MDAU-Net: A Liver and Liver Tumor Segmentation Method Combining an Attention Mechanism and Multi-Scale Features
Abstract
:1. Introduction
- We redesigned the jump connection and introduced a double-flow linear pooling enhancement unit (DLE) to improve the interaction ability between deep and shallow features, which helped to narrow the semantic gap.
- To better realize the extraction and reuse of useful features, we proposed a cascaded adaptive feature extraction unit (CAE) as a substitute for the bottleneck layer. It was based on an multi-head attention mechanism and a series of dense connections.
- We designed a cross-level information interaction mechanism (CII). It used bidirectional residual connections and was placed in the skip connection to overcome the problem of forgetting a priori knowledge in the learning process.
- We proposed a residual encoder to bolster the preservation of original features and supply additional initial information for the segmentation task.
2. Related Works
2.1. Medical Image Segmentation Methods
2.2. Atrous Spatial Pyramid Pooling
2.3. Multi-Head Attention Mechanism
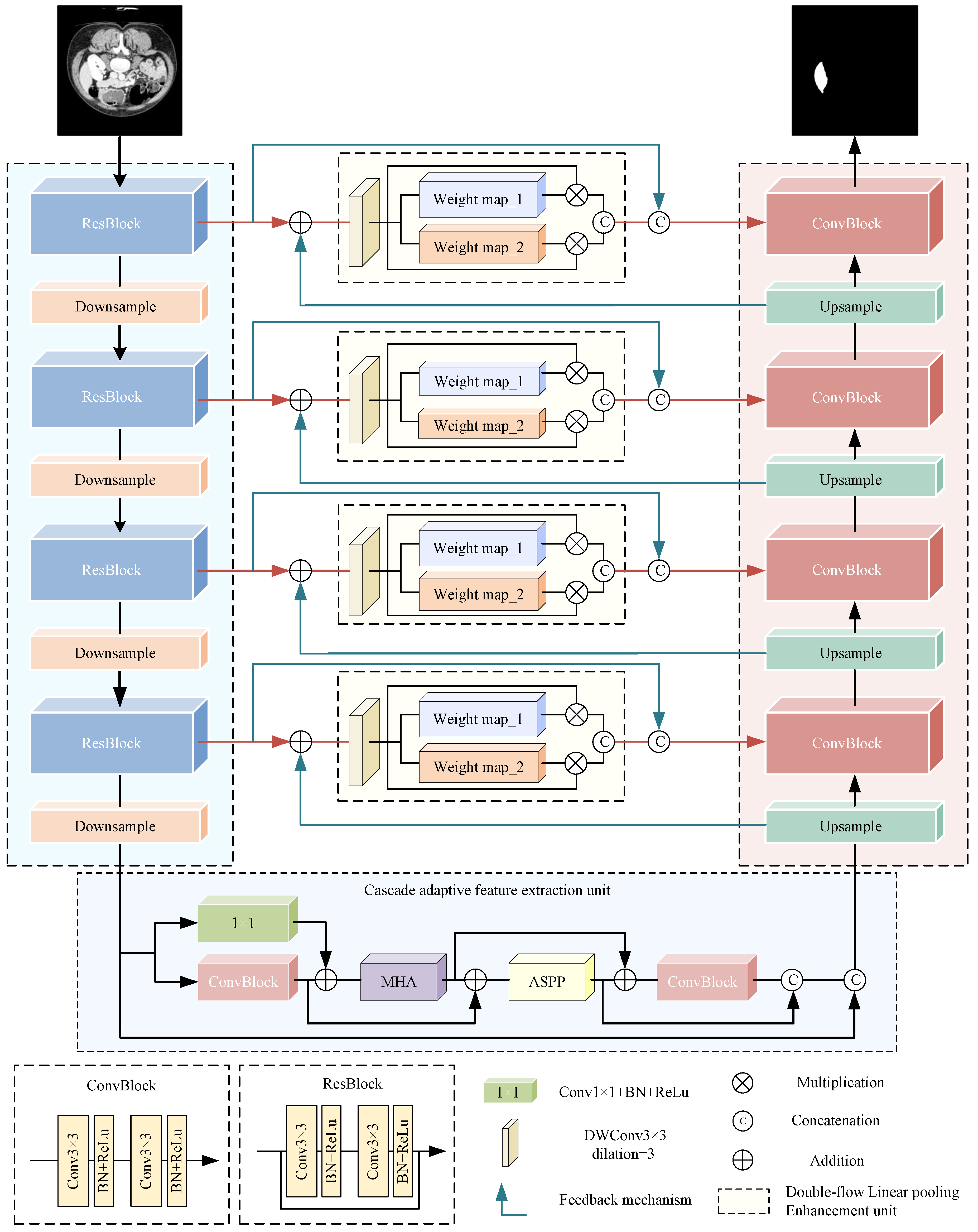
3. Proposed Method
3.1. Overall Architecture
| Algorithm 1: MDAU-Net |
| Data: Dataset X, mask L, module parameters Result: Segmentation result Y 1 for to N do 2 Preprocessing and enhancement of image . 3 for to 4 do 4 Encode as using ResBlock and MaxPooling. 5 Obtain the feature map for each encoder layer. 6 end 7 Adaptive feature extraction by CAE module, obtain . 8 for to 4 do 9 Calculate the DLE by and , obtain the feature map . 10 Decode as using bilinear interpolation and ConvBlock. 11 Obtain the feature map for each decoder layer. 12 Obtain the segmentation result of image as . 13 end 14 end 15 Output the segmentation result . |
3.2. Residual Encoder
3.3. Cascaded Adaptive Feature Extraction Unit
3.4. Double-Flow Linear Pooling Enhancement Unit
3.5. Cross-Level Information Interaction
4. Results
4.1. Implementation Details
4.1.1. Dataset
4.1.2. Data Preprocessing and Enhancement
4.1.3. Loss Function
4.1.4. Evaluation Metrics
- Dice coefficient (Dice)
- Precision
- Recall
- Volumetric overlap error (VOE)
- Relative volume error (RVD)
4.2. Loss Function Comparison Experiment
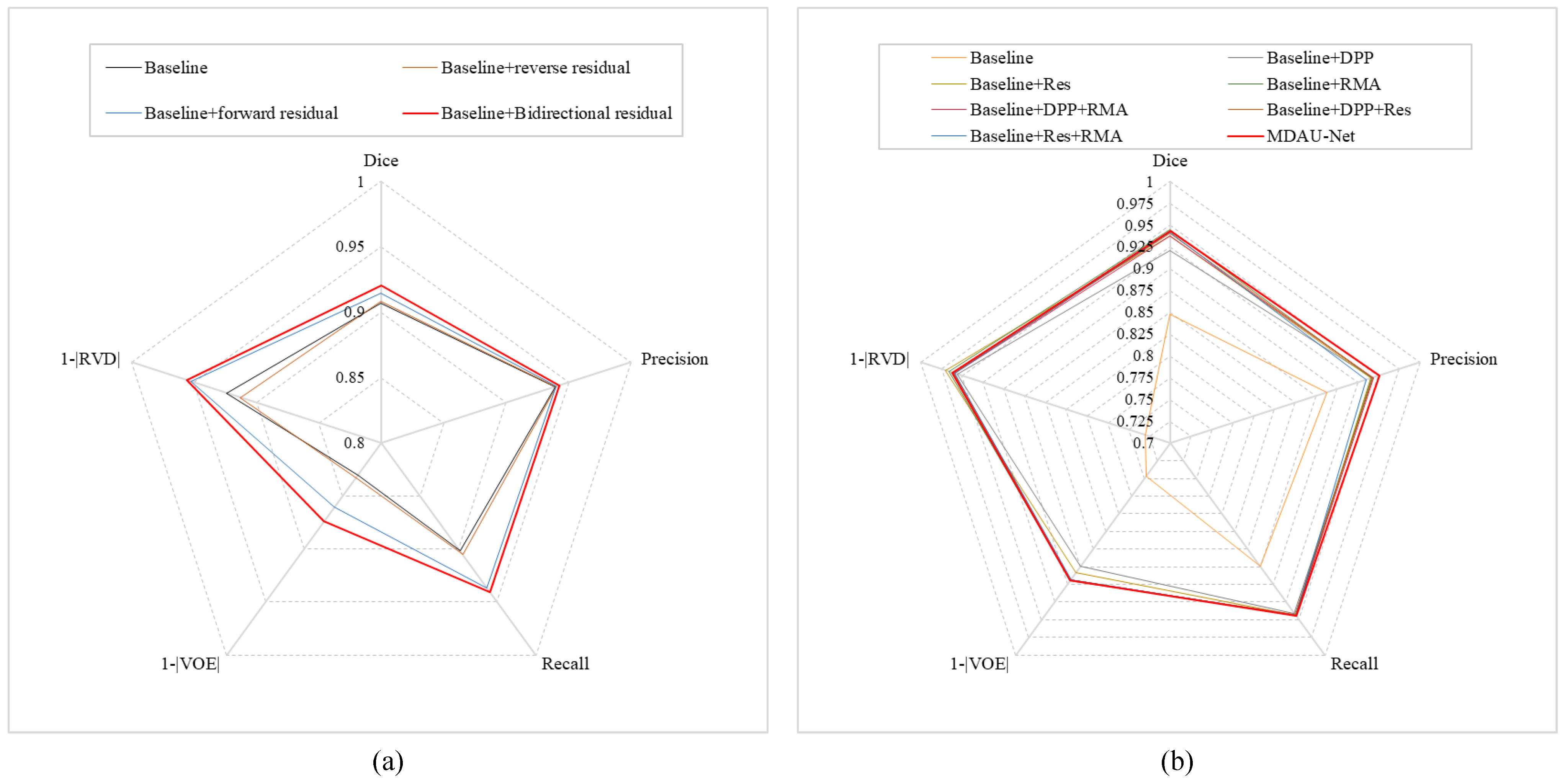
4.3. Validity Experiment of Cross-Level Information Interaction
4.4. Ablation Results
5. Discussion
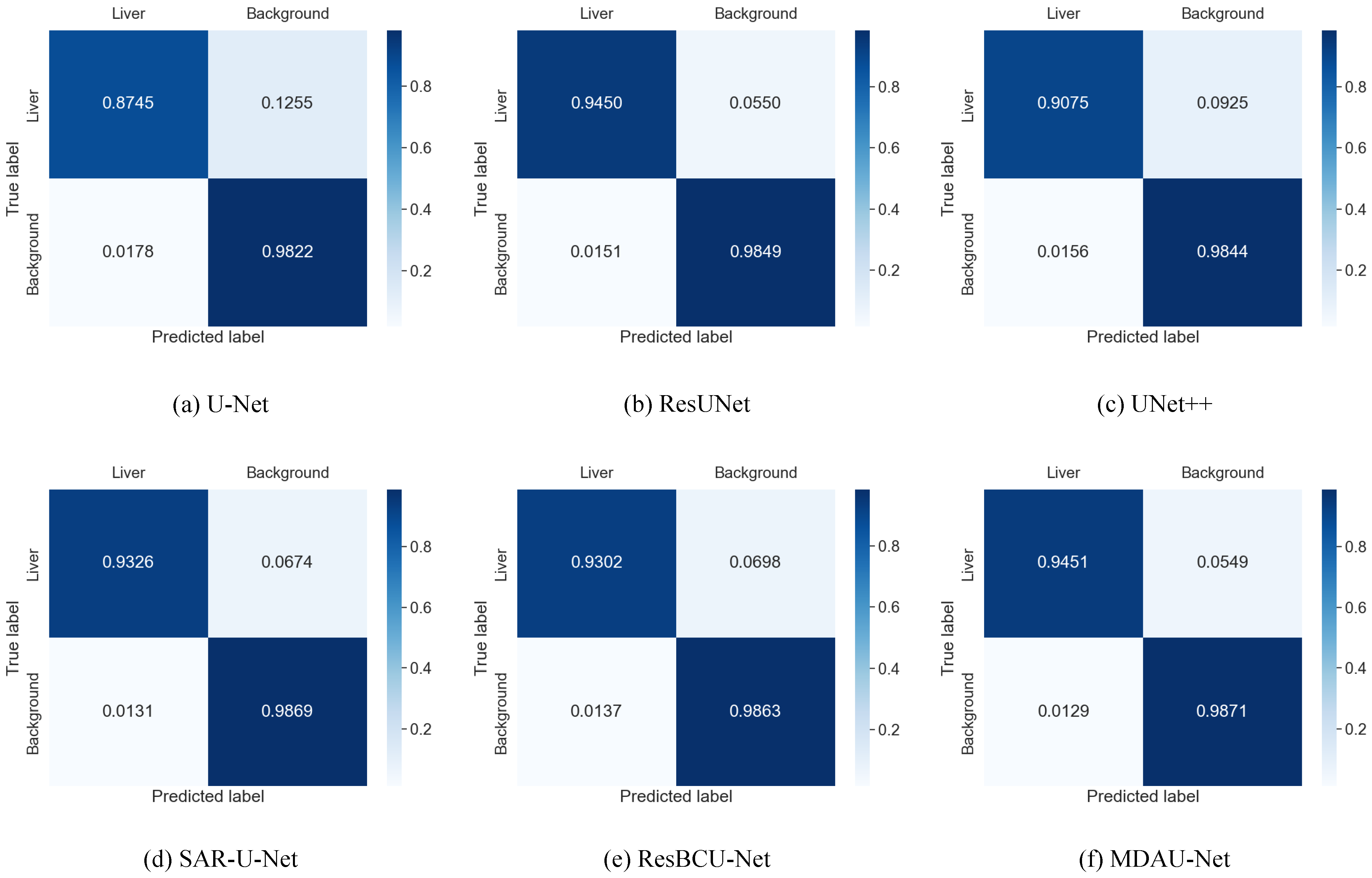
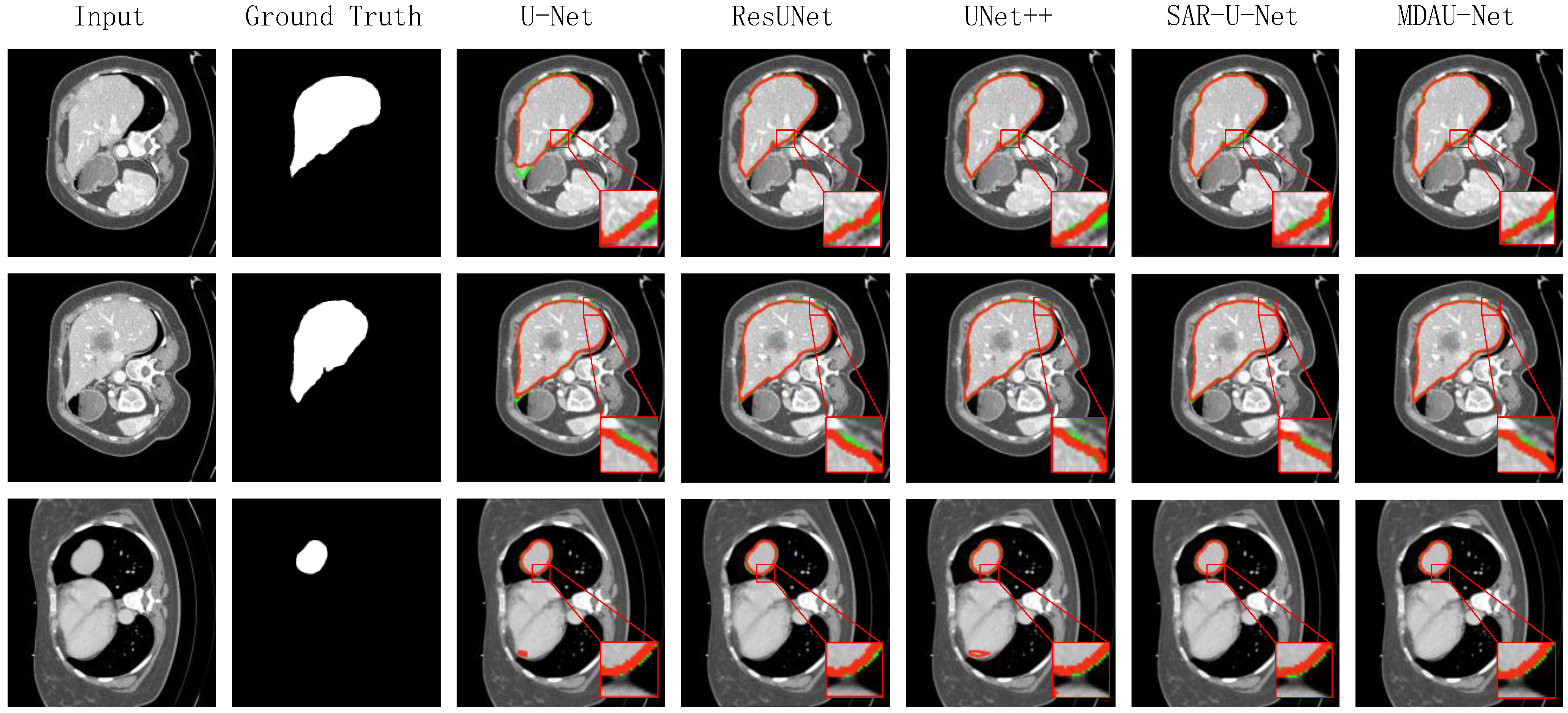
5.1. Quantitative Analysis of Liver Segmentation
5.2. Quantitative Analysis of Liver Tumor Segmentation
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hinton, G.E.; Osindero, S.; Teh, Y.W. A fast learning algorithm for deep belief nets. Neural Comput. 2006, 18, 1527–1554. [Google Scholar] [CrossRef] [PubMed]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, Munich, Germany, 5–9 October 2015, Proceedings, Part III 18; Springer: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar]
- Dickson, J.; Lincely, A.; Nineta, A. A Dual Channel Multiscale Convolution U-Net Methodfor Liver Tumor Segmentation from Abdomen CT Images. In Proceedings of the 2022 International Conference on Sustainable Computing and Data Communication Systems (ICSCDS), Erode, India, 7–9 April 2022; pp. 1624–1628. [Google Scholar]
- Sabir, M.W.; Khan, Z.; Saad, N.M.; Khan, D.M.; Al-Khasawneh, M.A.; Perveen, K.; Qayyum, A.; Azhar Ali, S.S. Segmentation of Liver Tumor in CT Scan Using ResU-Net. Appl. Sci. 2022, 12, 8650. [Google Scholar]
- Deng, Y.; Hou, Y.; Yan, J.; Zeng, D. ELU-net: An efficient and lightweight U-net for medical image segmentation. IEEE Access 2022, 10, 35932–35941. [Google Scholar]
- Seong, W.; Kim, J.H.; Kim, E.J.; Park, J.W. Segmentation of abnormal liver using adaptive threshold in abdominal CT images. In Proceedings of the IEEE Nuclear Science Symposuim & Medical Imaging Conference, Knoxville, TN, USA, 30 October–6 November 2010; pp. 2372–2375. [Google Scholar]
- Chen, Y.; Wang, Z.; Zhao, W.; Yang, X. Liver segmentation from CT images based on region growing method. In Proceedings of the 2009 3rd International Conference on Bioinformatics and Biomedical Engineering, Beijing, China, 11–13 June 2009; pp. 1–4. [Google Scholar]
- Gambino, O.; Vitabile, S.; Re, G.L.; La Tona, G.; Librizzi, S.; Pirrone, R.; Ardizzone, E.; Midiri, M. Automatic volumetric liver segmentation using texture based region growing. In Proceedings of the 2010 International Conference on Complex, Intelligent and Software Intensive Systems, Krakow, Poland, 15–18 February 2010; pp. 146–152. [Google Scholar]
- Okada, T.; Shimada, R.; Hori, M.; Nakamoto, M.; Chen, Y.W.; Nakamura, H.; Sato, Y. Automated segmentation of the liver from 3D CT images using probabilistic atlas and multilevel statistical shape model. Acad. Radiol. 2008, 15, 1390–1403. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Rahman Siddiquee, M.M.; Tajbakhsh, N.; Liang, J. Unet++: A nested u-net architecture for medical image segmentation. In Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support: 4th International Workshop, DLMIA 2018, and 8th International Workshop, ML-CDS 2018, Held in Conjunction with MICCAI 2018, Granada, Spain, 20 September 2018, Proceedings 4; Springer: Cham, Switzerland, 2018; pp. 3–11. [Google Scholar]
- Huang, H.; Lin, L.; Tong, R.; Hu, H.; Zhang, Q.; Iwamoto, Y.; Han, X.; Chen, Y.W.; Wu, J. Unet 3+: A full-scale connected unet for medical image segmentation. In Proceedings of the ICASSP 2020-2020 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Barcelona, Spain, 4–8 May 2020; pp. 1055–1059. [Google Scholar]
- Bi, R.; Ji, C.; Yang, Z.; Qiao, M.; Lv, P.; Wang, H. Residual based attention-Unet combing DAC and RMP modules for automatic liver tumor segmentation in CT. Math. Biosci. Eng. 2022, 19, 4703–4718. [Google Scholar] [PubMed]
- Kushnure, D.T.; Talbar, S.N. HFRU-Net: High-level feature fusion and recalibration unet for automatic liver and tumor segmentation in CT images. Comput. Methods Programs Biomed. 2022, 213, 106501. [Google Scholar]
- Zhou, Y.; Kong, Q.; Zhu, Y.; Su, Z. MCFA-UNet: Multiscale cascaded feature attention U-Net for liver segmentation. IRBM 2023, 44, 100789. [Google Scholar] [CrossRef]
- Chen, L.C.; Zhu, Y.; Papandreou, G.; Schroff, F.; Adam, H. Encoder-decoder with atrous separable convolution for semantic image segmentation. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 801–818. [Google Scholar]
- Meng, T.; Ghiasi, G.; Mahjorian, R.; Le, Q.V.; Tan, M. Revisiting Multi-Scale Feature Fusion for Semantic Segmentation. arXiv 2022, arXiv:2203.12683. [Google Scholar]
- Zhang, D.; Zhang, H.; Tang, J.; Wang, M.; Hua, X.; Sun, Q. Feature pyramid transformer. In Computer Vision–ECCV 2020: 16th European Conference, Glasgow, UK, 23–28 August 2020, Proceedings, Part XXVIII 16; Springer: Cham, Switzerland, 2020; pp. 323–339. [Google Scholar]
- Chen, L.C.; Papandreou, G.; Schroff, F.; Adam, H. Rethinking atrous convolution for semantic image segmentation. arXiv 2017, arXiv:1706.05587. [Google Scholar]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-excitation networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 7132–7141. [Google Scholar]
- Woo, S.; Park, J.; Lee, J.Y.; Kweon, I.S. Cbam: Convolutional block attention module. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 3–19. [Google Scholar]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is all you need. arXiv 2017, arXiv:1706.03762. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- Salehi, S.S.M.; Erdogmus, D.; Gholipour, A. Tversky loss function for image segmentation using 3D fully convolutional deep networks. In Machine Learning in Medical Imaging: 8th International Workshop, MLMI 2017, Held in Conjunction with MICCAI 2017, Quebec City, QC, Canada, 10 September 2017, Proceedings 8; Springer: Cham, Switzerland, 2017; pp. 379–387. [Google Scholar]
- Milletari, F.; Navab, N.; Ahmadi, S.A. V-net: Fully convolutional neural networks for volumetric medical image segmentation. In Proceedings of the 2016 fourth international conference on 3D vision (3DV), Stanford, CA, USA, 25–28 October 2016; pp. 565–571. [Google Scholar]
- Lin, T.Y.; Goyal, P.; Girshick, R.; He, K.; Dollár, P. Focal loss for dense object detection. In Proceedings of theIEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 2980–2988. [Google Scholar]
- Alom, M.Z.; Hasan, M.; Yakopcic, C.; Taha, T.M.; Asari, V.K. Recurrent residual convolutional neural network based on u-net (r2u-net) for medical image segmentation. arXiv 2018, arXiv:1802.06955. [Google Scholar]
- Xiao, X.; Lian, S.; Luo, Z.; Li, S. Weighted res-unet for high-quality retina vessel segmentation. In Proceedings of the 2018 9th International Conference on Information Technology in Medicine and Education (ITME), Hangzhou, China, 19–21 October 2018; pp. 327–331. [Google Scholar]
- Oktay, O.; Schlemper, J.; Folgoc, L.L.; Lee, M.; Heinrich, M.; Misawa, K.; Mori, K.; McDonagh, S.; Hammerla, N.Y.; Kainz, B.; et al. Attention u-net: Learning where to look for the pancreas. arXiv 2018, arXiv:1804.03999. [Google Scholar]
- Wang, J.; Lv, P.; Wang, H.; Shi, C. SAR-U-Net: Squeeze-and-excitation block and atrous spatial pyramid pooling based residual U-Net for automatic liver segmentation in Computed Tomography. Comput. Methods Programs Biomed. 2021, 208, 106268. [Google Scholar] [CrossRef]
- Badshah, N.; Ahmad, A. ResBCU-net: Deep learning approach for segmentation of skin images. Biomed. Signal Process. Control 2022, 71, 103137. [Google Scholar] [CrossRef]
- Khan, R.A.; Luo, Y.; Wu, F.X. RMS-UNet: Residual multi-scale UNet for liver and lesion segmentation. Artif. Intell. Med. 2022, 124, 102231. [Google Scholar] [CrossRef]
- Ge, R.; Cai, H.; Yuan, X.; Qin, F.; Huang, Y.; Wang, P.; Lyu, L. MD-UNET: Multi-input dilated U-shape neural network for segmentation of bladder cancer. Comput. Biol. Chem. 2021, 93, 107510. [Google Scholar] [CrossRef]











| Loss | Dice | Precision | Recall | VOE | RVD |
|---|---|---|---|---|---|
| Dice loss | 0.9420 | 0.9490 | 0.9393 | 0.1076 | 0.0205 |
| Focal loss | 0.9044 | 0.9662 | 0.9116 | 0.1745 | 0.1872 |
| Tversky loss | 0.9433 | 0.9515 | 0.9451 | 0.1053 | 0.0383 |
| BCE loss | 0.9328 | 0.9486 | 0.9396 | 0.1239 | 0.0189 |
| Method | Dice | Precision | Recall | VOE | RVD |
|---|---|---|---|---|---|
| Baseline | 0.9067 | 0.9392 | 0.9019 | 0.1694 | 0.0759 |
| Baseline + reverse residual | 0.9080 | 0.9399 | 0.9054 | 0.1674 | 0.0872 |
| Baseline + forward residual | 0.9145 | 0.9403 | 0.9366 | 0.1398 | 0.0478 |
| Baseline + bidirectional residual | 0.9204 | 0.9424 | 0.9408 | 0.1262 | 0.0446 |
| Method | Dice | Precision | Recall | VOE | RVD |
|---|---|---|---|---|---|
| Baseline | 0.8481 | 0.8879 | 0.8745 | 0.2536 | 0.2698 |
| Baseline + DLE | 0.9204 | 0.9424 | 0.9408 | 0.1262 | 0.0446 |
| Baseline + ResBlock | 0.9375 | 0.9409 | 0.9437 | 0.1161 | 0.0293 |
| Baseline + CAE | 0.9447 | 0.9422 | 0.9445 | 0.1064 | 0.0339 |
| Baseline + DLE + CAE | 0.9371 | 0.9437 | 0.9436 | 0.1062 | 0.0412 |
| Baseline + DAE + ResBlock | 0.9407 | 0.9425 | 0.9431 | 0.1056 | 0.0395 |
| Baseline + ResBlock + CAE | 0.9419 | 0.9354 | 0.9443 | 0.1070 | 0.0407 |
| MDAU-Net | 0.9433 | 0.9515 | 0.9451 | 0.1053 | 0.0383 |
| Method | Dice | Precision | Recall | VOE | RVD |
|---|---|---|---|---|---|
| U-Net | 0.8481 | 0.8879 | 0.8745 | 0.2536 | 0.2698 |
| RU-Net [27] | 0.8614 | 0.8902 | 0.8807 | 0.2415 | 0.2501 |
| ResUNet [28] | 0.9220 | 0.9263 | 0.9450 | 0.1427 | 0.0599 |
| Attention U-net [29] | 0.9197 | 0.9189 | 0.9236 | 0.1463 | 0.0575 |
| UNet++ [10] | 0.9106 | 0.9173 | 0.9075 | 0.1591 | 0.0818 |
| SAR-U-Net [30] | 0.9378 | 0.9504 | 0.9326 | 0.1142 | 0.0736 |
| ResBCU-Net [31] | 0.9359 | 0.9428 | 0.9302 | 0.1810 | 0.0587 |
| RMS-UNet [32] | 0.9171 | 0.9227 | 0.9157 | 0.1492 | 0.0646 |
| MD-UNET [33] | 0.9338 | 0.9433 | 0.9331 | 0.1224 | 0.0604 |
| MDAU-Net (our model) | 0.9433 | 0.9515 | 0.9451 | 0.1053 | 0.0383 |
| Method | Dice | Precision | Recall | VOE | RVD |
|---|---|---|---|---|---|
| U-Net | 0.8568 | 0.9606 | 0.9588 | 0.1501 | 0.1619 |
| RU-Net [27] | 0.9032 | 0.9617 | 0.9546 | 0.1012 | 0.0523 |
| ResUNet [28] | 0.9697 | 0.9693 | 0.9740 | 0.0591 | 0.0184 |
| Attention U-net [29] | 0.9617 | 0.9501 | 0.9749 | 0.0733 | −0.0254 |
| UNet++ [10] | 0.9703 | 0.9696 | 0.9515 | 0.0574 | −0.0117 |
| SAR-U-Net [30] | 0.9655 | 0.9672 | 0.9746 | 0.0664 | −0.0184 |
| ResBCU-Net [31] | 0.9658 | 0.9647 | 0.9723 | 0.0610 | −0.0229 |
| RMS-UNet [32] | 0.9673 | 0.9601 | 0.9755 | 0.0591 | −0.0238 |
| MD-UNET [33] | 0.9679 | 0.9732 | 0.9746 | 0.0601 | −0.0162 |
| MDAU-Net (our model) | 0.9706 | 0.9743 | 0.9757 | 0.0569 | −0.0095 |
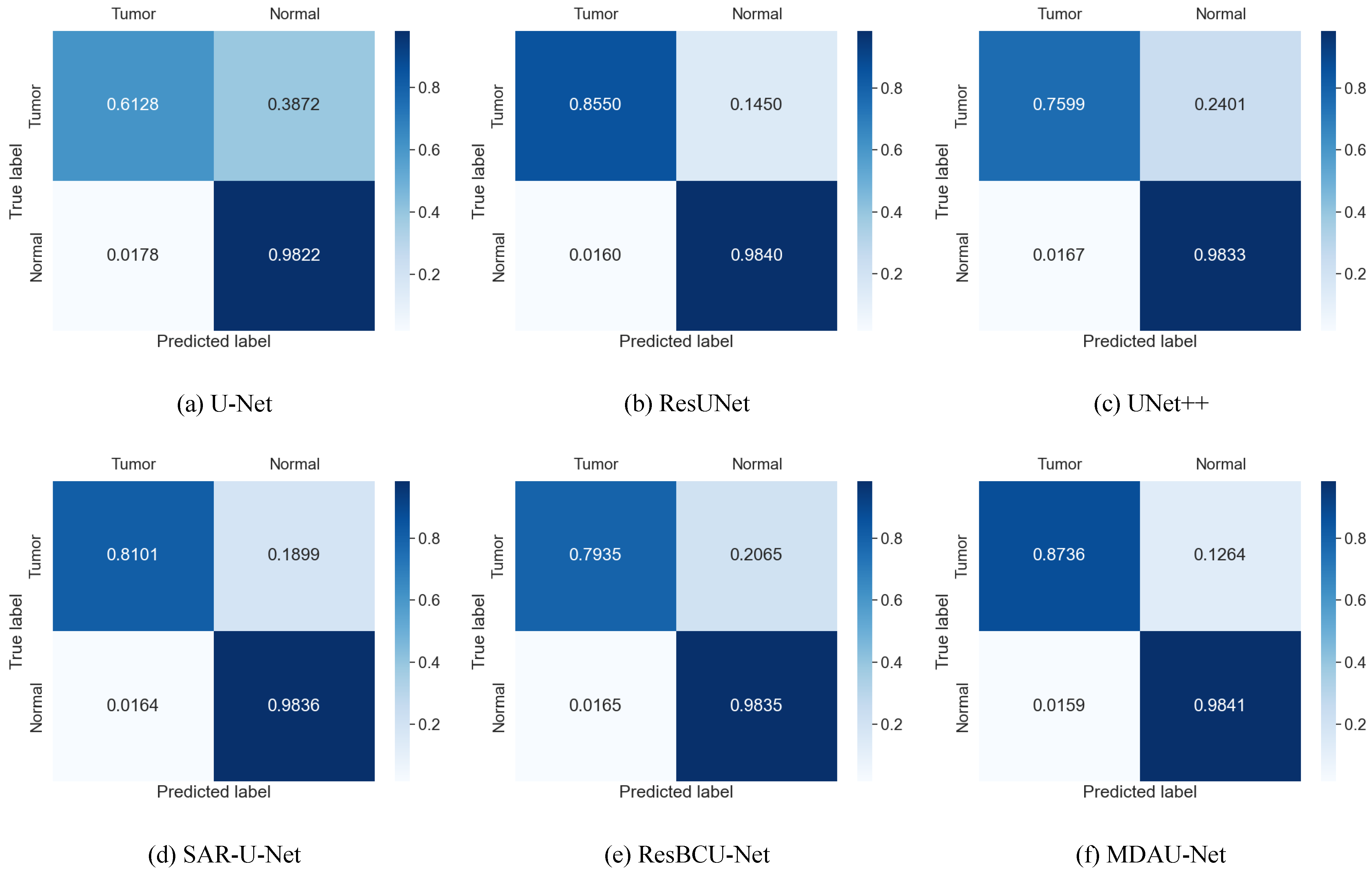
| Method | Dice | Precision | Recall | VOE | RVD |
|---|---|---|---|---|---|
| U-Net | 0.6257 | 0.6013 | 0.6128 | 0.4597 | −0.2672 |
| RU-Net [27] | 0.6528 | 0.6233 | 0.6657 | 0.3926 | −0.2519 |
| ResUNet [28] | 0.8254 | 0.8027 | 0.8550 | 0.2874 | −0.0798 |
| Attention U-net [29] | 0.6683 | 0.6620 | 0.6807 | 0.3819 | −0.0818 |
| UNet++ [10] | 0.7397 | 0.9340 | 0.7599 | 0.3995 | −0.1930 |
| SAR-U-Net [30] | 0.8096 | 0.8317 | 0.8101 | 0.3495 | −0.0770 |
| ResBCU-Net [31] | 0.6818 | 0.6243 | 0.7935 | 0.4588 | −0.2278 |
| RMS-UNet [32] | 0.6712 | 0.6258 | 0.7829 | 0.4031 | −0.2517 |
| MD-UNET [33] | 0.7838 | 0.7289 | 0.8593 | 0.3447 | −0.1596 |
| MDAU-Net (our model) | 0.8387 | 0.8211 | 0.8736 | 0.2699 | −0.0743 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, J.; Xia, M.; Ma, Z.; Jiu, Z. MDAU-Net: A Liver and Liver Tumor Segmentation Method Combining an Attention Mechanism and Multi-Scale Features. Appl. Sci. 2023, 13, 10443. https://doi.org/10.3390/app131810443
Ma J, Xia M, Ma Z, Jiu Z. MDAU-Net: A Liver and Liver Tumor Segmentation Method Combining an Attention Mechanism and Multi-Scale Features. Applied Sciences. 2023; 13(18):10443. https://doi.org/10.3390/app131810443
Chicago/Turabian StyleMa, Jinlin, Mingge Xia, Ziping Ma, and Zhiqing Jiu. 2023. "MDAU-Net: A Liver and Liver Tumor Segmentation Method Combining an Attention Mechanism and Multi-Scale Features" Applied Sciences 13, no. 18: 10443. https://doi.org/10.3390/app131810443
APA StyleMa, J., Xia, M., Ma, Z., & Jiu, Z. (2023). MDAU-Net: A Liver and Liver Tumor Segmentation Method Combining an Attention Mechanism and Multi-Scale Features. Applied Sciences, 13(18), 10443. https://doi.org/10.3390/app131810443







