2008, 49, 1621–1639. [Google Scholar] [CrossRef]
- Fernandes, C.M.; Goldman, G.H.; Del Poeta, M. Biological Roles Played by Sphingolipids in Dimorphic and Filamentous Fungi. mBio 2018, 9, e00642-18. [Google Scholar] [CrossRef] [PubMed]
- Hannich, J.T.; Umebayashi, K.; Riezman, H. Distribution and Functions of Sterols and Sphingolipids. Cold Spring Harb. Perspect. Biol. 2011, 3, a004762. [Google Scholar] [CrossRef]
- Ishibashi, Y.; Kohyama-Koganeya, A.; Hirabayashi, Y. New Insights on Glucosylated Lipids: Metabolism and Functions. Biochim. Biophys. Acta-Mol. Cell Biol. Lipids 2013, 1831, 1475–1485. [Google Scholar] [CrossRef]
- Guimaratildes, L.L.; Toledo, M.S.; Ferreira, F.A.S.; Straus, A.H.; Takahashi, H.K. Structural Diversity and Biological Significance of Glycosphingolipids in Pathogenic and Opportunistic Fungi. Front. Cell. Infect. Microbiol. 2014, 4, 138. [Google Scholar]
- Gault, C.R.; Obeid, L.M.; Hannun, Y.A. An Overview of Sphingolipid Metabolism: From Synthesis to Breakdown. Adv. Exp. Med. Biol. 2010, 688, 1–23. [Google Scholar] [PubMed]
- Merrill, A.H. Sphingolipid and Glycosphingolipid Metabolic Pathways in the Era of Sphingolipidomics. Chem. Rev. 2011, 111, 6387–6422. [Google Scholar] [CrossRef] [PubMed]
- Han, X. Lipidomics for Studying Metabolism. Nat. Rev. Endocrinol. 2016, 12, 668–679. [Google Scholar] [CrossRef]
- de Haan, N.; Yang, S.; Cipollo, J.; Wuhrer, M. Glycomics Studies Using Sialic Acid Derivatization and Mass Spectrometry. Nat. Rev. Chem. 2020, 4, 229–242. [Google Scholar] [CrossRef]
- Schnaar, R.L.; Gerardy-Schahn, R.; Hildebrandt, H. Sialic Acids in the Brain: Gangliosides and Polysialic Acid in Nervous System Development, Stability, Disease, and Regeneration. Physiol. Rev. 2014, 94, 461–518. [Google Scholar] [CrossRef] [PubMed]
- Szlasa, W.; Zendran, I.; Zalesińska, A.; Tarek, M.; Kulbacka, J. Lipid Composition of the Cancer Cell Membrane. J. Bioenerg. Biomembr. 2020, 52, 321–342. [Google Scholar] [CrossRef]
- Krengel, U.; Bousquet, P.A. Molecular Recognition of Gangliosides and Their Potential for Cancer Immunotherapies. Front. Immunol. 2014, 5, 352. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, N.; Chiba, H.; Kuronuma, K.; Go, S.; Hasegawa, Y.; Takahashi, M.; Gasa, S.; Watanabe, A.; Hasegawa, T.; Kuroki, Y.; et al. Detection of N-Glycolyated Gangliosides in Non-Small-Cell Lung Cancer Using GMR8 Monoclonal Antibody. Cancer Sci. 2013, 104, 43–47. [Google Scholar] [CrossRef] [PubMed]
- Blanco, R.; Domínguez, E.; Morales, O.; Blanco, D.; Martínez, D.; Rengifo, C.E.; Viada, C.; Cedeño, M.; Rengifo, E.; Carr, A. Prognostic Significance of N-Glycolyl GM3 Ganglioside Expression in Non-Small Cell Lung Carcinoma Patients: New Evidences. Patholog. Res. Int. 2015, 2015, 132326. [Google Scholar] [CrossRef] [PubMed]
- Blanco, R.; Rengifo, C.E.; Cedeño, M.; Frómeta, M.; Rengifo, E.; Carr, A. Immunoreactivity of the 14F7 Mab (Raised against N-Glycolyl GM3 Ganglioside) as a Positive Prognostic Factor in Non-Small-Cell Lung Cancer. Patholog. Res. Int. 2012, 2012, 235418. [Google Scholar] [CrossRef] [PubMed]
- van Cruijsen, H.; Ruiz, M.; van der Valk, P.; de Gruijl, T.D.; Giaccone, G. Tissue Micro Array Analysis of Ganglioside N-Glycolyl GM3 Expression and Signal Transducer and Activator of Transcription (STAT)-3 Activation in Relation to Dendritic Cell Infiltration and Microvessel Density in Non-Small Cell Lung Cancer. BMC Cancer 2009, 9, 180. [Google Scholar] [CrossRef]
- Oliva, J.P.; Valdés, Z.; Casacó, A.; Pimentel, G.; González, J.; Álvarez, I.; Osorio, M.; Velazco, M.; Figueroa, M.; Ortiz, R.; et al. Clinical Evidences of GM3 (NeuGc) Ganglioside Expression in Human Breast Cancer Using the 14F7 Monoclonal Antibody Labelled with 99mTc. Breast Cancer Res. Treat. 2006, 96, 115–121. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.; Wu, Y.; Li, C.; Tang, J.; Wang, X.; Ren, G.; Carr, A.; Pérez, R.; Guo, W. N-Glycolyl GM3 Ganglioside Immunoexpression in Oral Mucosal Melanomas of Chinese. Oral Dis. 2012, 18, 741–747. [Google Scholar] [CrossRef] [PubMed]
- Blanco, R.; Quintana, Y.; Blanco, D.; Cedeño, M.; Rengifo, C.E.; Frómeta, M.; Ríos, M.; Rengifo, E.; Carr, A. Tissue Reactivity of the 14F7 Mab Raised against N-Glycolyl GM3 Ganglioside in Tumors of Neuroectodermal, Mesodermal, and Epithelial Origin. J. Biomarkers 2013, 2013, 602417. [Google Scholar] [CrossRef] [PubMed]
- Blanco, R.; Rengifo, E.; Cedeño, M.; Rengifo, C.E.; Alonso, D.F.; Carr, A. Immunoreactivity of the 14F7 Mab Raised against N-Glycolyl GM3 Ganglioside in Epithelial Malignant Tumors from Digestive System. ISRN Gastroenterol. 2011, 2011, 645641. [Google Scholar] [CrossRef]
- Blanco, R.; Cedeño, M.; Escobar, X.; Blanco, D.; Rengifo, C.E.; Frómeta, M.; Alvarez, R.I.; Rengifo, E.; Carr, A. Immunorecognition of the 14F7 Mab Raised against N-Glycolyl GM3 Ganglioside in Some Normal and Malignant Tissues from Genitourinary System. ISRN Pathol. 2011, 2011, 953803. [Google Scholar] [CrossRef][Green Version]
- Blanco, R.; Blanco, D.; Quintana, Y.; Escobar, X.; Rengifo, C.E.; Osorio, M.; Gutiérrez, Z.; Lamadrid, J.; Cedeño, M.; Frómeta, M.; et al. Immunoreactivity of the 14F7 Mab Raised against N-Glycolyl GM3 Ganglioside in Primary Lymphoid Tumors and Lymph Node Metastasis. Patholog. Res. Int. 2013, 2013, 920972. [Google Scholar] [CrossRef]
- Scursoni, A.M.; Galluzzo, L.; Camarero, S.; Lopez, J.; Lubieniecki, F.; Sampor, C.; Segatori, V.I.; Gabri, M.R.; Alonso, D.F.; Chantada, G.; et al. Detection of N-Glycolyl GM3 Ganglioside in Neuroectodermal Tumors by Immunohistochemistry: An Attractive Vaccine Target for Aggressive Pediatric Cancer. Clin. Dev. Immunol. 2011, 2011, 245181. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sampor, C.; Guthmann, M.D.; Scursoni, A.; Cacciavillano, W.; Torbidoni, A.; Galluzzo, L.; Camarero, S.; Lopez, J.; de Dávila, M.T.G.; Fainboim, L.; et al. Immune Response to Racotumomab in a Child with Relapsed Neuroblastoma. Front. Oncol. 2012, 2, 195. [Google Scholar] [CrossRef]
- Scursoni, A.M.; Galluzzo, L.; Camarero, S.; Pozzo, N.; Gabri, M.R.; De Acosta, C.M.; Vázquez, A.M.; Alonso, D.F.; De Davila, M.T.G. Detection and Characterization of N-Glycolyated Gangliosides in Wilms Tumor by Immunohistochemistry. Pediatr. Dev. Pathol. 2010, 13, 18–23. [Google Scholar] [CrossRef] [PubMed]
- Sites, J.B.C.A. Carcinoma Cells: CHEMICAL MAMMALS Identification of 2-Keto-3-Deoxy-D-Glycero-D-Galacto Nononic Acid Acid) Residues in Mammalian Tissues and Human Lung. Histochemistry 1996, 549, 2–6. [Google Scholar]
- Go, S.; Sato, C.; Yin, J.; Kannagi, R.; Kitajima, K. Hypoxia-Enhanced Expression of Free Deaminoneuraminic Acid in Human Cancer Cells. Biochem. Biophys. Res. Commun. 2007, 357, 537–542. [Google Scholar] [CrossRef] [PubMed]
- Inoue, S.; Lin, S.L.; Chang, T.; Wu, S.H.; Yao, C.W.; Chu, T.Y.; Troy, F.A.; Inoue, Y. Identification of Free Deaminated Sialic Acid (2-Keto-3-Deoxy-D-Glycero-D-Galacto-Nononic Acid) in Human Red Blood Cells and Its Elevated Expression in Fetal Cord Red Blood Cells and Ovarian Cancer Cells. J. Biol. Chem. 1998, 273, 27199–27204. [Google Scholar] [CrossRef]
- Yu, R.K.; Tsai, Y.T.; Ariga, T.; Yanagisawa, M. Structures, Biosynthesis, and Functions of Gangliosides-an Overview. J. Oleo Sci. 2011, 60, 537–544. [Google Scholar] [CrossRef]
- Hakomori, S. Structure, Organization, and Function of Glycosphingolipids in Membrane. Curr. Opin. Hematol. 2003, 10, 16–24. [Google Scholar] [CrossRef] [PubMed]
- Hakomori, S.; Ishizuka, I. Glycolipid: Animal. In Encyclopedia of Life Sciences; Nature Publishing Group: London, UK, 2001. [Google Scholar]
- Levery, S.B.; Nudelman, E.D.; Hakomori, S. Novel Modification of Glycosphingolipids by Long-Chain Cyclic Acetals: Isolation and Characterization of Plasmalocerebroside from Human Brain. Biochemistry 1992, 31, 5335–5340. [Google Scholar] [CrossRef]
- Kolter, T. Ganglioside Biochemistry. ISRN Biochem. 2012, 2012, 506160. [Google Scholar] [CrossRef] [PubMed]
- Mlinac, K.; Fabris, D.; Vukelić, Ž.; Rožman, M.; Heffer, M.; Bognar, S.K. Structural Analysis of Brain Ganglioside Acetylation Patterns in Mice with Altered Ganglioside Biosynthesis. Carbohydr. Res. 2013, 382, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Chester, M.A. IUPAC-IUB Joint Commission on Biochemical Nomenclature (JCBN). Nomenclature of Glycolipids--Recommendations 1997. Eur. J. Biochem. 1998, 257, 293–298. [Google Scholar] [PubMed]
- Svennerholm, L. The Gangliosides. J. Lipid Res. 1964, 5, 145–155. [Google Scholar] [CrossRef] [PubMed]
- Svennerholm, L. Chromatographic Separation of Human Brain Gangliosides. J. Neurochem. 1963, 10, 613–623. [Google Scholar] [CrossRef]
- Liebisch, G.; Fahy, E.; Aoki, J.; Dennis, E.A.; Durand, T.; Ejsing, C.S.; Fedorova, M.; Feussner, I.; Griffiths, W.J.; Köfeler, H.; et al. Update on LIPID MAPS Classification, Nomenclature, and Shorthand Notation for MS-Derived Lipid Structures. J. Lipid Res. 2020, 61, 1539–1555. [Google Scholar] [CrossRef]
- Domon, B.; Costello, C.E. A Systematic Nomenclature for Carbohydrate Fragmentations in FAB-MS/MS Spectra of Glycoconjugates. Glycoconj. J. 1988, 5, 379–409. [Google Scholar] [CrossRef]
- Ann, Q.; Adams, J. Structure Determination of Ceramides and Neutral Glycosphingolipids by Collisional Activation of [M + Li]+ Ions. J. Am. Soc. Mass Spectrom. 1992, 3, 260–263. [Google Scholar] [CrossRef] [PubMed]
- Costello, C.E.; Vath, J.E. Tandem Mass Spectrometry of Glycolipids. Methods Enzymol. 1990, 193, 738–768. [Google Scholar]
- Hsu, F.F.; Turk, J. Structural Determination of Glycosphingolipids as Lithiated Adducts by Electrospray Ionization Mass Spectrometry Using Low-Energy Collisional-Activated Dissociation on a Triple Stage Quadrupole Instrument. J. Am. Soc. Mass Spectrom. 2001, 12, 61–79. [Google Scholar] [CrossRef] [PubMed]
- Harvey, D.J. Matrix-Assisted Laser Desorption/Ionisation Mass Spectrometry of Oligosaccharides and Glycoconjugates. J. Chromatogr. A 1996, 720, 429–446. [Google Scholar] [CrossRef]
- Hillenkamp, F.; Karas, M. The MALDI Process and Method. In MALDI MS: A Practical Guide to Instrumentation, Methods and Applications; John Wiley & Sons: Hoboken, NJ, USA, 2007. [Google Scholar]
- Krutchinsky, A.N.; Chait, B.T. On the Nature of the Chemical Noise in MALDI Mass Spectra. J. Am. Soc. Mass Spectrom. 2002, 13, 129–134. [Google Scholar] [CrossRef]
- Zarei, M.; Bindila, L.; Souady, J.; Dreisewerd, K.; Berkenkamp, S.; Müthing, J.; Peter-Katalinić, J. A Sialylation Study of Mouse Brain Gangliosides by MALDI A-TOF and o-TOF Mass Spectrometry. J. Mass Spectrom. 2008, 43, 716–725. [Google Scholar] [CrossRef]
- Bode, L.; Beermann, C.; Mank, M.; Kohn, G.; Boehm, G. Human and Bovine Milk Gangliosides Differ in Their Fatty Acid Composition. J. Nutr. 2004, 134, 3016–3020. [Google Scholar] [CrossRef] [PubMed]
- Tajiri, M.; Takeuchi, T.; Wada, Y. Distinct Features of Matrix-Assisted 6 Μm Infrared Laser Desorption/Ionization Mass Spectrometry in Biomolecular Analysis. Anal. Chem. 2009, 81, 6750–6755. [Google Scholar] [CrossRef]
- O’Connor, P.B.; Mirgorodskaya, E.; Costello, C.E. High Pressure Matrix-Assisted Laser Desorption/Ionization Fourier Transform Mass Spectrometry for Minimization of Ganglioside Fragmentation. J. Am. Soc. Mass Spectrom. 2002, 13, 402–407. [Google Scholar] [CrossRef] [PubMed]
- Penn, S.G.; Cancilla, M.T.; Green, M.K.; Lebrilla, C.B. Direct Comparison of Matrix-Assisted Laser Desorption/Ionisation and Electrospray Ionisation in the Analysis of Gangliosides by Fourier Transform Mass Spectrometry. Eur. J. Mass Spectrom. 1997, 3, 67–79. [Google Scholar] [CrossRef]
- Lee, H.; An, H.J.; Lerno, L.A.; German, J.B.; Lebrilla, C.B. Rapid Profiling of Bovine and Human Milk Gangliosides by Matrix-Assisted Laser Desorption/Ionization Fourier Transform Ion Cyclotron Resonance Mass Spectrometry. Int. J. Mass Spectrom. 2011, 305, 138–150. [Google Scholar] [CrossRef]
- Sugiyama, E.; Hara, A.; Uemura, K.I.; Taketomi, T. Application of Matrix-Assisted Laser Desorption Ionization Time-of-Flight: Mass Spectrometry with Delayed Ion Extraction to Ganglioside Analyses. Glycobiology 1997, 7, 719–724. [Google Scholar] [CrossRef] [PubMed]
- Mank, M.; Stahl, B.; Boehm, G. 2,5-Dihydroxybenzoic Acid Butylamine and Other Ionic Liquid Matrixes for Enhanced MALDI-MS Analysis of Biomolecules. Anal. Chem. 2004, 76, 2938–2950. [Google Scholar] [CrossRef] [PubMed]
- Colsch, B.; Woods, A.S. Localization and Imaging of Sialylated Glycosphingolipids in Brain Tissue Sections by MALDI Mass Spectrometry. Glycobiology 2010, 20, 661–667. [Google Scholar] [CrossRef]
- Ruh, H.; Sandhoff, R.; Meyer, B.; Gretz, N.; Hopf, C. Quantitative Characterization of Tissue Globotetraosylceramides in a Rat Model of Polycystic Kidney Disease by PrimaDrop Sample Preparation and Indirect High-Performance Thin Layer Chromatography-Matrix-Assisted Laser Desorption/Ionization-Time-of-Flight. Anal. Chem. 2013, 85, 6233–6240. [Google Scholar] [CrossRef]
- Zarei, M.; Kirsch, S.; Müthing, J.; Bindila, L.; Peter-Katalinić, J. Automated Normal Phase Nano High Performance Liquid Chromatography/Matrix Assisted Laser Desorption/Ionization Mass Spectrometry for Analysis of Neutral and Acidic Glycosphingolipids. Anal. Bioanal. Chem. 2008, 391, 289–297. [Google Scholar] [CrossRef]
- Taketomi, T.; Hara, A.; Uemura, K.I.; Kurahashi, H.; Sugiyama, E. Preparation of Various Lysogangliosides Including Lyso-Fucosyl GM1 and Delayed Extraction Matrix-Assisted Laser Desorption Ionization Time-of-Flight Mass Spectrometric Analysis. J. Biochem. 1997, 121, 264–269. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.; Cha, S. 5-Methoxysalicylic Acid Matrix for Ganglioside Analysis with Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2015, 26, 522–525. [Google Scholar] [CrossRef] [PubMed]
- Krutchinsky, A.N.; Kalkum, M.; Chait, B.T. Automatic Identification of Proteins with a MALDI-Quadrupole Ion Trap Mass Spectrometer. Anal. Chem. 2001, 73, 5066–5077. [Google Scholar] [CrossRef]
- Ivleva, V.B.; Elkin, Y.N.; Budnik, B.A.; Moyer, S.C.; O’Connor, P.B.; Costello, C.E. Coupling Thin-Layer Chromatography with Vibrational Cooling Matrix-Assisted Laser Desorption/Ionization Fourier Transform Mass Spectrometry for the Analysis of Ganglioside Mixtures. Anal. Chem. 2004, 76, 1552–1560. [Google Scholar] [CrossRef]
- Kamimiya, H.; Suzuki, Y.; Mathew, A.; Kabayama, K.; Kojima, H.; Kushi, Y. Simple and Rapid Removal of the Interference in Gangliosides Extracted from HPTLC Spot on MALDI-TOF MS Analysis. Anal. Methods 2013, 5, 6617–6621. [Google Scholar]
- Mechref, Y.; Novotny, M.V. Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry of Acidic Glycoconjugates Facilitated by the Use of Spermine as a Co-Matrix. J. Am. Soc. Mass Spectrom. 1998, 9, 1293–1302. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gizaw, S.T.; Ohashi, T.; Tanaka, M.; Hinou, H.; Nishimura, S.I. Glycoblotting Method Allows for Rapid and Efficient Glycome Profiling of Human Alzheimer’s Disease Brain, Serum and Cerebrospinal Fluid towards Potential Biomarker Discovery. Biochim. Biophys. Acta-Gen. Subj. 2016, 1860, 1716–1727. [Google Scholar]
- Holst, S.; Stavenhagen, K.; Balog, C.I.A.; Koeleman, C.A.M.; McDonnell, L.M.; Mayboroda, O.A.; Verhoeven, A.; Mesker, W.E.; Tollenaar, R.A.E.M.; Deelder, A.M.; et al. Investigations on Aberrant Glycosylation of Glycosphingolipids in Colorectal Cancer Tissues Using Liquid Chromatography and Matrix-Assisted Laser Desorption Time-of- Flight Mass Spectrometry (MALDI-TOF-MS). Mol. Cell. Proteomics 2013, 12, 3081–3093. [Google Scholar] [CrossRef]
- Du, H.; Yu, H.; Ma, T.; Yang, F.; Jia, L.; Zhang, C.; Zhang, J.; Niu, L.; Yang, J.; Zhang, Z.; et al. Analysis of Glycosphingolipid Glycans by Lectin Microarrays. Anal. Chem. 2019, 91, 10663–10671. [Google Scholar] [CrossRef] [PubMed]
- Fujiwaki, T.; Yamaguchi, S.; Sukegawa, K.; Taketomi, T. Application of Delayed Extraction Matrix-Assisted Laser Desorption Ionization Time-of-Flight Mass Spectrometry for Analysis of Sphingolipids in Tissues from Sphingolipidosis Patients. J. Chromatogr. B Biomed. Sci. Appl. 1999, 731, 45–52. [Google Scholar] [CrossRef]
- Jackson, S.N.; Wang, H.Y.J.; Woods, A.S. Direct Profiling of Lipid Distribution in Brain Tissue Using MALDI-TOFMS. Anal. Chem. 2005, 77, 4523–4527. [Google Scholar] [CrossRef]
- Jackson, S.N.; Colsch, B.; Egan, T.; Lewis, E.K.; Schultz, J.A.; Woods, A.S. Gangliosides’ Analysis by MALDI-Ion Mobility MS. Analyst 2011, 136, 463–466. [Google Scholar] [CrossRef]
- Ito, E.; Tominaga, A.; Waki, H.; Miseki, K.; Tomioka, A.; Nakajima, K.; Kakehi, K.; Suzuki, M.; Taniguchi, N.; Suzuki, A. Structural Characterization of Monosialo-, Disialo- and Trisialo-Gangliosides by Negative Ion AP-MALDI-QIT-TOF Mass Spectrometry with MS n Switching. Neurochem. Res. 2012, 37, 1315–1324. [Google Scholar] [CrossRef] [PubMed]
- Powell, A.K.; Harvey, D.J. Stabilization of Sialic Acids in N-Linked Oligosaccharides and Gangliosides for Analysis by Positive Ion Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry. Rapid Commun. Mass Spectrom. 1996, 10, 1027–1032. [Google Scholar] [CrossRef]
- Taketomi, T.; Hara, A.; Uemura, K.I.; Sugiyama, E. Matrix-Assisted Laser Desorption Ionization Time-of-Flight Mass Spectrometric Analysis of Glycosphingolipids Including Gangliosides. Acta Biochim. Pol. 1998, 45, 987–999. [Google Scholar] [CrossRef] [PubMed]
- Park, W.; Lee, D.; Cha, S. Differentiation of Ganglioside Isomers by MALDI Tandem Mass Spectrometry with Alkali Earth Metal Additives. Bull. Korean Chem. Soc. 2015, 36, 2272–2277. [Google Scholar] [CrossRef]
- Fuchs, B.; Schiller, J. MALDI-TOF MS Analysis of Lipids from Cells, Tissues and Body Fluids. Subcell. Biochem. 2008, 49, 541–565. [Google Scholar] [PubMed]
- Fuchs, B.; Müller, K.; Göritz, F.; Blottner, S.; Schiller, J. Characteristic Oxidation Products of Choline Plasmalogens Are Detectable in Cattle and Roe Deer Spermatozoa by MALDI-TOF Mass Spectrometry. Lipids 2007, 42, 991–998. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, B.; Schiller, J.; Cross, M.A. Apoptosis-Associated Changes in the Glycerophospholipid Composition of Hematopoietic Progenitor Cells Monitored by 31P NMR Spectroscopy and MALDI-TOF Mass Spectrometry. Chem. Phys. Lipids 2007, 150, 229–238. [Google Scholar] [CrossRef]
- Fuchs, B.; Schiller, J.; Wagner, U.; Häntzschel, H.; Arnold, K. The Phosphatidylcholine/Lysophosphatidylcholine Ratio in Human Plasma Is an Indicator of the Severity of Rheumatoid Arthritis: Investigations by 31P NMR and MALDI-TOF MS. Clin. Biochem. 2005, 38, 925–933. [Google Scholar] [CrossRef]
- Li, D.; Yi, J.; Han, G.; Qiao, L. MALDI-TOF Mass Spectrometry in Clinical Analysis and Research. ACS Meas. Sci. Au 2022, 2, 385–404. [Google Scholar] [CrossRef]
- Hanamatsu, H.; Nishikaze, T.; Miura, N.; Piao, J.; Okada, K.; Sekiya, S.; Iwamoto, S.; Sakamoto, N.; Tanaka, K.; Furukawa, J.I. Sialic Acid Linkage Specific Derivatization of Glycosphingolipid Glycans by Ring-Opening Aminolysis of Lactones. Anal. Chem. 2018, 90, 13193–13199. [Google Scholar] [CrossRef]
- Suzuki, A.; Suzuki, M.; Ito, E.; Nitta, T.; Inokuchi, J. ichi Mass Spectrometry of Gangliosides. Methods Mol. Biol. 2018, 1804, 207–221. [Google Scholar]
- Nakamura, K.; Suzuki, Y.; Goto-Inoue, N.; Yoshida-Noro, C.; Suzuki, A. Structural Characterization of Neutral Glycosphingolipids by Thin-Layer Chromatography Coupled to Matrix-Assisted Laser Desorption/Ionization Quadrupole Ion Trap Time-of-Flight MS/MS. Anal. Chem. 2006, 78, 5736–5743. [Google Scholar] [CrossRef]
- Torretta, E.; Fania, C.; Vasso, M.; Gelfi, C. HPTLC-MALDI MS for (Glyco)Sphingolipid Multiplexing in Tissues and Blood: A Promising Strategy for Biomarker Discovery and Clinical Applications. Electrophoresis 2016, 37, 2036–2049. [Google Scholar] [CrossRef]
- QRueda, R.; Puente, R. New data on content and distribution of gangliosides in human milk. Biol. Chem. 1995, 376, 723–727. [Google Scholar]
- Ma, L.; Fong, B.Y.; MacGibbon, A.K.H.; Norris, G. Qualitative and Quantitative Study of Glycosphingolipids in Human Milk and Bovine Milk Using High Performance Liquid Chromatography–Data-Dependent Acquisition–Mass Spectrometry. Molecules 2020, 25, 4024. [Google Scholar] [CrossRef] [PubMed]
- Ballard, O.; Morrow, A.L. Human Milk Composition. Nutrients and Bioactive Factors. Pediatr. Clin. North Am. 2013, 60, 49–74. [Google Scholar] [CrossRef]
- Sipione, S.; Monyror, J. Gangliosides in the brain: Physiology, pathophysiology and therapeutic applications. Front. Neurosci. 2020, 14, 572965. [Google Scholar] [CrossRef]
- Ariga, T.; McDonald, M. Role of ganglioside metabolism in the pathogenesis of Alzheimer’s disease–A review. J. Lip. Res. 2008, 49, 1157–1175. [Google Scholar] [CrossRef]
- Taki, T. An approach to glycobiology from glycolipidomics: Ganglioside molecular scanning in the brains of patients with Alzheimer’s disease by TLC-Blot/Matrix assisted laser desorption/ionization-time of flight MS. Biol. Pharm. Bull. 2012, 35, 1642–1647. [Google Scholar] [CrossRef] [PubMed]
- Yu, R.; Tsai, Y.T. Functional roles of gangliosides in neurodevelopment: An overview of recent advances. Neurochem. Res. 2012, 37, 1230–1244. [Google Scholar] [CrossRef]
- Zaprianova, E.; Deleva, D. Serum ganglioside patterns in multiple sclerosis. Nuerochem. Res. 2001, 26, 95–100. [Google Scholar] [CrossRef] [PubMed]
- Hakomori, S. Tumor malignancy defined by aberrant glycosylation and sphingo(glyco)lipid metabolism. Cancer Res. 1996, 56, 5309–5318. [Google Scholar] [PubMed]
- Taketomi, T.; Sugiyama, E. Extraction and analysis of multiple sphingolipids from a single sample. Meth Enzym. 2000, 312, 80–101. [Google Scholar] [PubMed]
- Miller-Podraza, H.; Mansson, J.E. Pentasialogangliosides of human brain. Fed. Europ. Biochem. Soc. 1991, 288, 212–214. [Google Scholar] [CrossRef] [PubMed]
- Miller-Podraza, H.; Mansson, J.E. Isolation of complex gangliosides from human brain. Biochim. Biophys. Acta 1992, 1124, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Prokazova, N.V.; Samovilova, N.N.; Gracheva, E.V.; Golovanova, N.K. Ganglioside GM3 and Its Biological Functions. Biochemistry 2009, 74, 235–249. [Google Scholar] [CrossRef] [PubMed]
- Ngamukote, S.; Yanagisawa, M.; Ariga, T.; Ando, S.; Yu, R.K. Developmental Changes of Glycosphingolipids and Expression of Glycogenes in Mouse Brains. J. Neurochem. 2007, 103, 2327–2341. [Google Scholar] [CrossRef] [PubMed]
- Tettamanti, G.; Bonali, F.; Marchesini, S.; Zambotti, V. A New Procedure for the Extraction, Purification and Fractionation of Brain Gangliosides. Biochim. Biophys. Acta (BBA)/Lipids Lipid Metab. 1973, 296, 160–170. [Google Scholar] [CrossRef]
- Kracun, I.; Rosner, H.; Drnovsek, V.; Heffer-Lauc, M.; Cosovic, C.; Lauc, G. Human Brain Gangliosides in Development, Aging and Disease. Int. J. Dev. Biol. 1991, 35, 289–295. [Google Scholar] [PubMed]
- Svennerholm, L.; Boström, K.; Jungbjer, B.; Olsson, L. Membrane Lipids of Adult Human Brain: Lipid Composition of Frontal and Temporal Lobe in Subjects of Age 20 to 100 Years. J. Neurochem. 1994, 63, 1802–1811. [Google Scholar] [CrossRef]
- Huebecker, M.; Moloney, E.B.; Van Der Spoel, A.C.; Priestman, D.A.; Isacson, O.; Hallett, P.J.; Platt, F.M. Reduced Sphingolipid Hydrolase Activities, Substrate Accumulation and Ganglioside Decline in Parkinson’s Disease. Mol. Neurodegener. 2019, 14, 40. [Google Scholar] [CrossRef]
- Cartron, J.P.; Colin, Y. Structural and functional diversity of blood group antigens. Transus. Clin. Biol. 2001, 8, 163–199. [Google Scholar] [CrossRef] [PubMed]
- Arab, S.; Russel, E.; Chapman, W.B.; Rosen, B.; Lingwood, C.A. Expression of the Verotoxin Receptor Glycolipid, Globotriaosylceramide, in Ovarian Hyperplasias. Oncol. Res. 1997, 9, 553–563. [Google Scholar] [PubMed]
- Tyler, A.; Johansson, A.; Karlsson, T.; Gudey, S.K.; Brännström, T.; Grankvist, K.; Behnam-Motlagh, P. Targeting Glucosylceramide Synthase Induction of Cell Surface Globotriaosylceramide (Gb3) in Acquired Cisplatin-Resistance of Lung Cancer and Malignant Pleural Mesothelioma Cells. Exp. Cell Res. 2015, 336, 23–32. [Google Scholar] [CrossRef]
- Ferroni, P.; Lenti, L.; Martini, F.; Ciatti, F.; Pontieri, G.M.; Gazzaniga, P.P. Ganglioside Content of Human Platelets-Differences in Resting and Activated Platelets. Thromb. Haemost. 1997, 77, 548–554. [Google Scholar] [CrossRef] [PubMed]
- Mazurov, A.V.; Prokazova, N.V.; Mikhailenko, I.A.; Mukjin, D.N.; Repin, V.S.; Bergelson, L.D. Stimulation of Platelet Adhesion and Activation by Ganglioside GD3 Adsorbed to Plastic. BBA-Mol. Cell Res. 1988, 968, 167–171. [Google Scholar] [CrossRef]
- Kyogashima, M.; Taketomi, T. Lipids from Human Platelets in Primary Thrombocythemia. Jpn. J. Exp. Med. 1986, 56, 113–118. [Google Scholar] [PubMed]
- Kushi, Y.; Arita, M.; Ishizuka, I.; Kasama, T.; Fredman, P.; Handa, S. Sulfatide Is Expressed in Both Erythrocytes and Platelets of Bovine Origin. Biochim. Biophys. Acta-Lipids Lipid Metab. 1996, 1304, 254–262. [Google Scholar] [CrossRef]
- Zdebska, E.; Soszyńska, B.; Dobrowolski, Z.; Kościelak, J. The Levels of Glycosphingolipids, Ceramides, Sialic Acid and Glycogen Are Changed in Plasma Membranes of Rat Platelets Harvested during Recovery from Immune-Mediated Thrombocytopenia. Acta Biochim. Pol. 1996, 43, 547–556. [Google Scholar] [CrossRef]
- Liu, L.; Zhang, K.; Tan, L.; Chen, Y.H.; Cao, Y.P. Alterations in Cholesterol and Ganglioside GM1 Content of Lipid Rafts in Platelets from Patients with Alzheimer Disease. Alzheimer Dis. Assoc. Disord. 2015, 29, 63–69. [Google Scholar] [CrossRef]
- Cooling, L.L.W.; Walker, K.E.; Gille, T.; Koerner, T.A.W. Shiga Toxin Binds Human Platelets via Globotriaosylceramide (P(K) Antigen) and a Novel Platelet Glycosphingolipid. Infect. Immun. 1998, 66, 4355–4366. [Google Scholar] [CrossRef]
- Cooling, L.L.W.; Zhang, D.S.; Koerner, T.A.W. Human Platelets Express Gangliosides with LKE Activity and ABH Blood Group Activity. Transfusion 2001, 41, 504–516. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, N.; Toyoda, M. Vascular diseases and gangliosides. Int. J. Mol. Sci. 2019, 20, 6362. [Google Scholar] [CrossRef]
- Miller-Podraza, H.; Johanssons, L. A strain of human influenza A virus binds to extended but not short gangliosides as assayed by thin-layer chromatography overlay. Glycobiology 2000, 10, 975–982. [Google Scholar] [CrossRef]
- Nimrichter, L.; Burdick, M.M.; Aoki, K.; Laroy, W.; Fierro, M.A.; Hudson, S.A.; Von Seggern, C.E.; Cotter, R.J.; Bochner, B.S.; Tiemeyer, M.; et al. E-Selectin Receptors on Human Leukocytes. Blood 2008, 112, 3744–3752. [Google Scholar] [CrossRef] [PubMed]
- Hakomori, S. Glycosphingolipids in cellular interaction, differentiation, and oncogenesis. Ann. Rev. Biochem. 1981, 50, 733–764. [Google Scholar] [CrossRef]
- Filho, J.A.F.; Shapiro, B.E. Tay-Sachs Disease. Arch. Neurol. 2004, 61, 1466–1468. [Google Scholar] [CrossRef] [PubMed]
- Vanier, M.T. Niemann–Pick Disease Type C–Mini Review. Clin Genet 2003, 64, 269–281. [Google Scholar] [CrossRef] [PubMed]
- Kohlschütter, A.; Eichler, F. Childhood Leukodystrophies: A Clinical Perspective. Expert Rev. Neurother. 2011, 11, 1485–1496. [Google Scholar] [CrossRef]
- Nagral, A. Gaucher Disease. J. Clin. Exp. Hepatol. 2014, 4, 37–50. [Google Scholar] [CrossRef]
- Ehlert, K.; Frosch, M.; Fehse, N.; Zander, A.; Roth, J.; Vormoor, J. Farber Disease: Clinical Presentation, Pathogenesis and a New Approach to Treatment. Pediatr. Rheumatol. 2007, 5, 15. [Google Scholar] [CrossRef] [PubMed]
- Clarke, J.T.R. Narrative Review: Fabry Disease. Ann. Intern. Med. 2007, 146, 425–433. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, K.; Hamamura, K.; Aixinjueluo, W.; Furukawa, K. Biosignals Modulated by Tumor-Associated Carbohydrate Antigens: Novel Targets for Cancer Therapy. Ann. N. Y. Acad. Sci. 2006, 1086, 185–198. [Google Scholar] [CrossRef]
- Groux-Degroote, S.; Guérardel, Y.; Delannoy, P. Gangliosides: Structures, Biosynthesis, Analysis, and Roles in Cancer. ChemBioChem 2017, 18, 1146–1154. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Cordon-Cardo, C.; Zhang, H.S.; Reuter, V.E.; Adluri, S.; Hamilton, W.B.; Lloyd, K.O.; Livingiston, P.O. Selection of Tumor Antigens as Targets for Immune Attack Using Immunohistochemistry: I. Focus on Gangliosides. Int. J. Cancer 1997, 73, 42–49. [Google Scholar] [CrossRef]
- Ho, M.Y.; Yu, A.L.; Yu, J. Glycosphingolipid Dynamics in Human Embryonic Stem Cell and Cancer: Their Characterization and Biomedical Implications. Glycoconj. J. 2017, 34, 765–777. [Google Scholar] [CrossRef] [PubMed]
- Yu, A.L.; Hung, J.T.; Ho, M.Y.; Yu, J. Alterations of Glycosphingolipids in Embryonic Stem Cell Differentiation and Development of Glycan-Targeting Cancer Immunotherapy. Stem Cells Dev. 2016, 25, 1532–1548. [Google Scholar] [CrossRef]
- Liang, Y.J.; Ding, Y.; Levery, S.B.; Lobaton, M.; Handa, K.; Hakomori, S.I. Differential Expression Profiles of Glycosphingolipids in Human Breast Cancer Stem Cells vs. Cancer Non-Stem Cells. Proc. Natl. Acad. Sci. USA 2013, 110, 4968–4973. [Google Scholar] [CrossRef]
- Nazha, B.; Inal, C.; Owonikoko, T.K. Disialoganglioside GD2 Expression in Solid Tumors and Role as a Target for Cancer Therapy. Front. Oncol. 2020, 10, 1000. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zheng, X.; Pang, X.; Li, L.; Wang, J.; Yang, C.; Du, G. Ganglioside GD3 Synthase (GD3S), a Novel Cancer Drug Target. Acta Pharm. Sin. B 2018, 8, 713–720. [Google Scholar] [CrossRef] [PubMed]
- Ramos, R.I.; Bustos, M.A.; Wu, J.; Jones, P.; Chang, S.C.; Kiyohara, E.; Tran, K.; Zhang, X.; Stern, S.L.; Izraely, S.; et al. Upregulation of Cell Surface GD3 Ganglioside Phenotype Is Associated with Human Melanoma Brain Metastasis. Mol. Oncol. 2020, 14, 1760–1778. [Google Scholar] [CrossRef]
- Chang, W.W.; Chien, H.L.; Lee, P.; Lin, J.; Hsu, C.W.; Hung, J.T.; Lin, J.J.; Yu, J.C.; Shao, L.E.; Yu, J.; et al. Expression of Globo H and SSEA3 in Breast Cancer Stem Cells and the Involvement of Fucosyl Transferases 1 and 2 in Globo H Synthesis. Proc. Natl. Acad. Sci. USA 2008, 105, 11667–11672. [Google Scholar] [CrossRef]
- Kuo, H.H.; Lin, R.J.; Hung, J.T.; Hsieh, C.B.; Hung, T.H.; Lo, F.Y.; Ho, M.Y.; Yeh, C.T.; Huang, Y.L.; Yu, J.; et al. High Expression FUT1 and B3GALT5 Is an Independent Predictor of Postoperative Recurrence and Survival in Hepatocellular Carcinoma. Sci. Rep. 2017, 7, 10750. [Google Scholar] [CrossRef] [PubMed]
- Steenackers, A.; Vanbeselaere, J.; Cazet, A.; Bobowski, M.; Rombouts, Y.; Colomb, F.; Le Bourhis, X.; Guérardel, Y.; Delannoy, P. Accumulation of Unusual Gangliosides G Q3 and G P3 in Breast Cancer Cells Expressing the G D3 Synthase. Molecules 2012, 17, 9559–9572. [Google Scholar] [CrossRef] [PubMed]
- Zhuo, D.; Li, X.; Guan, F. Biological Roles of Aberrantly Expressed Glycosphingolipids and Related Enzymes in Human Cancer Development and Progression. Front. Physiol. 2018, 9, 466. [Google Scholar] [CrossRef]
- Yu, J.; Hung, J.T.; Wang, S.H.; Cheng, J.Y.; Yu, A.L. Targeting Glycosphingolipids for Cancer Immunotherapy. FEBS Lett. 2020, 594, 3602–3618. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Zhang, H.S.; Reuter, V.E.; Slovin, S.F.; Scher, H.I.; Livingston, P.O. Expression of Potential Target Antigens for Immunotherapy on Primary and Metastatic Prostate Cancers. Clin. Cancer Res. 1998, 4, 295–302. [Google Scholar]
- Lou, Y.W.; Wang, P.Y.; Yeh, S.C.; Chuang, P.K.; Li, S.T.; Wu, C.Y.; Khoo, K.H.; Hsiao, M.; Hsu, T.L.; Wong, C.H. Stage-Specific Embryonic Antigen-4 as a Potential Therapeutic Target in Glioblastoma Multiforme and Other Cancers. Proc. Natl. Acad. Sci. USA 2014, 111, 2482–2487. [Google Scholar] [CrossRef]
- Korekane, H.; Tsuji, S.; Noura, S.; Ohue, M.; Sasaki, Y.; Imaoka, S.; Miyamoto, Y. Novel Fucogangliosides Found in Human Colon Adenocarcinoma Tissues by Means of Glycomic Analysis. Anal. Biochem. 2007, 364, 37–50. [Google Scholar] [CrossRef]
- Kohla, G.; Stockfleth, E.; Schauer, R. Gangliosides with O-Acetylated Sialic Acids in Tumors of Neuroectodermal Origin. Neurochem. Res. 2002, 27, 583–592. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, T.; Watanabe, M.; Katsumata, T.; Teramoto, T.; Kitajima, M. Expression of Sialyl Lewisa as a New Prognostic Factor for Patients with Advanced Colorectal Carcinoma. Cancer 1995, 75, 2051–2056. [Google Scholar] [CrossRef] [PubMed]
- Kannagi, R. Molecular Mechanism for Cancer-Associated Induction of Sialyl Lewis X and Sialyl Lewis A Expression-The Warburg Effect Revisited. Glycoconj. J. 2003, 20, 353–364. [Google Scholar] [CrossRef]
- Nudelman, E.; Fukushi, Y.; Levery, S.B.; Higuchi, T.; Hakomori, S. Novel Fucolipids of Human Adenocarcinoma: Disialosyl Lea Antigen (III4FucIII6NeuAcIV3NeuAcLc4) of Human Colonic Adenocarcinoma and the Monoclonal Antibody (FH7) Defining This Structure. J. Biol. Chem. 1986, 261, 5487–5495. [Google Scholar] [CrossRef]
- Miyazaki, K.; Ohmori, K.; Izawa, M.; Koike, T.; Kumamoto, K.; Furukawa, K.; Ando, T.; Kiso, M.; Yamaji, T.; Hashimoto, Y.; et al. Loss of Disialyl Lewisa, the Ligand for Lymphocyte Inhibitory Receptor Sialic Acid-Binding Immunoglobulin-like Lectin-7 (Siglec-7) Associated with Increased Sialyl Lewisa Expression on Human Colon Cancers. Cancer Res. 2004, 64, 4498–4505. [Google Scholar] [CrossRef] [PubMed]
- Tsuchida, A.; Okajima, T.; Furukawa, K.; Ando, T.; Ishida, H.; Yoshida, A.; Nakamura, Y.; Kannagi, R.; Kiso, M.; Furukawa, K. Synthesis of Disialyl Lewis a (Lea) Structure in Colon Cancer Cell Lines by a Sialyltransferase, ST6GalNAc VI, Responsible for the Synthesis of α-Series Gangliosides. J. Biol. Chem. 2003, 278, 22787–22794. [Google Scholar] [CrossRef]
- Hakomori, S.I. Aberrant Glycosylation in Cancer Cell Membranes as Focused on Glycolipids: Overview and Perspectives. Cancer Res. 1985, 45, 2405–2414. [Google Scholar] [PubMed]
- Serb, A.F.; Novaconi, C. Preliminary analysis of the glycolipid profile in secondary brain tumors. Biomed Res. Int. 2022, 2022, 4293172. [Google Scholar] [CrossRef]
- Yabu, M.; Korekane, H. Occurrence of free deaminoneuraminic acid (KDN)-containing complex-type N-glycans in human prostate cancers. Glycobiology 2013, 23, 634–642. [Google Scholar] [CrossRef]
- Wang, F.; Xie, B. LC-MS/MS glycomic analyses of free and conjugated forms of the sialic acids, Neu5Ac, Neu5Gc and KDN in human throat cancers. Glycobiology 2015, 25, 1362–1374. [Google Scholar] [CrossRef]
- Hakomori, S. itiroh Functional Role of Glycosphingolipids in Tumor Progression. Tohoku J. Exp. Med. 1992, 168, 211–222. [Google Scholar] [CrossRef] [PubMed]
- Patwardhan, G.A.; Liu, Y.Y. Sphingolipids and Expression Regulation of Genes in Cancer. Prog. Lipid Res. 2011, 50, 104–114. [Google Scholar] [CrossRef]
- Yin, J.; Miyazaki, K.; Shaner, R.L.; Merrill, A.H.; Kannagi, R. Altered Sphingolipid Metabolism Induced by Tumor Hypoxia-New Vistas in Glycolipid Tumor Markers. FEBS Lett. 2010, 584, 1872–1878. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S. Sialic Acids and Sialoglycoconjugates in Cancer. In Sialic Acids and Sialoglycoconjugates in the Biology of Life, Health and Disease; Academic Press: Cambridge, MA, USA, 2020; pp. 211–245. [Google Scholar]
- Samraj, A.N.; Läubli, H.; Varki, N.; Varki, A. Involvement of a non-human sialic Acid in human cancer. Front. Oncol. 2014, 4, 33. [Google Scholar] [CrossRef] [PubMed]
- Reymond, N.; D’Água, B.B.; Ridley, A.J. Crossing the Endothelial Barrier during Metastasis. Nat. Rev. Cancer 2013, 13, 858–870. [Google Scholar] [CrossRef] [PubMed]
- Kannagi, R.; Izawa, M.; Koike, T.; Miyazaki, K.; Kimura, N. Carbohydrate-Mediated Cell Adhesion in Cancer Metastasis and Angiogenesis. Cancer Sci. 2004, 95, 377–384. [Google Scholar] [CrossRef] [PubMed]
- Belarbi, K.; Cuvelier, E. Glycosphingolipids and neuroinflammation in Parkinson’s disease. Mol. Neurodegen. 2020, 15, 59. [Google Scholar] [CrossRef]
- Savica, R.; Murray, M.E.; Persson, X.M.; Kantarci, K.; Parisi, J.E.; Dickson, D.W.; Petersen, R.C.; Ferman, T.J.; Boeve, B.F.; Mielke, M.M. Plasma Sphingolipid Changes with Autopsy-Confirmed Lewy Body or Alzheimer’s Pathology. Alzheimer’s Dement. Diagnosis Assess. Dis. Monit. 2016, 3, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Rozen, S.; Boyle, S.H.; Hellegers, C.; Cheng, H.; Burke, J.R.; Welsh-Bohmer, K.A.; Doraiswamy, P.M.; Kaddurah-Daouk, R. Metabolomics in Early Alzheimer’s Disease: Identification of Altered Plasma Sphingolipidome Using Shotgun Lipidomics. PLoS ONE 2011, 6, e21643. [Google Scholar] [CrossRef]
- Kim, M.; Nevado-Holgado, A.; Whiley, L.; Snowden, S.G.; Soininen, H.; Kloszewska, I.; Mecocci, P.; Tsolaki, M.; Vellas, B.; Thambisetty, M.; et al. Association between Plasma Ceramides and Phosphatidylcholines and Hippocampal Brain Volume in Late Onset Alzheimer’s Disease. J. Alzheimer’s Dis. 2017, 60, 809–817. [Google Scholar] [CrossRef]
- Matsuzaki, K. How Do Membranes Initiate Alzheimers Disease? Formation of Toxic Amyloid Fibrils by the Amyloid β-Protein on Ganglioside Clusters. Acc. Chem. Res. 2014, 47, 2397–2404. [Google Scholar] [CrossRef]
- Ke, P.C.; Sani, M.A.; Ding, F.; Kakinen, A.; Javed, I.; Separovic, F.; Davis, T.P.; Mezzenga, R. Implications of Peptide Assemblies in Amyloid Diseases. Chem. Soc. Rev. 2017, 46, 6492–6531. [Google Scholar] [CrossRef]
- Kotler, S.A.; Walsh, P.; Brender, J.R.; Ramamoorthy, A. Differences between Amyloid-β Aggregation in Solution and on the Membrane: Insights into Elucidation of the Mechanistic Details of Alzheimer’s Disease. Chem. Soc. Rev. 2014, 43, 6692–6700. [Google Scholar] [CrossRef]
- Matsuzaki, K. Formation of Toxic Amyloid Fibrils by Amyloid β-Protein on Ganglioside Clusters. Int. J. Alzheimers. Dis. 2011, 2011, 956104. [Google Scholar] [CrossRef]
- Korshavn, K.J.; Satriano, C.; Lin, Y.; Zhang, R.; Dulchavsky, M.; Bhunia, A.; Ivanova, M.I.; Lee, Y.H.; La Rosa, C.; Lim, M.H.; et al. Reduced Lipid Bilayer Thickness Regulates the Aggregation and Cytotoxicity of Amyloid-β. J. Biol. Chem. 2017, 292, 4638–4650. [Google Scholar] [CrossRef] [PubMed]
- Matsuzaki, K.; Kato, K.; Yanagisawa, K. Aβ Polymerization through Interaction with Membrane Gangliosides. Biochim. Biophys. Acta-Mol. Cell Biol. Lipids 2010, 1801, 868–877. [Google Scholar] [CrossRef]
- Sahoo, B.R.; Genjo, T.; Cox, S.J.; Stoddard, A.K.; Anantharamaiah, G.M.; Fierke, C.; Ramamoorthy, A. Nanodisc-Forming Scaffold Protein Promoted Retardation of Amyloid-Beta Aggregation. J. Mol. Biol. 2018, 430, 4230–4244. [Google Scholar] [CrossRef]
- Matsuzaki, K. Aβ–Ganglioside Interactions in the Pathogenesis of Alzheimer’s Disease. Biochim. Biophys. Acta-Biomembr. 2020, 1862, 183233. [Google Scholar] [CrossRef]
- Kumar, M.; Ivanova, M.I.; Ramamoorthy, A. Non-Micellar Ganglioside GM1 Induces an Instantaneous Conformational Change in Aβ42 Leading to the Modulation of the Peptide Amyloid-Fibril Pathway. Biophys. Chem. 2023, 301, 107091. [Google Scholar] [CrossRef] [PubMed]
- Matsuzaki, K.; Noguch, T.; Wakabayashi, M.; Ikeda, K.; Okada, T.; Ohashi, Y.; Hoshino, M.; Naiki, H. Inhibitors of Amyloid β-Protein Aggregation Mediated by GM1-Containing Raft-like Membranes. Biochim. Biophys. Acta-Biomembr. 2007, 1768, 122–130. [Google Scholar] [CrossRef]
- Kreutz, F.; Scherer, E.B.; Ferreira, A.G.K.; Petry, F.D.S.; Pereira, C.L.; Santana, F.; De Souza Wyse, A.T.; Salbego, C.G.; Trindade, V.M.T. Alterations on Na+,K+-ATPase and Acetylcholinesterase Activities Induced by Amyloid-β Peptide in Rat Brain and GM1 Ganglioside Neuroprotective Action. Neurochem. Res. 2013, 38, 2342–2350. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Wang, Q.; Min, L.; Sui, R.; Li, J.; Liu, X. Monosialoanglioside Improves Memory Deficits and Relieves Oxidative Stress in the Hippocampus of Rat Model of Alzheimer’s Disease. Neurol. Sci. 2013, 34, 1447–1451. [Google Scholar] [CrossRef]
- Huang, M.; Hu, M.; Song, Q.; Song, H.; Huang, J.; Gu, X.; Wang, X.; Chen, J.; Kang, T.; Feng, X.; et al. GM1-Modified Lipoprotein-like Nanoparticle: Multifunctional Nanoplatform for the Combination Therapy of Alzheimer’s Disease. ACS Nano 2015, 9, 10801–10816. [Google Scholar] [CrossRef]
- Farrall, A.J.; Wardlaw, J.M. Blood-Brain Barrier: Ageing and Microvascular Disease-Systematic Review and Meta-Analysis. Neurobiol. Aging 2009, 30, 337–352. [Google Scholar] [CrossRef]
- Ledeen, R.; Chowdhury, S. Gangliosides in Neurodegenerative Diseases. In Advances in Neurobiology; Springer International Publishing: Berlin/Heidelberg, Germany, 2023; Volume 29, pp. 391–418. [Google Scholar]
- Akram, M.; Iqbal, M. Awareness and current knowledge of breast cancer. Biol. Res. 2017, 50, 33. [Google Scholar] [CrossRef] [PubMed]
- Giussani, P.; Prinetti, A.; Tringali, C. The Role of Sphingolipids in Cancer Immunotherapy. Int. J. Mol. Sci. 2021, 22, 6492. [Google Scholar] [CrossRef]
- Marquina, G.; Waki, H.; Fernandez, L.E.; Kon, K.; Carr, A.; Valiente, O.; Perez, R.; Ando, S. Gangliosides Expressed in Human Breast Cancer. Cancer Res. 1996, 56, 5165–5171. [Google Scholar] [PubMed]
- Ruckhäberle, E.; Karn, T.; Rody, A.; Hanker, L.; Gätje, R.; Metzler, D.; Holtrich, U.; Kaufmann, M. Gene Expression of Ceramide Kinase, Galactosyl Ceramide Synthase and Ganglioside GD3 Synthase Is Associated with Prognosis in Breast Cancer. J. Cancer Res. Clin. Oncol. 2009, 135, 1005–1013. [Google Scholar] [CrossRef]
- Battula, V.L.; Shi, Y.; Evans, K.W.; Wang, R.Y.; Spaeth, E.L.; Jacamo, R.O.; Guerra, R.; Sahin, A.A.; Marini, F.C.; Hortobagyi, G.; et al. Ganglioside GD2 Identifies Breast Cancer Stem Cells and Promotes Tumorigenesis. J. Clin. Investig. 2012, 122, 2066–2078. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.J. Glycosphingolipids in Human Embryonic Stem Cells and Breast Cancer Stem Cells, and Potential Cancer Therapy Strategies Based on Their Structures and Functions. Glycoconj. J. 2022, 39, 177–195. [Google Scholar] [CrossRef]
- Shao, C.; Anand, V.; Andreeff, M.; Battula, V.L. Ganglioside GD2: A Novel Therapeutic Target in Triple-Negative Breast Cancer. Ann. N. Y. Acad. Sci. 2022, 1508, 35–53. [Google Scholar] [CrossRef]
- Cheng, J.Y.; Hung, J.T.; Lin, J.; Lo, F.Y.; Huang, J.R.; Chiou, S.P.; Wang, Y.H.; Lin, R.J.; Wu, J.C.; Yu, J.; et al. O-Acetyl-GD2 as a Therapeutic Target for Breast Cancer Stem Cells. Front. Immunol. 2022, 12, 791551. [Google Scholar] [CrossRef]
- Ly, S.; Anand, V.; El-Dana, F.; Nguyen, K.; Cai, Y.; Cai, S.; Piwnica-Worms, H.; Tripathy, D.; Sahin, A.A.; Andreeff, M.; et al. Anti-GD2 Antibody Dinutuximab Inhibits Triple-Negative Breast Tumor Growth by Targeting GD2+ Breast Cancer Stem-like Cells. J. Immunother. Cancer 2021, 9, e001197. [Google Scholar] [CrossRef]
- Seitz, C.M.; Schroeder, S.; Knopf, P.; Krahl, A.C.; Hau, J.; Schleicher, S.; Martella, M.; Quintanilla-Martinez, L.; Kneilling, M.; Pichler, B.; et al. GD2-Targeted Chimeric Antigen Receptor T Cells Prevent Metastasis Formation by Elimination of Breast Cancer Stem-like Cells. Oncoimmunology 2020, 9, 1683345. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Sun, M.; Yu, M.; Fu, Q.; Jiang, H.; Yu, G.; Li, G. Gangliosides Profiling in Serum of Breast Cancer Patient: GM3 as a Potential Diagnostic Biomarker. Glycoconj. J. 2019, 36, 419–428. [Google Scholar] [CrossRef] [PubMed]
- Cavdarli, S.; Dewald, J.H.; Yamakawa, N.; Guérardel, Y.; Terme, M.; Le Doussal, J.M.; Delannoy, P.; Groux-Degroote, S. Identification of 9-O-Acetyl-N-Acetylneuraminic Acid (Neu5,9Ac 2) as Main O-Acetylated Sialic Acid Species of GD2 in Breast Cancer Cells. Glycoconj. J. 2019, 36, 79–90. [Google Scholar] [CrossRef] [PubMed]
- Valdes-Zayas, A.; Gonzalez, Z.; Mulens, V.; Vega, A.M.; Perez, K.; Lorenzo-Luaces, P.; Rubio, M.C.; Estevez, A.; Curbelo, I.; Fernandez, L.E.; et al. Immunologic Response Elicited in Breast Cancer Patients Receiving a NeuGcGM3-Based Vaccine as Adjuvant Therapy. J. Immunother. 2017, 40, 289–301. [Google Scholar] [CrossRef] [PubMed]
- Orsi, G.; Barbolini, M.; Ficarra, G.; Tazzioli, G.; Manni, P.; Petrachi, T.; Mastrolia, I.; Orvieto, E.; Spano, C.; Prapa, M.; et al. GD2 Expression in Breast Cancer. Oncotarget 2017, 8, 31592–31600. [Google Scholar] [CrossRef]
- Sarkar, T.R.; Battula, V.L.; Werden, S.J.; Vijay, G.V.; Ramirez-Peña, E.Q.; Taube, J.H.; Chang, J.T.; Miura, N.; Porter, W.; Sphyris, N.; et al. GD3 Synthase Regulates Epithelial-Mesenchymal Transition and Metastasis in Breast Cancer. Oncogene 2015, 34, 2958–2967. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.J.; Wang, C.Y.; Wang, I.A.; Chen, Y.W.; Li, L.T.; Lin, C.Y.; Ho, M.Y.; Chou, T.L.; Wang, Y.H.; Chiou, S.P.; et al. Interaction of Glycosphingolipids GD3 and GD2 with Growth Factor Receptors Maintains Breast Cancer Stem Cell Phenotype. Oncotarget 2017, 8, 47454–47473. [Google Scholar] [CrossRef]
- Ha, S.H.; Lee, J.M.; Kwon, K.M.; Kwak, C.H.; Abekura, F.; Park, J.Y.; Cho, S.H.; Lee, K.; Chang, Y.C.; Lee, Y.C.; et al. Exogenous and Endogeneous Disialosyl Ganglioside GD1b Induces Apoptosis of MCF-7 Human Breast Cancer Cells. Int. J. Mol. Sci. 2016, 17, 652. [Google Scholar] [CrossRef]
- Cheung, S.K.C.; Chuang, P.K.; Huang, H.W.; Hwang-Verslues, W.W.; Cho, C.H.H.; Yang, W.; Shen, C.N.; Hsiao, M.; Hsu, T.L.; Chang, C.F.; et al. Stage-Specific Embryonic Antigen-3 (SSEA-3) and Β3GalT5 Are Cancer Specific and Significant Markers for Breast Cancer Stem Cells. Proc. Natl. Acad. Sci. USA 2016, 113, 960–965. [Google Scholar] [CrossRef] [PubMed]
- Vandermeersch, S.; Vanbeselaere, J.; Delannoy, C.P.; Drolez, A.; Mysiorek, C.; Guérardel, Y.; Delannoy, P.; Julien, S. Accumulation of GD1α Ganglioside in MDA-MB-231 Breast Cancer Cells Expressing ST6GalNAc V. Molecules 2015, 20, 6913–6924. [Google Scholar] [CrossRef]
- Cheng, J.Y.; Wang, S.H.; Lin, J.; Tsai, Y.C.; Yu, J.; Wu, J.C.; Hung, J.T.; Lin, J.J.; Wu, Y.Y.; Yeh, K.T.; et al. Globo-H Ceramide Shed from Cancer Cells Triggers Translin-Associated Factor X-Dependent Angiogenesis. Cancer Res. 2014, 74, 6856–6866. [Google Scholar] [CrossRef] [PubMed]
- Cavdarli, S.; Yamakawa, N. Profiling of O-acetylated gangliosides expressed in neurectoderm derived cells. Int. J. Mol. Sci. 2020, 21, 370. [Google Scholar] [CrossRef] [PubMed]
- Cazet, A.; Bobowski, M. The ganglioside GD2 induces the constitutive activation of c-Met in MDA-MB-231 breast cancer cells expressing GD3 synthase. Glycobiology 2012, 22, 806–816. [Google Scholar] [CrossRef]
- Alvarez-Rueda, N.; Desselle, A. A monoclonal antibody to O-acetyl-GD2 ganglioside and not to GD2 shows potent anti-tumor activity without peripheral nervous system cross-reactivity. PLoS ONE 2011, 6, e25220. [Google Scholar] [CrossRef]
- Cochonneau, D.; Terme, M. Cell cycle arrest and apoptosis induced by O-acetyl-GD2-specific monoclonal antibody 8B6 inhibits tumor growth in vitro and in vivo. Cancer Lett. 2013, 333, 194–204. [Google Scholar] [CrossRef] [PubMed]
- Dewald, J.; Cavdarli, S. TNF differentially regulates ganglioside biosynthesis and expression in breast cancer cell lines. PLoS ONE 2018, 13, e0196369. [Google Scholar] [CrossRef] [PubMed]
- Bobowski, M.; Cazet, A.; Steenackers, A.; Delannoy, P. Role of complex gangliosides in cancer progression. Carbohydr. Chem. 2012, 37, 1–20. [Google Scholar]
- Kniep, B.; Kniep, E.; Özkucur, N.; Barz, S.; Bachmann, M.; Malisan, F.; Testi, R.; Rieber, E.P. 9-O-Acetyl GD3 Protects Tumor Cells from Apoptosis. Int. J. Cancer 2006, 119, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Malisan, F.; Franchi, L.; Tomassini, B.; Ventura, N.; Condò, I.; Rippo, M.R.; Rufini, A.; Liberati, L.; Nachtigall, C.; Kniep, B.; et al. Acetylation Suppresses the Proapoptotic Activity of GD3 Ganglioside. J. Exp. Med. 2002, 196, 1535–1541. [Google Scholar] [CrossRef] [PubMed]
- Kristal, B.S.; Brown, A.M. Apoptogenic Ganglioside GD3 Directly Induces the Mitochondrial Permeability Transition. J. Biol. Chem. 1999, 274, 23169–23175. [Google Scholar] [CrossRef]
- Sa, G.; Das, T.; Moon, C.; Hilston, C.M.; Rayman, P.A.; Rini, B.I.; Tannenbaum, C.S.; Finke, J.H. GD3 an Overexpressed Tumor-Derived Ganglioside, Mediatesthe Apoptosis of Activated but Not Resting T Cells. Cancer Res. 2009, 69, 3095–3104. [Google Scholar] [CrossRef] [PubMed]
- Terme, M.; Dorvillius, M.; Cochonneau, D.; Chaumette, T.; Xiao, W.; Diccianni, M.B.; Barbet, J.; Yu, A.L.; Paris, F.; Sorkin, L.S.; et al. Chimeric Antibody c.8B6 to O-Acetyl-GD2 Mediates the Same Efficient Anti-Neuroblastoma Effects as Therapeutic Ch14.18 Antibody to GD2 without Antibody Induced Allodynia. PLoS ONE 2014, 9, e87210. [Google Scholar] [CrossRef] [PubMed]
- Johnsen, J.I.; Dyberg, C. Neuroblastoma–A neural crest derived embryonal malignancy. Front. Mol. Neurosci. 2019, 12, 9. [Google Scholar] [CrossRef] [PubMed]
- Mehta, A.; Herrera, H.; Block, T. Glycosylation and Liver Cancer. In Advances in Cancer Research; Elsevier: Amsterdam, The Netherlands, 2015; Volume 126, pp. 257–279. [Google Scholar]
- Zhu, J.; Wang, Y.; Yu, Y.; Wang, Z.; Zhu, T.; Xu, X.; Liu, H.; Hawke, D.; Zhou, D.; Li, Y. Aberrant Fucosylation of Glycosphingolipids in Human Hepatocellular Carcinoma Tissues. Liver Int. 2014, 34, 147–160. [Google Scholar] [CrossRef]
- Feng, X.; Zhao, L.; Gao, S.; Song, X.; Dong, W.; Zhao, Y.; Zhou, H.; Cheng, L.; Miao, X.; Jia, L. Increased Fucosylation Has a Pivotal Role in Multidrug Resistance of Breast Cancer Cells through MiR-224-3p Targeting FUT4. Gene 2016, 578, 232–241. [Google Scholar] [CrossRef] [PubMed]
- Tan, K.P.; Ho, M.Y.; Cho, H.C.; Yu, J.; Hung, J.T.; Yu, A.L.T. Fucosylation of LAMP-1 and LAMP-2 by FUT1 Correlates with Lysosomal Positioning and Autophagic Flux of Breast Cancer Cells. Cell Death Dis. 2016, 7, e2347. [Google Scholar] [CrossRef]
- Lu, Y.C.; Chen, C.N.; Chu, C.Y.; Lu, J.H.; Wang, B.J.; Chen, C.H.; Huang, M.C.; Lin, T.H.; Pan, C.C.; Chen, S.S.A.; et al. Calreticulin Activates Β1 Integrin via Fucosylation by Fucosyltransferase 1 in J82 Human Bladder Cancer Cells. Biochem. J. 2014, 460, 69–78. [Google Scholar] [CrossRef]
- Palumberi, D.; Aldi, S.; Ermini, L.; Ziche, M.; Finetti, F.; Donnini, S.; Rosati, F. RNA-Mediated Gene Silencing of FUT1 and FUT2 Influences Expression and Activities of Bovine and Human Fucosylated Nucleolin and Inhibits Cell Adhesion and Proliferation. J. Cell. Biochem. 2010, 111, 229–238. [Google Scholar] [CrossRef]
- Mathieu, S.; Prorok, M.; Benoliel, A.M.; Uch, R.; Langlet, C.; Bongrand, P.; Gerolami, R.; El-Battari, A. Transgene Expression of α(1,2)-Fucosyltransferase-I (FUT1) in Tumor Cells Selectively Inhibits Sialyl-Lewis x Expression and Binding to E-Selectin without Affecting Synthesis of Sialyl-Lewis a or Binding to P-Selectin. Am. J. Pathol. 2004, 164, 371–383. [Google Scholar] [CrossRef] [PubMed]
- Hutchinson, W.L.; Du, M.-Q.; Johnson, P.J.; Williams, R. Fucosyltransferases: Differential Plasma and Tissue Alterations in Hepatocellular Carcinoma and Cirrhosis. Hepatology 1991, 13, 683–688. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.; Xu, L.; Xu, X.; Wang, Z.; Zhu, J.; Xie, Q.; Zhang, B.; Wang, Y.; Ju, L.; He, Y.; et al. Analysis of Breast Cancer-Associated Glycosphingolipids Using Electrospray Ionization-Linear Ion Trap Quadrupole Mass Spectrometry. Carbohydr. Res. 2015, 402, 189–199. [Google Scholar] [CrossRef] [PubMed]
- Lobasso, S.; Tanzarella, P. Lipid profiling of parkin-mutant skin fibroblasts. J. Cell. Physiol. 2017, 12, 3540–3551. [Google Scholar] [CrossRef] [PubMed]
- Bond, M.R.; Whitman, C.M. Matabolically incorporated photocrosslinking sialic acid covalently captures a ganglioside-protein complex. Mol. BioSyst. 2010, 6, 1796–1799. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.J.; Kuo, H.H. Switching of the core structures of glycosphingolipids from globo- and lacto- to ganglio-series upon human embryonic stem cell differentiation. Proc. Natl. Acad. Sci. USA 2010, 107, 22564–22569. [Google Scholar] [CrossRef] [PubMed]
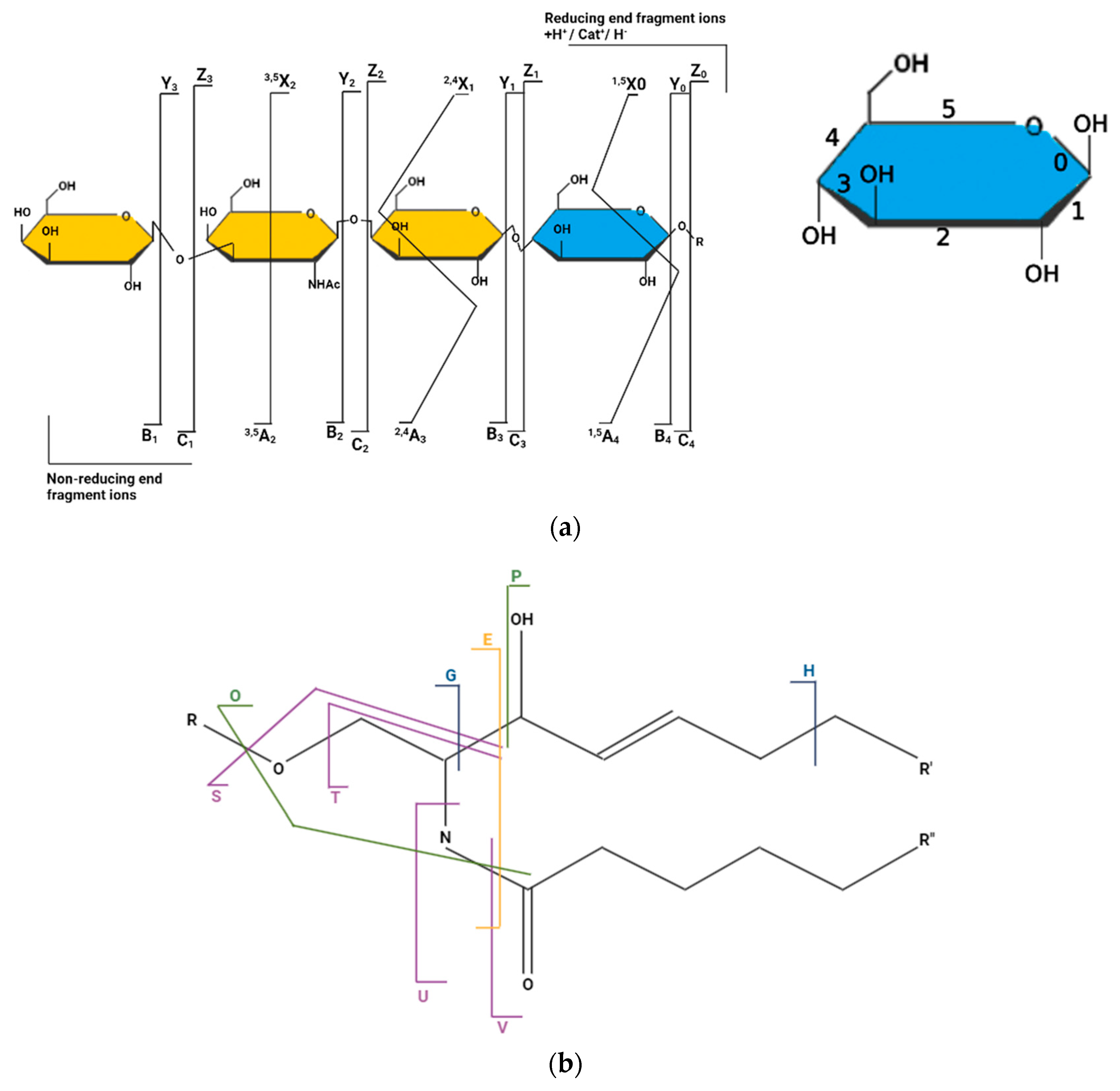
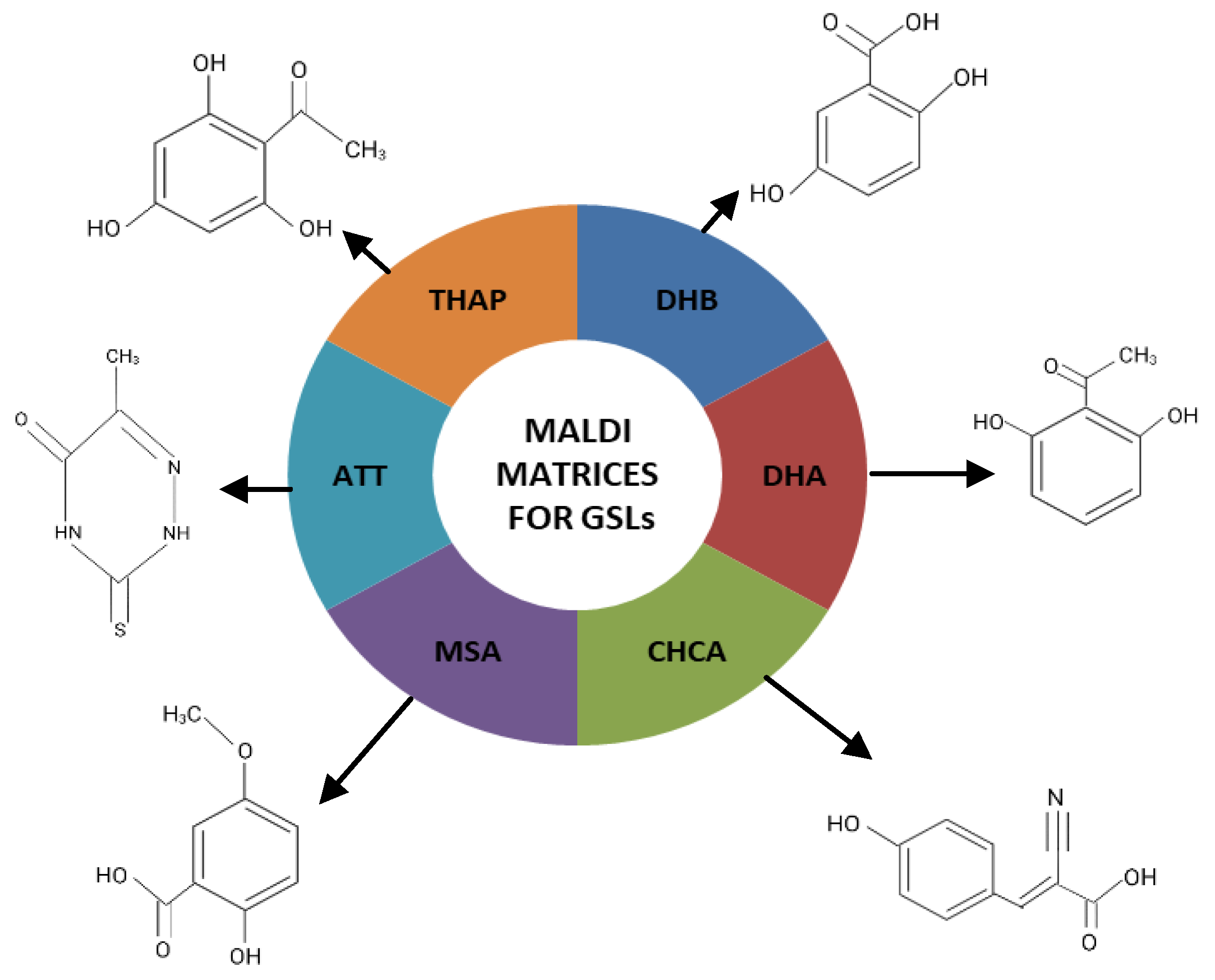
| GSL Series (Symbol) | Core Carbohydrate Structure | Tissue Specific Distribution |
|---|---|---|
| Ganglio (Gg) | Galß1-3GalNAcß1-4Galß1-4Glcß1-1′Cer | In the brain (vertebrates) |
| Isoganglio (iGg) | Galß1-3GalNAcß1-3Galß1-4Glcß1-1′Cer | |
| Lacto (Lc) | Galß1-3GlcNAcß1-3Galß1-4Glcß1-1′Cer | In secretory organs (vertebrates) |
| Neolacto (nLc) | Galß1-4GlcNAcß1-3Galß1-4Glcß1-1′Cer | In certain hematopoietic cells, including leukocytes (vertebrates) |
| Globo (Gb) | GalNAcβ1-3Galα1-4Galß1-4Glcß1-1′Cer | In erythrocytes (vertebrates) |
| Isoglobo (iGb) | GalNAcβ1-3Galα1-3Galβ1-4Glcß1-1′Cer | In vertebrates |
| Mollu (Mu) | GlcNAcβ1-2Manα1-3Manβ1-4Glcß1-1′Cer | In invertebrates |
| Arthro (At) | GalNAcβ1-4GlcNAcβ1-3Manβ1-4Glcß1-1′Cer | In invertebrates |
| Svennerholm Nomenclature | IUPAC-IUB Nomenclature |
|---|---|
| LacCer | Galb4Glcb1Cer |
| GA2, Gg3Cer | GalNAcb4Galb4Glcb1Cer |
| GA, Gg4Cer | Galb3GalNAcb4Galb4Glcb1Cer |
| nLc4Cer | Galb4GlcNAcb3Galb4Glcb1Cer |
| Lc4Cer | Galb3GlcNAcb3Galb4Glcb1Cer |
| GM4 | I3-a-Neu5Ac-GalCer |
| GM4 | I3-a-Neu5Ac-GlcCer |
| GM3 | II3-a-Neu5Ac-LacCer |
| GM3 | II3-a-Neu5Gc-LacCer |
| GD3 | II3-a-(Neu5Ac)2-LacCer |
| GD3 | II3-a-(Neu5Gc)2-LacCer |
| GD3 | II3-a-(Neu5Ac, Neu5Gc)2-LacCer |
| GT3 | II3-a-(Neu5Ac)3-LacCer |
| GM2 | II3-a-Neu5Ac-Gg3Cer |
| GD2 | II3-a-(Neu5Ac)2-Gg3Cer |
| GM1a | II3-a-Neu5Ac-Gg4Cer |
| GM1b | IV3-a-Neu5Ac-Gg4Cer |
| GalNAc-GM1b | IV3-a-Neu5Ac-Gg5Cer |
| LM1 | IV3-a-Neu5Ac-Lc4Cer |
| LM1 | IV3-a-Neu5Gc-Lc4Cer |
| GD1a | IV3-a-Neu5Ac,II3-a-Neu5Ac-Gg4Cer |
| GD1b | II3-a-(Neu5Ac)2-Gg4Cer |
| LD1 | IV3-a-(Neu5Ac)2-Lc4Cer |
| GT1a | IV3-a-(Neu5Ac)2,II3-a-Neu5Ac-Gg4Cer |
| GT1b | IV3-a-Neu5Ac,II3-a-(Neu5Ac)2-Gg4Cer |
| GT1c | II3-a-(Neu5Ac)3-Gg4Cer |
| GQ1b | IV3-a-(Neu5Ac)2,II3-a-(Neu5Ac)2-Gg4Cer |
| GQ1c | IV3-a-Neu5Ac,II3-a-(Neu5Ac)3-Gg4Cer |
| GP1b | IV3-a-(Neu5Ac)3,II3-a-(Neu5Ac)2-Gg4Cer |
| GP1c | IV3-a-(Neu5Ac)2,II3-a-(Neu5Ac)3-Gg4Cer |
| Matrix | Advantage | Reference | Disadvantage | Reference |
|---|---|---|---|---|
| DHB | Higher signal-to-noise ratio (S/N) | [116] | Resolution is considerably low | [117,118,119,120] |
| Decreased fragmentation in negative ion mode compared to positive ion mode | [121,122] | Altered sample geometry | [117,118,119,120] | |
| Minimizes loss of sialic acid | [123] | Increased fragmentation the higher the molecular mass is | [117,118,119,120] | |
| Minimizes loss of CO2 | [121] | |||
| Addition of butylamine reduces fragmentation | [124] | |||
| DHA | Increase in resolution | [125] | Decrease in the signal-to-noise (S/N) ratio | [125] |
| Negative ion mode improved detection of O-acetylated GT1 forms | [125] | |||
| THAP | High-quality Gb4Cer spectra | [126] | Low-quality spectra | [126] |
| ATT | Enhanced crystal formation by adding lithium citarte | [127] | Inhomogeneous distribution of crystals | [126] |
| Superior S/N ratio (in positive ion mode) | [117] | |||
| Significantly reduced fragmentation at high pressure | [120] | |||
| CHCA | Formation of homogeneous crystals | [117] | Considerable fragmentation | [117] |
| Differentiation of GD1a compared to GD1b | [115] | Trisialogangliosides undetectable as molecular ions | [117] | |
| Lysogangliosides are analyzed better | [128] | |||
| MSA | High-quality ganglioside profiles (in positive ion mode) | [129] | ||
| Minimal loss of sialic acid (in positive ion mode) | [129] | |||
| Detects the isomer specific fragment ions for both GD1a and GD1b | [129] |
| Cell Lines | Glycan Structures |
|---|---|
| All cell lines | [Galβ1 − 3Galα1 − 4Galβ1 − 4Glc], [GalNAcβ1 − 4(NeuAcα2 − 3)Galβ1 − 4Glc], [Galβ1 − 3GlcNAcβ1 − 3Galβ1 − 4Glc], [Galβ1 − 3(Fucα1 − 4)GlcNAcβ1 − 3Galβ1 − 4Glc and/or Fucα1 − 2Galβ1 − 3GlcNAcβ1 − 3Galβ1 − 4Glc], [Galβ1 − 3GalNAcβ1 − 4(NeuAcα2 − 3)Galβ1 − 4Glc], [Fucα1 − 2Galβ1 − 3GalNAcβ1 − 3Galα1 − 4Galβ1 − 4Glc] |
| Normal liver cell line HL-7702 | [Galβ1 − 3GlcNAcβ1 − 3Galβ1 − 3(Fucα1 − 4)GlcNAcβ1 − 3Galβ1 − 4Glc] |
| Hepatocarcinoma cell lines | [NeuGcα2 − 8NeuAcα2 − 3Galβ1 − 4Glc], [GalNAcα1 − 3(Fucα1 − 2)Galβ1 − 3GlcNAcβ1 − 3Galβ1 − 4Glc], [Galβ1 − 3GlcNAcβ1 − 3Galβ1 − 3GlcNAcβ1 − 3Galβ1 − 4Glc], [Galβ1 − 3GlcNAcβ1 − 3Galβ1 − 3GlcNAcβ1 − 3Galβ1 − 4Glc], [Galα1 − 3(Fucα1 − 2)-Galβ1 − 4(Fucα1 − 3)GlcNAcβ1 − 3Galβ1 − 4Glc], [GalNAcα1 − 3(Fucα1 − 2)Galβ1 − 4(Fucα1 − 3)GlcNAcβ1 − 3Galβ1 − 4Glc], [NeuAcα2 − 3SulfGalβ1 − 4(Fucα1 − 3)GlcNAcβ1 − 3Galβ1 − 4Glc], [Fucα1 − 2Galβ1 − 4-(Fucα1 − 3)GlcNAcβ1 − 3Galβ1 − 4(Fucα1 − 3)GlcNAcβ1 − 3Galβ1 − 4Glc], and [Galα1 − 3(Fucα1 − 2)Galβ1 − 4(Fucα1 − 3)GlcNAcβ1 − 3Galβ1 − 4(Fucα1 − 3)GlcNAcβ1 − 3Galβ1 − 4Glc] |
| High metastatic potential cell lines MHCC97H and HCCLM3 | [GalNAcα1 − 3(Fucα1 − 2)Galβ1 − 3GlcNAcβ1 − 3Galβ1 − 4Glc], [Galβ1 − 3GlcNAcβ1 − 3Galβ1 − 3GlcNAcβ1 − 3Galβ1 − 4Glc], [Galα1 − 3- (Fucα1 − 2)Galβ1 − 4(Fucα1 − 3)GlcNAcβ1 − 3Galβ1 − 4Glc], [GalNAcα1 − 3(Fucα1 − 2)Galβ1 − 4(Fucα1 − 3)- GlcNAcβ1 − 3Galβ1 − 4Glc], [NeuAcα2 − 3SulfGalβ1 − 4(Fucα1 − 3)GlcNAcβ1 − 3Galβ1 − 4Glc], [Fucα1 − 2Galβ1 − 4(Fucα1 − 3)GlcNAcβ1 − 3Galβ1 − 4- (Fucα1 − 3)GlcNAcβ1 − 3Galβ1 − 4Glc], [Galα1 − 3(Fucα1 − 2)Galβ1 − 4(Fucα1 − 3)GlcNAcβ1 − 3Galβ1 − 4(Fucα1 − 3)GlcNAcβ1 − 3Galβ1 − 4Glc] |
| High metastatic potential cell line HCCLM3 | [GalNAcα1 − 3(Fucα1 − 2)Galβ1 − 3GlcNAcβ1 − 3Galβ1 − 4Glc], [Galβ1 − 3GlcNAcβ1 − 3Galβ1 − 3GlcNAcβ1 − 3Galβ1 − 4Glc], [Galα1 − 3(Fucα1 − 2)- Galβ1 − 4(Fucα1 − 3)GlcNAcβ1 − 3Galβ1 − 4Glc], [GalNAcα1 − 3(Fucα1 − 2)Galβ1 − 4(Fucα1 − 3)GlcNAcβ1 − 3Galβ1 − 4Glc], [NeuAcα2 − 3SulfGalβ1 − 4-(Fucα1 − 3)GlcNAcβ1 − 3Galβ1 − 4Glc] |
| Hepatocarcinoma cell line with low metastatic potential MHCC97L | [Galβ1 − 3GlcNAcβ1 − 3Galβ1 − 3GlcNAcβ1 − 3Galβ1 − 4Glc] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Novaconi, C.R.; Onulov, R.; Serb, A.F.; Sisu, E.; Dinca, N.; Pascariu, M.-C.; Georgescu, M. Assessing Glycosphingolipid Profiles in Human Health and Disease Using Non-Imaging MALDI Mass Spectrometry. Appl. Sci. 2023, 13, 9922. https://doi.org/10.3390/app13179922
Novaconi CR, Onulov R, Serb AF, Sisu E, Dinca N, Pascariu M-C, Georgescu M. Assessing Glycosphingolipid Profiles in Human Health and Disease Using Non-Imaging MALDI Mass Spectrometry. Applied Sciences. 2023; 13(17):9922. https://doi.org/10.3390/app13179922
Chicago/Turabian StyleNovaconi, Cristina Ramona, Robert Onulov, Alina Florina Serb, Eugen Sisu, Nicolae Dinca, Mihai-Cosmin Pascariu, and Marius Georgescu. 2023. "Assessing Glycosphingolipid Profiles in Human Health and Disease Using Non-Imaging MALDI Mass Spectrometry" Applied Sciences 13, no. 17: 9922. https://doi.org/10.3390/app13179922
APA StyleNovaconi, C. R., Onulov, R., Serb, A. F., Sisu, E., Dinca, N., Pascariu, M.-C., & Georgescu, M. (2023). Assessing Glycosphingolipid Profiles in Human Health and Disease Using Non-Imaging MALDI Mass Spectrometry. Applied Sciences, 13(17), 9922. https://doi.org/10.3390/app13179922





.png)

