Discrimination of Healthy and Cancerous Colon Cells Based on FTIR Spectroscopy and Machine Learning Algorithms
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture and Preparation
2.2. FTIR Measurements
2.3. Spectra Analysis
3. Results and Discussion
- ✓
- CN2-RI: ordered rules, exclusive covering, entropy evaluation with beam width equal to 5 for rule searching, minimum rule coverage of one, and maximum rule length equal to 5;
- ✓
- LR: non-regularization type;
- ✓
- CT: a binary tree, with minimum two samples per leaf; subsets were not split if they contained fewer than five samples and the maximal tree depth was equal to 100;
- ✓
- SVM: radial basis function (RBF) kernel, SVM with cost 1.0 and regression loss epsilon 0.1, tolerance 0.001, and maximum 100 iterations;
- ✓
- kNN: the number of neighbours equal to four for LWR and two for HWr, by using an Euclidean metric and weights by distances;
- ✓
- NN: 95 neurons in the hidden layer, ReLu activation, Adam solver, and 300 maximum iterations.
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Available online: https://www.who.int/news-room/fact-sheets/detail/cancer (accessed on 31 July 2023).
- Nierengarten, M.B. Colonoscopy remains the gold standard for screening despite recent tarnish. Cancer 2023, 129, 330–331. [Google Scholar] [CrossRef] [PubMed]
- Rashmi, R.; Prasad, K.; Udupa, C.B.K. Breast histopathological image analysis using image processing techniques for diagnostic purposes: A methodological review. J. Med. Syst. 2022, 46, 7. [Google Scholar] [CrossRef] [PubMed]
- Baker, M.; Trevisan, J.; Bassan, P.; Bhargava, R.; Butler, H.J.; Dorling, K.M.; Fielden, P.R.; Fogarty, S.W.; Fullwood, N.J.; Martin, F.L.; et al. Using Fourier transform IR spectroscopy to analyze biological materials. Nat. Protoc. 2014, 9, 1771–1791. [Google Scholar] [CrossRef] [PubMed]
- Errico, S.; Moggio, M.; Diano, N.; Portaccio, M.; Lepore, M. Different experimental approaches for Fourier-transform infrared spectroscopy applications in biology and biotechnology: A selected choice of representative results. Biotechnol. Appl. Biochem. 2022, 70, 937–961. [Google Scholar] [CrossRef]
- De Bruyne, S.; Speeckaert, M.M.; Delanghe, J.R. Applications of mid-infrared spectroscopy in the clinical laboratory setting. Crit. Rev. Clin. Lab. Sci. 2018, 55, 1. [Google Scholar] [CrossRef]
- Gardner, P.; Lyang, F.; Gazi, E.; Moss, D. (Eds.) Preparation of Tissues and Cells for Infrared and Raman Spectroscopy and Imaging. In Synchrotron Radiation Infrared Microscopy: A Practical Approach, 1st ed.; Royal Society of Chemistry: London, UK, 2010; pp. 145–191. [Google Scholar]
- Song, C.L.; Kazarian, S.G. Micro ATR-FTIR spectroscopic imaging of colon biopsies with a large area Ge crystal. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2020, 228, 117695. [Google Scholar] [CrossRef]
- Kaznowska, E.; Depciuch, J.; Szmuc, K.; Cebulski, J. Use of FTIR spectroscopy and PCA-LDC analysis to identify cancerous lesions within the human colon. J. Pharm. Biomed. Anal. 2017, 134, 259–268. [Google Scholar] [CrossRef]
- Tiwari, S.; Falahkheirkhah, K.; Cheng, G.; Bhargava, R. Colon Cancer Grading Using Infrared Spectroscopic Imaging-Based Deep Learning. Appl. Spectrosc. 2022, 76, 475–484. [Google Scholar] [CrossRef]
- Muniz, F.B.; de Freitas Oliveira Baffa, M.; Garcia, S.B.; Bachmann, L.; Felipe, J.C. Histopathological diagnosis of colon cancer using micro-FTIR hyperspectral imaging and deep learning. Comput. Methods Programs Biomed. 2023, 231, 107388. [Google Scholar] [CrossRef]
- Piva, J.A.D.A.C.; Silva, J.L.R.; Raniero, L.J.; Lima, C.S.P.; Arisawa, E.A.L.; Oliveira, C.D.; Canevari, R.D.A.; Ferreira, J.; Martin, A.A. Biochemical imaging of normal, adenoma, and colorectal adenocarcinoma tissues by fourier transform infrared spectroscopy (FTIR) and morphological correlation by histopathological analysis: Preliminary results. Rev. Bras. Eng. Biomed. 2015, 31, 10–18. [Google Scholar] [CrossRef]
- Khanmohammadi, M.; Garmarudi, A.B.; Ghasemi, K.; Jaliseh, H.K.; Kaviani, A. Diagnosis of colon cancer by attenuated total reflectance-Fourier transform infrared microspectroscopy and soft independent modeling of class analogy. Med Oncol. 2009, 26, 292–297. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, Q.B.; Zhang, G.J.; Xu, Y.Z.; Sun, X.J.; Shi, J.S.; Zhang, Y.F.; Wu, J.G. Identification of colitis and cancer in colon biopsies by Fourier Transform Infrared spectroscopy and chemometrics. Sci. World J. 2012, 2012, 936149. [Google Scholar] [CrossRef] [PubMed]
- Inan Genç, A.; Gok, S.; Banerjee, S.; Severcan, F. Valdecoxib recovers the lipid composition, order and dynamics in colon cancer cell lines independent of COX-2 expression: An ATR-FTIR spectroscopy study. Appl. Spectrosc. 2017, 71, 105–117. [Google Scholar] [CrossRef] [PubMed]
- Perna, G.; Capozzi, V.; Lasalvia, M. Classification of Healthy and Cancer Colon Cells Grown on Glass Coverslip by Means of Fourier Transform Infrared Spectroscopy and Multivariate Methods. Photonics 2023, 10, 481. [Google Scholar] [CrossRef]
- Varmuza, K.; Filzmoser, P. Introduction to Multivariate Statistical Analysis in Chemometrics; CRC Press: Boca Raton, FL, USA, 2009. [Google Scholar]
- Gautam, R.; Vanga, S.; Ariese, F.; Umapathy, S. Review of multidimensional data processing approaches for Raman and infrared spectroscopy. EPJ Techn. Instrum. 2015, 2, 8. [Google Scholar] [CrossRef]
- Morais, C.L.M.; Lima, K.M.G.; Singh, M.; Martin, F.L. Tutorial: Multivariate classification for vibrational spectroscopy in biological samples. Nat. Protoc. 2020, 15, 2143–2162. [Google Scholar] [CrossRef]
- Guo, S.; Popp, J.; Bocklitz, T. Chemometric analysis in Raman spectroscopy from experimental design to machine learning–based modeling. Nat Protoc. 2021, 16, 5426–5459. [Google Scholar] [CrossRef]
- Demsar, J.; Curk, T.; Erjavec, A.; Gorup, C.; Hocevar, T.; Milutinovic, M.; Mozina, M.; Polajnar, M.; Toplak, M.; Staric, A.; et al. Orange: Data Mining Toolbox in Python. J. Mach. Learn. Res. 2013, 14, 2349–2353. [Google Scholar]
- Swe, S.M.; Sett, K.M. Approaching Rules Induction: CN2 Algorithm in Categorizing of Biodiversity. Int. J. Trend Sci. Res. Dev. 2019, 3, 1581–1584. [Google Scholar]
- Heymann, F.; Bessa, R.; Liebensteiner, M.; Parginos, K.; Hinojar, J.C.M.; Duenas, P. Scarcity events analysis in adequacy studies using CN2 rule mining. Energy AI 2022, 8, 100154. [Google Scholar] [CrossRef]
- Clark, P.; Boswell, R. Rule Induction with CN2: Some Recent Improvements. In Machine Learning, Proceedings of the Fifth European Conference (EWSL-91), Porto, Portugal, 6–8 March 1991; Springer: Berlin/Heidelberg, Germany, 1991; pp. 151–163. [Google Scholar]
- Asaduzzaman, S.; Ahmed, M.R.; Rehana, H.; Chakraborty, S.; Islam, M.S.; Bhuiyan, T. Machine learning to reveal an astute risk predictive framework for Gynecologic Cancer and its impact on women psychology: Bangladeshi perspective. BMC Bioinform. 2021, 22, 213. [Google Scholar] [CrossRef] [PubMed]
- Mencar, C.; Gallo, C.; Mantero, M.; Tarsia, P.; Carpagnano, G.E.; Foschino Barbaro, M.P.; Lacedonia, D. Application of machine learning to predict obstructive sleep apnea syndrome severity. Health Inform. J. 2020, 26, 298–317. [Google Scholar] [CrossRef] [PubMed]
- Negoiţă, C.; Praisler, M. Logistic regression classification model identifying drugs of abuse based on their ATR-FTIR spectra: Case study on LASSO and Ridge regularization methods. In Proceedings of the 2019 6th International Symposium on Electrical and Electronics Engineering (ISEEE), Galati, Romania, 18–20 October 2019; pp. 1–4. [Google Scholar]
- Arévalo, L.A.; Antonova, O.; O’Brien, S.A.; Singh, G.P.; Seifert, A. Detection of Alzheimer’s by machine learning-assisted vibrational spectroscopy in human cerebrospinal fluid. J. Phys. Conf. Ser. 2022, 2407, 012026. [Google Scholar] [CrossRef]
- Myles, A.J.; Feudale, R.N.; Liu, Y.; Woody, N.A.; Brown, S.D. An introduction to decision tree modeling. J. Chemom. 2004, 18, 275–285. [Google Scholar] [CrossRef]
- Li, H.; Wang, S.; Zeng, Q.; Chen, C.; Lv, X.; Ma, M.; Su, H.; Ma, B.; Chen, C.; Fang, J. Serum Raman spectroscopy combined with multiple classification models for rapid diagnosis of breast cancer. Photodiagnosis Photodyn. Ther. 2022, 40, 103115. [Google Scholar] [CrossRef] [PubMed]
- Cui, G.; Peng, W.; Liu, Y. Diagnosis of hepatocellular carcinoma by FTIR spectroscopy combined with classification tree—Proc. SPIE 11566. In Proceedings of the AOPC 2020: Optical Spectroscopy and Imaging; and Biomedical Optics, Beijing, China, 5 November 2020. [Google Scholar]
- Chen, Y.; Su, Y.; Ou, L.; Zou, C.; Chen, Z. Classification of nasopharyngeal cell lines (C666-1, CNE2, NP69) via Raman spectroscopy and decision tree. Vib. Spectrosc. 2015, 80, 24–29. [Google Scholar] [CrossRef]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Dawuti, W.; Zheng, X.; Liu, H.; Zhao, H.; Dou, J.; Sun, L.; Chu, J.; Lin, R.; Lü, G. Urine surface-enhanced Raman spectroscopy combined with SVM algorithm for rapid diagnosis of liver cirrhosis and hepatocellular carcinoma. Photodiagnosis Photodyn. Ther. 2022, 38, 102811. [Google Scholar] [CrossRef]
- Ali, S.; Naveed, A.; Hussain, I.; Qazi, J. Use of ATR-FTIR spectroscopy to differentiate between cirrhotic/non-cirrhotic HCV patients. Photodiagnosis Photodyn. Ther. 2023, 42, 103529. [Google Scholar] [CrossRef]
- Parlatan, U.; Inanc, M.T.; Ozgor, B.Y.; Oral, E.; Bastu, E.; Unlu, M.B.; Basar, G. Raman spectroscopy as a non-invasive diagnostic technique for endometriosis. Sci. Rep. 2019, 9, 19795. [Google Scholar] [CrossRef]
- Prakisya, N.P.T.; Liantoni, F.; Hatta, P.; Aristyagama, Y.H.; Setiawan, A. Utilization of K-nearest neighbor algorithm for classification of white blood cells in AML M4, M5, and M7. Open Eng. 2021, 11, 662–668. [Google Scholar] [CrossRef]
- Gallo, C. Artificial neural networks tutorial. In Encyclopedia of Information Science and Technology, 3rd ed.; IGI Global: Hershey, PA, USA, 2015; pp. 6369–6378. [Google Scholar]
- De Souza, N.M.P.; Machado, B.H.; Padoin, L.V.; Prá, D.; Fay, A.P.; de Arruda Tomaz, M.; Corbellini, V.A.; Rieger, A. Discrimination of molecular subtypes of breast cancer with ATR-FTIR spectroscopy in blood plasma coupled with partial least square-artificial neural network discriminant analysis (PLS-ANNDA). Chemom. Intell. Lab. Syst. 2023, 237, 104826. [Google Scholar] [CrossRef]
- Podshyvalov, A.; Sahu, R.K.; Mark, S.; Kantarovich, K.; Guterman, H.; Goldstein, J.; Jagannathan, R.; Argov, S.; Mordechai, S. Distinction of cervical cancer biopsies by use of infrared microspectroscopy and probabilistic neural networks. Appl. Opt. 2005, 44, 3725–3734. [Google Scholar] [CrossRef] [PubMed]
- Zeaiter, M.; Rutledge, D. Preprocessing methods. In Comprehensive Chemometrics: Chemical and Biochemical Data Analysis; Brown, S.D., Tauler, R., Walczak, B., Eds.; Elsevier: Amsterdam, The Netherlands, 2009; Volume 3, pp. 121–231. [Google Scholar]
- Talari, A.C.S.; Martinez, M.A.G.; Movasaghi, Z.; Rehman, S. Advances in Fourier transform infrared (FTIR) spectroscopy of biological tissues. Appl. Spectrosc. Rev. 2017, 52, 456–506. [Google Scholar] [CrossRef]
- Dong, L.; Sun, X.; Chao, Z.; Zhang, S.; Zheng, J.; Gurung, R.; Du, J.; Shi, J.; Xu, Y.; Zhang, Y.; et al. Evaluation of FTIR spectroscopy as diagnostic tool for colorectal cancer using spectral analysis. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2014, 122, 288–294. [Google Scholar] [CrossRef]
- De Santis, S.; Porcelli, F.; Sotgiu, G.; Crescenzi, A.; Ceccucci, A.; Verri, M.; Caricato, M.; Taffon, C.; Orsini, M. Identification of remodeled collagen fibers in tumor stroma by FTIR Micro-spectroscopy: A new approach to recognize the colon carcinoma. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 2022, 1868, 166279. [Google Scholar] [CrossRef]
- Brozek-Pluska, B. Statistics assisted analysis of Raman spectra and imaging of human colon cell lines—Label free, spectroscopic diagnostics of colorectal cancer. J. Mol. Struct. 2020, 1218, 128524. [Google Scholar] [CrossRef]
- Jia-Wei, T.; Qing-Hua, L.; Xiao-Cong, Y.; Ya-Cheng, P.; Peng-Bo, W.; Xin, L.; Xing-Xing, K.; Bing, G.; Zuo-Bin, Z.; Liang, W. Comparative Analysis of Machine Learning Algorithms on Surface Enhanced Raman Spectra of Clinical Staphylococcus Species. Front. Microbiol. 2021, 12, 696921. [Google Scholar]
- Fernández-Manteca, M.G.; Ocampo-Sosa, A.A.; de Alegría-Puig, C.R.; Roiz, M.P.; Rodríguez-Grande, J.; Madrazo, F.; Calvo, J.; Rodríguez-Cobo, L.; López-Higuera, J.M.; Fariñas, M.C.; et al. Automatic classification of Candida species using Raman spectroscopy and machine learning. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2023, 290, 122270. [Google Scholar] [CrossRef]
- Caixeta, D.C.; Carneiro, M.G.; Rodrigues, R.; Alves, D.C.T.; Goulart, L.R.; Cunha, T.M.; Espindola, F.S.; Vitorino, R.; Sabino-Silva, R. Salivary ATR-FTIR Spectroscopy Coupled with Support Vector Machine Classification for Screening of Type 2 Diabetes Mellitus. Diagnostics 2023, 13, 1396. [Google Scholar] [CrossRef]
- Uthamacumaran, A.; Elouatik, S.; Abdouh, M.; Berteau-Rainville, M.; Gao, Z.H.; Arena, G. Machine learning characterization of cancer patients-derived extracellular vesicles using vibrational spectroscopies: Results from a pilot study. Appl. Intell. 2022, 52, 12737–12753. [Google Scholar] [CrossRef]
- Du, Y.; Xie, F.; Wu, G.; Chen, P.; Yang, Y.; Yang, L.; Yin, L.; Wang, S. A classification model for detection of ductal carcinoma in situ by Fourier transform infrared spectroscopy based on deep structured semantic model. Anal. Chim. Acta 2023, 1251, 340991. [Google Scholar] [CrossRef]
- Zheng, Q.; Li, J.; Yang, L.; Zheng, B.; Wang, J.; Lv, N.; Luo, J.; Martin, F.L.; Liu, D.; He, J. Raman spectroscopy as a potential diagnostic tool to analyse biochemical alterations in lung cancer. Analyst 2019, 145, 385–392. [Google Scholar] [CrossRef] [PubMed]
- Ojala, M.; Garriga, G.C. Permutation tests for studying classifier performance. J. Mach. Learn. Res. 2010, 11, 6. [Google Scholar]
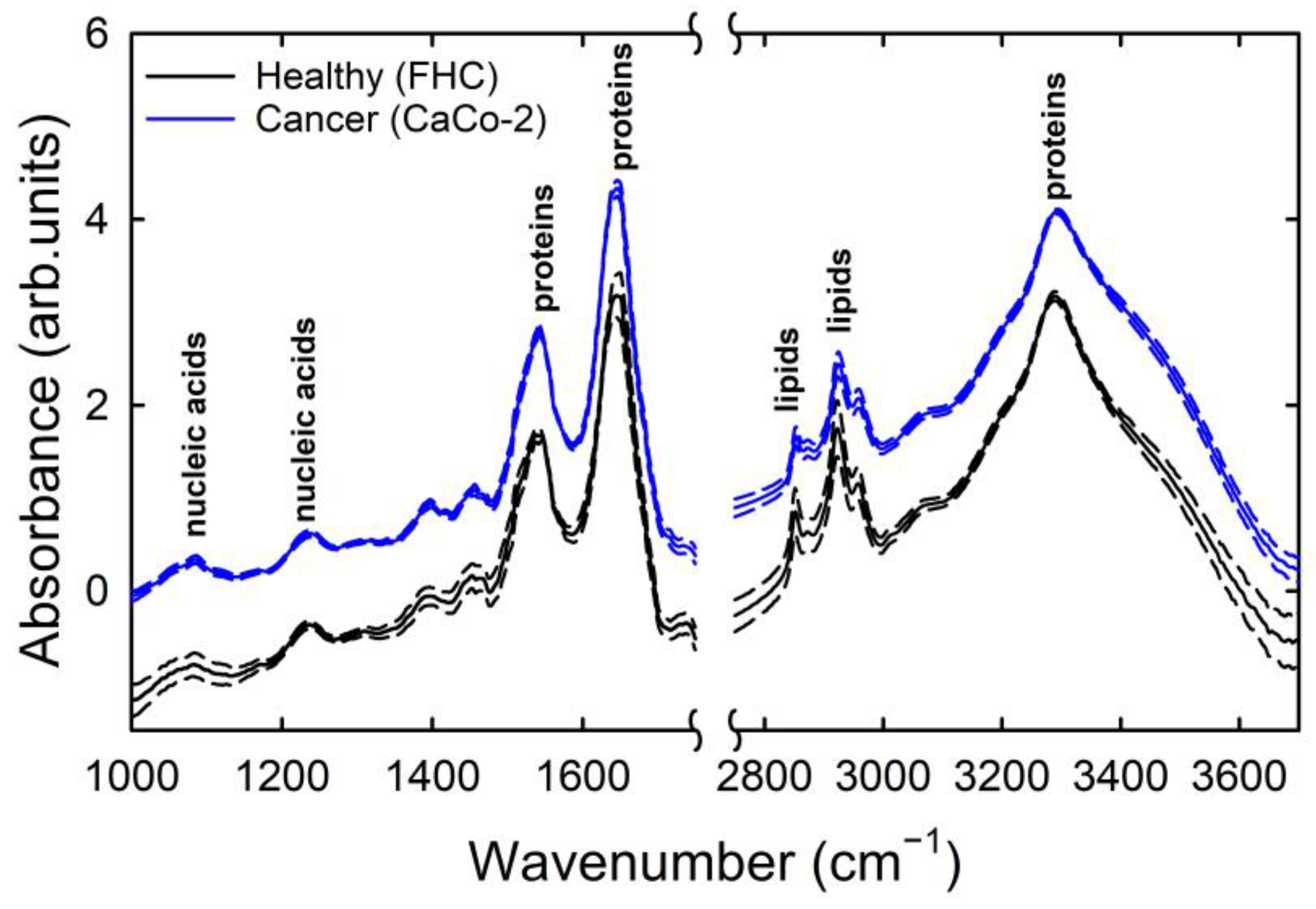
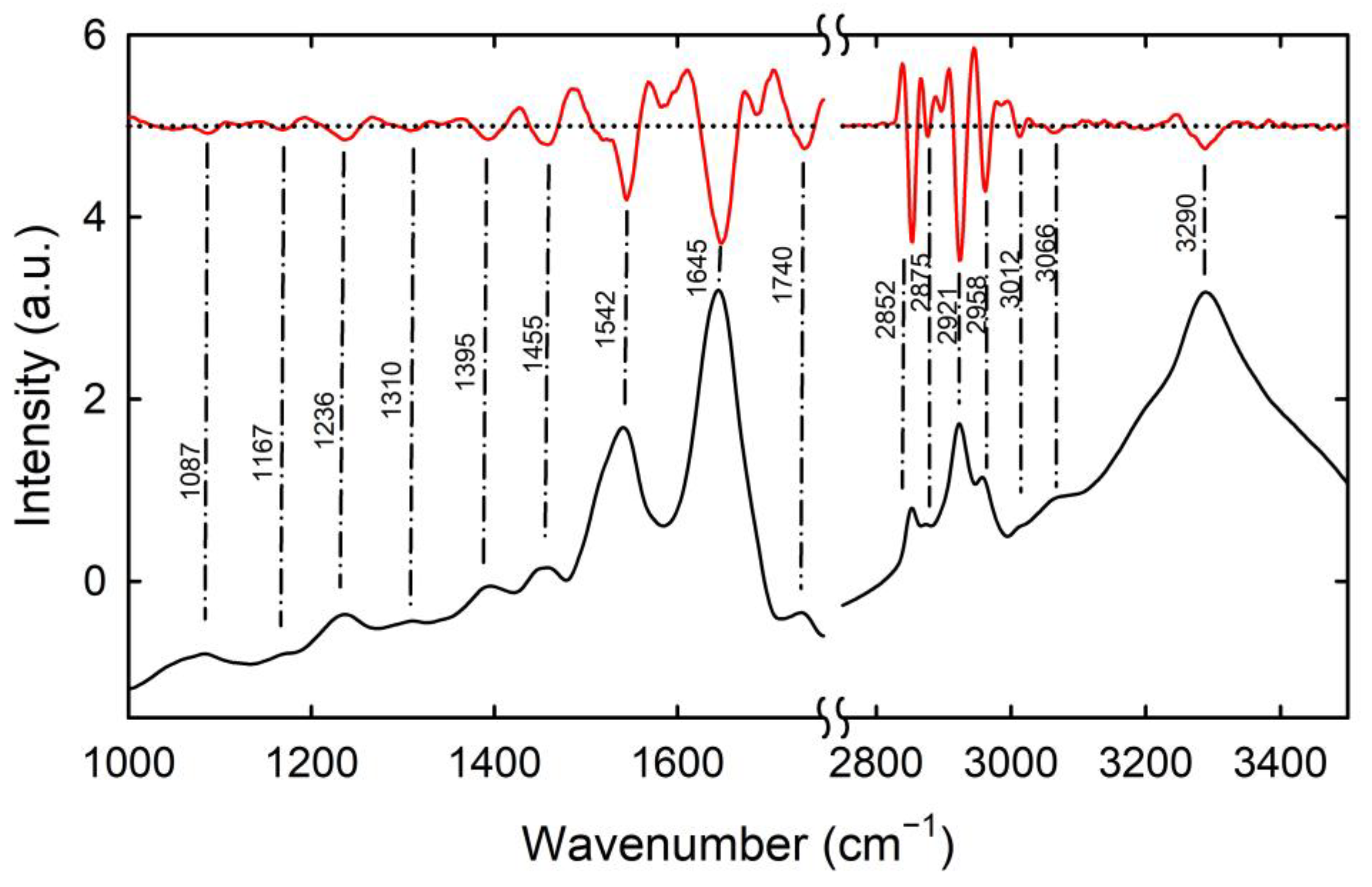
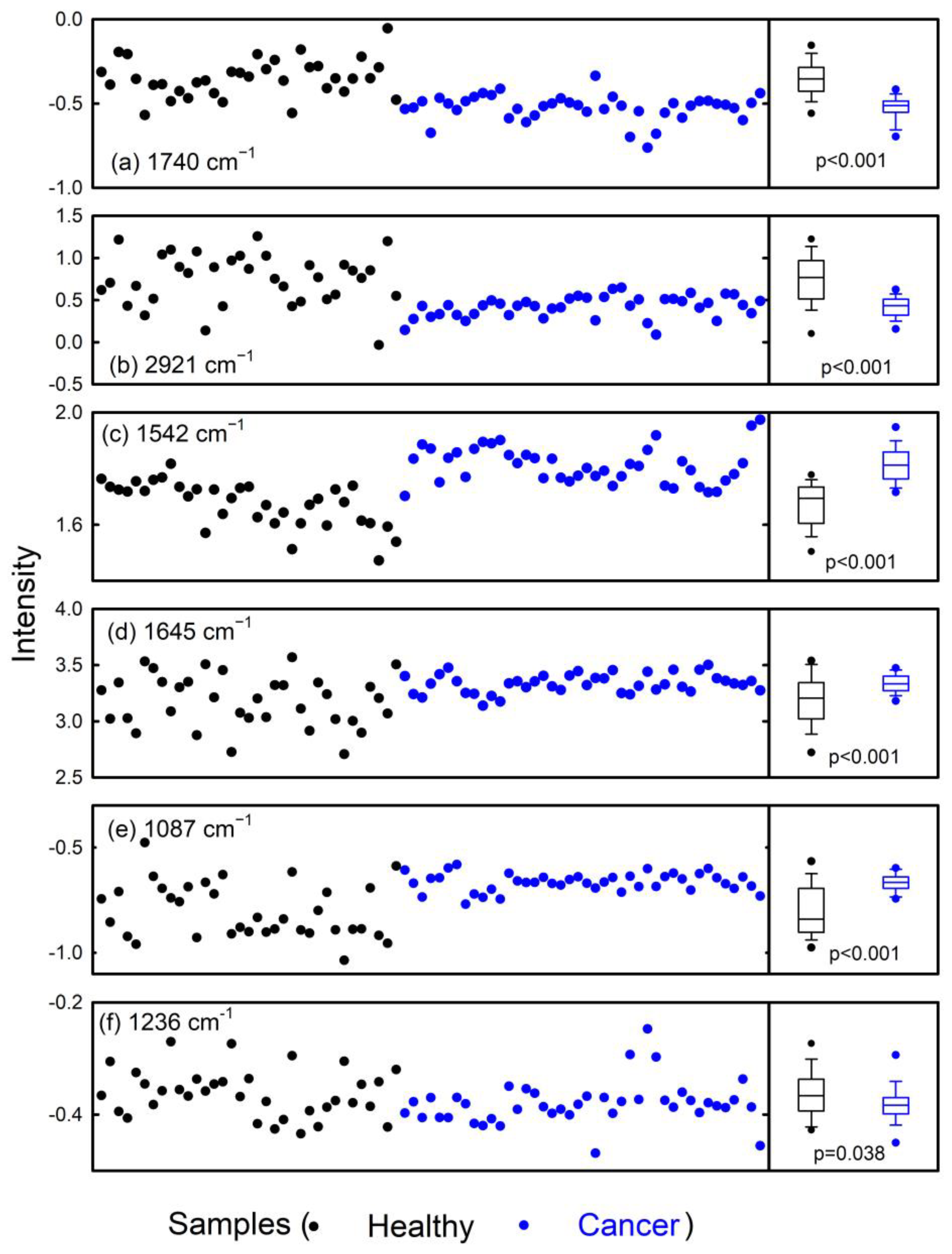
| Spectral Position (cm−1) | Assignment |
|---|---|
| 1087 | symmetric PO2− stretching of nucleic acids |
| 1167 | C-OH stretching of proteins |
| 1236 | asymmetric PO2− stretching of nucleic acids |
| 1310 | amide III of proteins |
| 1395 | COO− stretching of proteins and lipids |
| 1455 | CH3 bending of proteins and lipids |
| 1542 | amide II of proteins |
| 1645 | amide I of proteins |
| 1740 | C=O stretching of lipids |
| 2852 | symmetric CH2 stretching of lipids |
| 2875 | symmetric CH3 stretching of proteins and lipids |
| 2921 | asymmetric CH2 stretching of lipids |
| 2958 | asymmetric CH3 stretching of proteins and lipids |
| 3012 | CH stretching of lipids |
| 3066 | N-H stretching of amide B |
| 3290 | N-H stretching of amide A |
| Algorithm (Original Data) | Accuracy LWR (%) | Accuracy HWR (%) | Sensitivity LWR (%) | Sensitivity HWR (%) | Specificity LWR (%) | Specificity HWR (%) |
|---|---|---|---|---|---|---|
| kNN | 90.9 | 97.7 | 97.6 | 100.0 | 82.9 | 94.3 |
| LR | 94.8 | 93.5 | 95.2 | 95.2 | 94.3 | 91.4 |
| CT | 87.0 | 94.8 | 88.1 | 95.2 | 85.7 | 94.3 |
| CN2-RI | 90.9 | 89.6 | 95.2 | 95.2 | 85.7 | 82.9 |
| SVM | 100.0 | 97.4 | 100.0 | 97.6 | 100.0 | 97.1 |
| NN | 98.7 | 98.7 | 100.0 | 100.0 | 100.0 | 97.1 |
| Algorithm (Randomized Data) | Accuracy LWR (%) | Accuracy HWR (%) | Sensitivity LWR (%) | Sensitivity HWR (%) | Specificity LWR (%) | Specificity HWR (%) |
|---|---|---|---|---|---|---|
| kNN | 45.5 | 51.9 | 57.1 | 52.4 | 31.4 | 51.4 |
| LR | 54.5 | 50.6 | 64.3 | 64.3 | 42.9 | 34.3 |
| CT | 51.9 | 53.2 | 61.9 | 59.5 | 40.0 | 45.7 |
| CN2-RI | 49.4 | 46.8 | 54.8 | 50.0 | 42.9 | 42.9 |
| SVM | 51.9 | 54.5 | 73.8 | 78.6 | 25.7 | 25.7 |
| NN | 50.6 | 57.1 | 50.0 | 64.3 | 51.4 | 48.6 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lasalvia, M.; Gallo, C.; Capozzi, V.; Perna, G. Discrimination of Healthy and Cancerous Colon Cells Based on FTIR Spectroscopy and Machine Learning Algorithms. Appl. Sci. 2023, 13, 10325. https://doi.org/10.3390/app131810325
Lasalvia M, Gallo C, Capozzi V, Perna G. Discrimination of Healthy and Cancerous Colon Cells Based on FTIR Spectroscopy and Machine Learning Algorithms. Applied Sciences. 2023; 13(18):10325. https://doi.org/10.3390/app131810325
Chicago/Turabian StyleLasalvia, Maria, Crescenzio Gallo, Vito Capozzi, and Giuseppe Perna. 2023. "Discrimination of Healthy and Cancerous Colon Cells Based on FTIR Spectroscopy and Machine Learning Algorithms" Applied Sciences 13, no. 18: 10325. https://doi.org/10.3390/app131810325
APA StyleLasalvia, M., Gallo, C., Capozzi, V., & Perna, G. (2023). Discrimination of Healthy and Cancerous Colon Cells Based on FTIR Spectroscopy and Machine Learning Algorithms. Applied Sciences, 13(18), 10325. https://doi.org/10.3390/app131810325









