Stable Artificial Autopolyploids of the Zn/Cd Accumulator Arabidopsis arenosa—A Promising Genetic Resource for Phytoremediation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Obtaining Tetra- and Octaploid Plantlets
2.2.1. In Vitro and Hydroponic Culture
2.2.2. Acclimatization of Regenerated Plants
2.3. Genome Size and Endopolyploid Estimation by Flow Cytometry
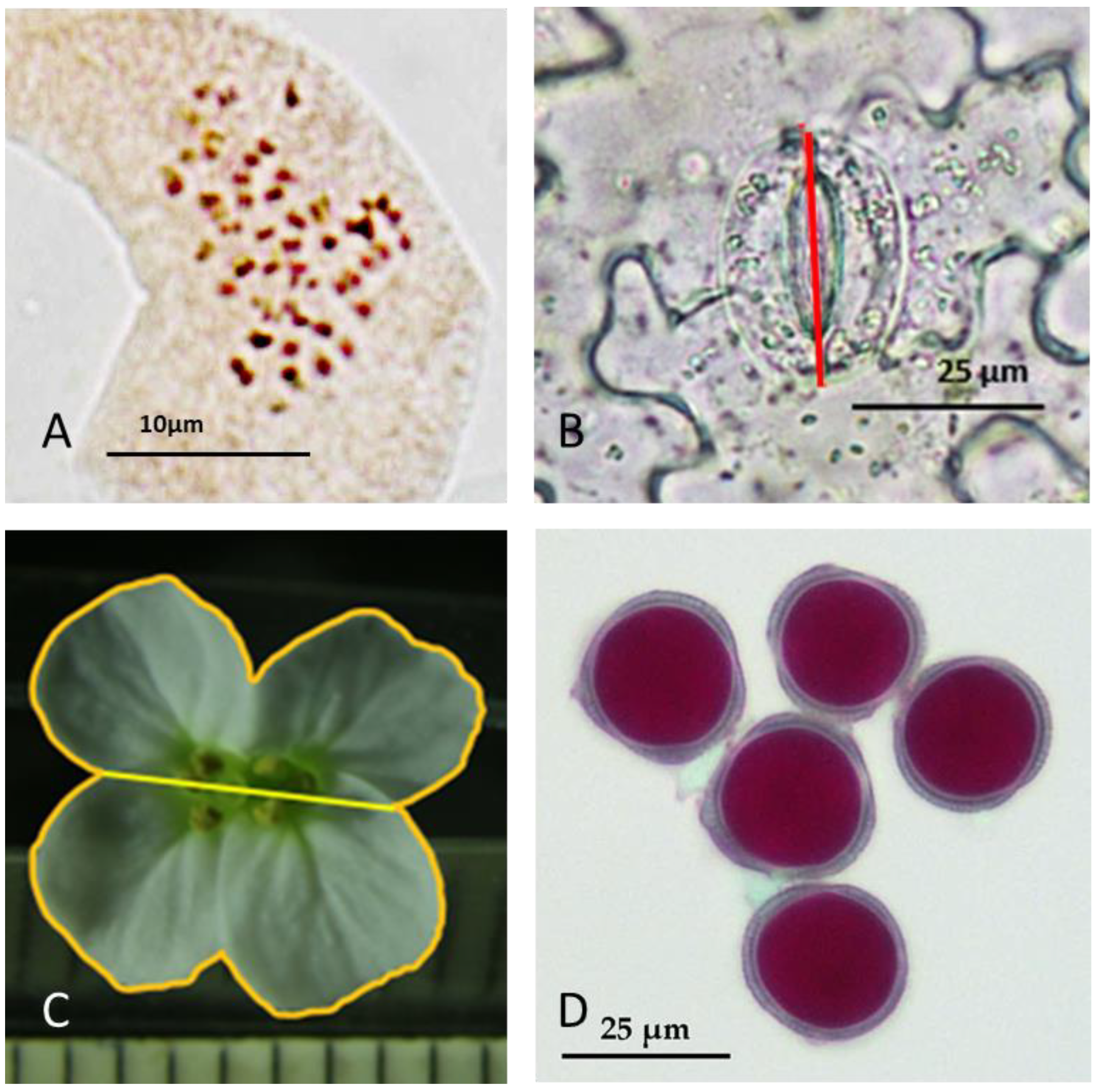
2.4. Chromosome Counting
2.5. Morpho-Anatomical and Physiological Parameter Measurements
2.5.1. Photosystem II Efficiency by a Fluorometer
2.5.2. Pollen Diameter and Viability by the Alexander Test
2.5.3. Length of Stomata
2.5.4. Width and Area of the Flowers
2.6. Seed Set in Tetra- and Octaploids and Germination of Octaploid Seeds
2.7. Micro- and Stereoscopy Usage
2.8. Statistical Analyses
3. Results
3.1. In Vitro Derivation and Ex Vitro Cultivation of Tetra- and Octaploids
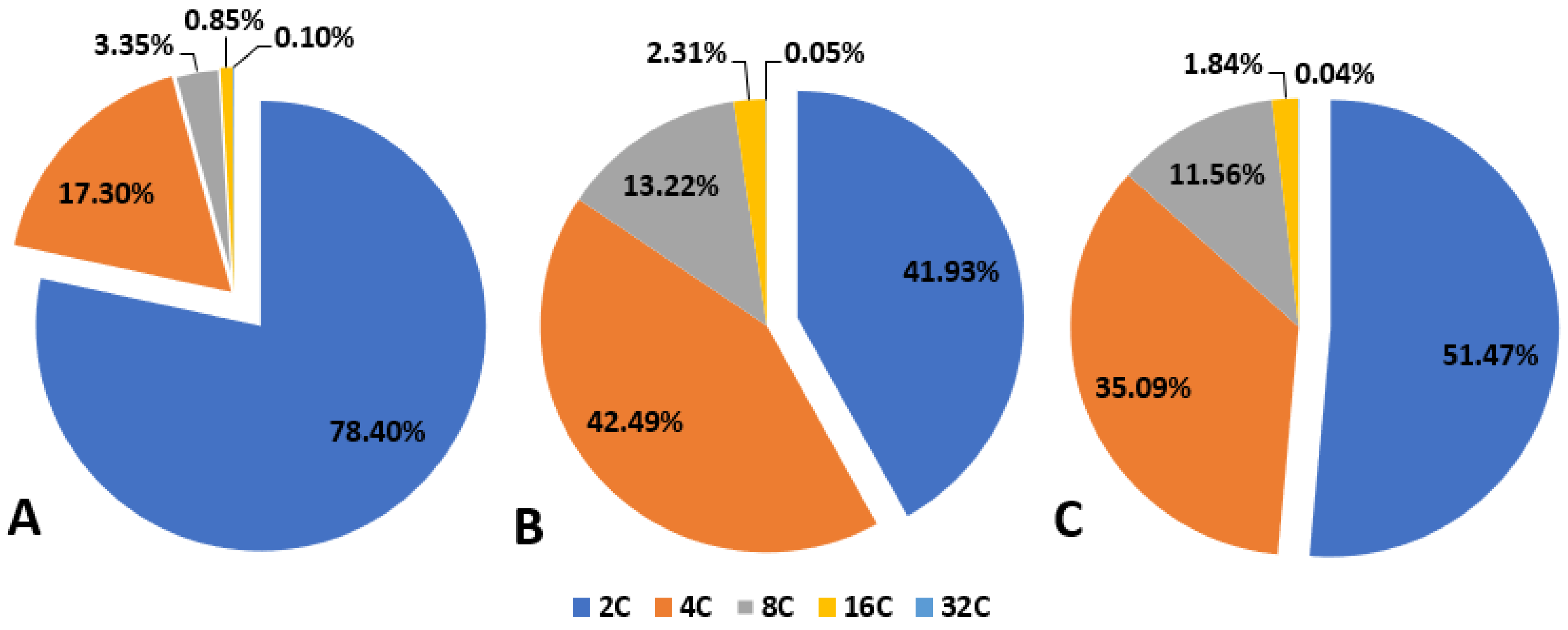
3.2. Genome Size, Endopolyploidy and Chromosome Number at Different Stages of Plant Culture
3.3. Morpho-Anatomical Characteristics of Regenerated Octaploids
4. Discussion
4.1. Octaploids Derived from Endopolyploid Callus Treated with TDZ
4.2. Genetic Stability of Octaploids
4.3. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Muszyńska, E.; Hanus-Fajerska, E.; Ciarkowska, K. Phytoremediation as an Antidote to Environmental Pollution. In Buckler Mustard (Biscutella laevigata L.) an Extraordinary Plant on Ordinary Mine Heaps Near Olkusz; Szarek-Łukaszewska, G., Ed.; W. Szafer Institute of Botany, Polish Academy of Sciences: Kraków, Poland, 2020; pp. 231–259. ISBN 978-83-62975-44-0. [Google Scholar]
- Baker, A.; Mcgrath, S.; Reeves, R.; Smith, J.A.C. Metal Hyperaccumulator Plants: A Review of the Ecology and Physiology of a Biological Resource for Phytoremediation of Metal-Polluted Soils. In Phytoremediation Contaminated Soil and Water; Terry, N., Banuelos, G., Eds.; Lewis Publishers: London, UK, 2000; pp. 85–107. ISBN 9780367803148. [Google Scholar]
- Mahar, A.; Wang, P.; Ali, A.; Awasthi, M.K.; Lahori, A.H.; Wang, Q.; Li, R.; Zhang, Z. Challenges and Opportunities in the Phytoremediation of Heavy Metals Contaminated Soils: A Review. Ecotoxicol. Environ. Saf. 2016, 126, 111–121. [Google Scholar] [CrossRef]
- Prasad, M.N.V.; Alves, L.J.; Nunes, F.C. Global Remediation Industry and Trends. In Handbook of Assisted and Amendment-Enhanced Sustainable Remediation Technology; Prasad, M.N.V., Ed.; Wiley: Chichester, UK, 2021; pp. 3–28. [Google Scholar]
- Koptsik, G. Problems and Prospects Concerning the Phytoremediation of Heavy Metal Polluted Soils: A Review. Eurasian Soil Sci. 2014, 47, 923–939. [Google Scholar] [CrossRef]
- Dalcorso, G.; Fasani, E.; Manara, A.; Visioli, G.; Furini, A. Heavy Metal Pollutions: State of the Art and Innovation in Phytoremediation. Int. J. Mol. Sci. 2019, 20, 3412. [Google Scholar] [CrossRef] [Green Version]
- Bhattacharyya, N.; Anand, U.; Kumar, R.; Ghorai, M.; Aftab, T.; Jha, N.; Rajapaksha, A.; Bundschuh, J.; Bontempi, E.; Dey, A. Phytoremediation and Sequestration of Soil Metals Using the CRISPR/Cas9 Technology to Modify Plants: A Review. Environ. Chem. Lett. 2022. [Google Scholar] [CrossRef]
- Venegas-Rioseco, J.; Ginocchio, R.; Ortiz-Calderón, C. Increase in Phytoextraction Potential by Genome Editing and Transformation: A Review. Plants 2021, 11, 86. [Google Scholar] [CrossRef]
- Yan, A.; Wang, Y.; Tan, S.N.; Mohd Yusof, M.L.; Ghosh, S.; Chen, Z. Phytoremediation: A Promising Approach for Revegetation of Heavy Metal-Polluted Land. Front. Plant Sci. 2020, 11, 359. [Google Scholar] [CrossRef]
- Koźmińska, A.; Wiszniewska, A.; Hanus-Fajerska, E.; Muszyńska, E. Recent Strategies of Increasing Metal Tolerance and Phytoremediation Potential Using Genetic Transformation of Plants. Plant Biotechnol. Rep. 2018, 12, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gunarathne, V.; Mayakaduwa, S.; Ashiq, A.; Weerakoon, S.R.; Biswas, J.K.; Vithanage, M. Transgenic Plants. In Transgenic Plant Technology for Remediation of Toxic Metals and Metalloids; Prasad, M.N.V., Ed.; Elsevier: Amsterdam, The Netherlands, 2019; pp. 89–102. ISBN 978-0-12-814389-6. [Google Scholar]
- Dietrich, C.C.; Tandy, S.; Murawska-Wlodarczyk, K.; Banaś, A.; Korzeniak, U.; Seget, B.; Babst-Kostecka, A. Phytoextraction Efficiency of Arabidopsis halleri Is Driven by the Plant and Not by Soil Metal Concentration. Chemosphere 2021, 285, 131437. [Google Scholar] [CrossRef] [PubMed]
- Szarek-Łukaszewska, G.; Grodzińska, K. Grasslands of a Zn-Pb Post-Mining Area (Olkusz Ore-Bearing Region, S Poland). Pol. Bot. J. 2010, 56, 245–260. [Google Scholar]
- Schmickl, R.; Paule, J.; Klein, J.; Marhold, K.; Koch, M.A. The Evolutionary History of the Arabidopsis arenosa Complex: Diverse Tetraploids Mask the Western Carpathian Center of Species and Genetic Diversity. PLoS ONE 2012, 7, e42691. [Google Scholar] [CrossRef]
- Hollister, J.D.; Arnold, B.J.; Svedin, E.; Xue, K.S.; Dilkes, B.P.; Bomblies, K. Genetic Adaptation Associated with Genome-Doubling in Autotetraploid Arabidopsis arenosa. PLoS Genet. 2012, 8, e1003093. [Google Scholar] [CrossRef]
- Yant, L.; Hollister, J.; Wright, K.; Arnold, B.; Higgins, J.; Franklin, F.C.H.; Bomblies, K. Meiotic Adaptation to Genome Duplication in Arabidopsis arenosa. Curr. Biol. 2013, 23, 2151–2156. [Google Scholar] [CrossRef] [Green Version]
- Konečná, V.; Bray, S.; Vlček, J.; Bohutínská, M.; Požárová, D.; Choudhury, R.R.; Bollmann-Giolai, A.; Flis, P.; Salt, D.E.; Parisod, C.; et al. Parallel Adaptation in Autopolyploid Arabidopsis arenosa Is Dominated by Repeated Recruitment of Shared Alleles. Nat. Commun. 2021, 12, 4979. [Google Scholar] [CrossRef]
- Baduel, P.; Arnold, B.; Weisman, C.M.; Hunter, B.; Bomblies, K. Habitat-Associated Life History and Stress-Tolerance Variation in Arabidopsis arenosa. Plant Physiol. 2016, 171, 437–451. [Google Scholar] [CrossRef] [Green Version]
- Monnahan, P.; Kolář, F.; Baduel, P.; Sailer, C.; Koch, J.; Horvath, R.; Laenen, B.; Schmickl, R.; Paajanen, P.; Šrámková, G.; et al. Pervasive Population Genomic Consequences of Genome Duplication in Arabidopsis arenosa. Nat. Ecol. Evol. 2019, 3, 457–468. [Google Scholar] [CrossRef]
- Rostański, A.; Myśliwiec, I.; Siwińska, D. Variability of Cardaminopsis arenosa [L.] Hayek Populations in Areas Polluted with Heavy Metals. Seria Biologiczna. Uniwersytet im. Adama Mickiewicza w Poznaniu 2005, 72, 457–461. [Google Scholar]
- Turisova, I.; Strba, T.; Aschenbrenner, S.; Andráš, P. Arabidopsis arenosa (L.) Law. on Metalliferous and Non-Metalliferous Sites in Central Slovakia. Bull. Environ. Contam. Toxicol. 2013, 91, 469–474. [Google Scholar] [CrossRef]
- Przedpełska, E.; Wierzbicka, M. Arabidopsis arenosa (Brassicaceae) from a Lead–Zinc Waste Heap in Southern Poland–a Plant with High Tolerance to Heavy Metals. Plant Soil 2007, 299, 43–53. [Google Scholar] [CrossRef]
- Słomka, A.; Kuta, E. Fiołek trójbarwny-Viola tricolor. In Ekotoksykologia. Rośliny, Gleby, Metale; Wierzbicka, M., Ed.; Wydawnictwo Uniwersytetu Warszawskiego: Warszawa, Poland, 2015; pp. 392–410. [Google Scholar]
- Preite, V.; Sailer, C.; Syllwasschy, L.; Bray, S.; Ahmadi, H.; Kraemer, U.; Yant, L. Convergent Evolution in Arabidopsis halleri and Arabidopsis arenosa on Calamine Metalliferous Soils. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2019, 374, 20180243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szopiński, M.; Sitko, K.; Rusinowski, S.; Zieleźnik-Rusinowska, P.; Corso, M.; Rostański, A.; Rojek-Jelonek, M.; Verbruggen, N.; Małkowski, E. Different Strategies of Cd Tolerance and Accumulation in Arabidopsis halleri and Arabidopsis arenosa. Plant Cell Environ. 2020, 43, 3002–3019. [Google Scholar] [CrossRef] [PubMed]
- Gieroń, Ż.; Sitko, K.; Zieleźnik-Rusinowska, P.; Szopiński, M.; Rojek-Jelonek, M.; Rostański, A.; Rudnicka, M.; Małkowski, E. Ecophysiology of Arabidopsis arenosa, a New Hyperaccumulator of Cd and Zn. J. Hazard. Mater. 2021, 412, 125052. [Google Scholar] [CrossRef]
- Gieroń, Ż.; Sitko, K.; Małkowski, E. The Different Faces of Arabidopsis arenosa—A Plant Species for a Special Purpose. Plants 2021, 10, 1342. [Google Scholar] [CrossRef] [PubMed]
- Bothe, H.; Słomka, A. Divergent Biology of Facultative Heavy Metal Plants. J. Plant Physiol. 2017, 219, 45–61. [Google Scholar] [CrossRef] [PubMed]
- Słomka, A.; Gubernat, M.; Pliszko, A.; Bothe, H. The Unusual Property of the Sand Violet, Viola rupestris, to Cope with Heavy Metal Toxicity. Flora 2020, 271, 151663. [Google Scholar] [CrossRef]
- VanBuren, R.; Pardo, J.; Man Wai, C.; Evans, S.; Bartels, D. Massive Tandem Proliferation of ELIPs Supports Convergent Evolution of Desiccation Tolerance across Land Plants. Plant Physiol. 2019, 179, 1040–1049. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roux, C.; Castric, V.; Pauwels, M.; Wright, S.I.; Saumitou-Laprade, P.; Vekemans, X. Does Speciation between Arabidopsis halleri and Arabidopsis lyrata Coincide with Major Changes in a Molecular Target of Adaptation? PLoS ONE 2011, 6, e26872. [Google Scholar] [CrossRef] [Green Version]
- Mallet, J. Hybrid Speciation. Nature 2007, 446, 279–283. [Google Scholar] [CrossRef]
- Selvi, F.; Vivona, L. Polyploidy in Odontarrhena bertolonii (Brassicaceae) in Relation to Seed Germination Performance and Plant Phenotype, with Taxonomic Implications. Plant Biosyst. 2022, 156, 959–968. [Google Scholar] [CrossRef]
- Meeus, S.; Šemberová, K.; De Storme, N.; Geelen, D.; Vallejo-Marín, M. Effect of Whole-Genome Duplication on the Evolutionary Rescue of Sterile Hybrid Monkeyflowers. Plant Commun. 2020, 1, 100093. [Google Scholar] [CrossRef]
- Simón-Porcar, V.I.; Silva, J.L.; Meeus, S.; Higgins, J.D.; Vallejo-Marín, M. Recent Autopolyploidization in a Naturalized Population of Mimulus guttatus (Phrymaceae). Bot. J. Linn. Soc. 2017, 185, 189–209. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.T.; Coate, J.E.; Meru, G. Editorial: Artificial Polyploidy in Plants. Front. Plant Sci. 2020, 11, 621849. [Google Scholar] [CrossRef] [PubMed]
- Gantait, S.; Mukherjee, E. Induced Autopolyploidy—A Promising Approach for Enhanced Biosynthesis of Plant Secondary Metabolites: An Insight. J. Genet. Eng. Biotechnol. 2021, 19, 4. [Google Scholar] [CrossRef] [PubMed]
- Cabała, J. Metale Ciężkie w Środowisku Glebowym Olkuskiego Rejonu Eksploatacji rud Zn-Pb; Jankowski, A.T., Ed.; Wydawnictwo Uniwersytetu Śląskiego Katowice: Katowice, Poland, 2009. [Google Scholar]
- Epstein, E. Mineral Nutrition of Plants: Principles and Perspectives; Wiley: New York, NY, USA, 1972. [Google Scholar]
- Sliwinska, E.; Thiem, B. Genome Size Stability in Six Medicinal Plant Species Propagated in Vitro. Biologia Plant. 2007, 51, 556–558. [Google Scholar] [CrossRef]
- Galbraith, D.W.; Harkins, K.R.; Maddox, J.M.; Ayres, N.M.; Sharma, D.P.; Firoozabady, E. Rapid Flow Cytometric Analysis of the Cell Cycle in Intact Plant Tissues. Science 1983, 220, 1049–1051. [Google Scholar] [CrossRef] [PubMed]
- Doležel, J.; Sgorbati, S.; Lucretti, S. Comparison of Three DNA Fluorochromes for Flow Cytometric Estimation of Nuclear DNA Content in Plants. Physiol. Plant. 1992, 85, 625–631. [Google Scholar] [CrossRef]
- Sliwinska, E.; Loureiro, J.; Leitch, I.J.; Šmarda, P.; Bainard, J.; Bureš, P.; Chumová, Z.; Horová, L.; Koutecký, P.; Lučanová, M.; et al. Application-based Guidelines for Best Practices in Plant Flow Cytometry. Cytom. Part A 2022, 101, 749–781. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.J. Plant Cytogenetics, 2nd ed.; CRC Press: Boca Raton, FL, USA, 2003. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Maxwell, K.; Johnson, G.N. Chlorophyll Fluorescence—a Practical Guide. J. Exp. Bot. 2000, 51, 659–668. [Google Scholar] [CrossRef]
- Muszyńska, E.; Hanus-Fajerska, E. In Vitro Multiplication of Dianthus carthusianorum Calamine Ecotype with the Aim to Revegetate and Stabilize Polluted Wastes. Plant Cell Tissue Organ Cult. 2017, 128, 631–640. [Google Scholar] [CrossRef] [Green Version]
- Muszyńska, E.; Hanus-Fajerska, E.; Koźmińska, A. Differential Tolerance to Lead and Cadmium of Micropropagated Gypsophila fastigiata Ecotype. Water Air Soil Pollut. 2018, 229, 42. [Google Scholar] [CrossRef] [Green Version]
- Wiszniewska, A.; Hanus-Fajerska, E.; Smoleń, S.; Muszyńska, E. In Vitro Selection for Lead Tolerance in Shoot Culture of Daphne Species. Acta Sci. Pol. Hortorum Cultus 2015, 14, 129–142. [Google Scholar]
- Sychta, K.; Słomka, A.; Sliwinska, E.; Migdałek, G.; Kuta, E. From Cells Highly Tolerant to Zn and Pb to Fully Fertile Plants – Selection of Tolerant Lines with in Vitro Culture. Plant Physiol. Biochem. 2020, 146, 231–237. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Wang, Q.; Meng, Q.; Liu, Y.; Pan, F.; Luo, S.; Wu, Y.; Ma, L.; Yang, X. Chromosome Doubling of Sedum alfredii Hance: A Novel Approach for Improving Phytoremediation Efficiency. J. Environ. Sci. 2019, 86, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Haberer, G.; Matthes, M.; Rattei, T.; Mayer, K.F.X.; Gierl, A.; Torres-Ruiz, R.A. Impact of Natural Genetic Variation on the Transcriptome of Autotetraploid Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2010, 107, 17809–17814. [Google Scholar] [CrossRef] [PubMed]
- Orzechowska, M.; Gurdek, S.; Siwinska, D.; Piekarska-Stachowiak, A. Cytogenetic Characterization of the Arabidopsis thaliana Natural Tetraploid Ecotype Warschau Stability during in Vitro Regeneration. Plant Cell Tissue Organ Cult. 2016, 126, 553–560. [Google Scholar] [CrossRef] [Green Version]
- Guo, B.; Abbasi, B.H.; Zeb, A.; Xu, L.L.; Wei, Y.H. Thidiazuron: A Multi-Dimensional Plant Growth Regulator. Afr. J. Biotechnol. 2011, 10, 8984–9000. [Google Scholar] [CrossRef] [Green Version]
- Murthy, B.N.S.; Murch, S.J.; Saxena, P.K. Thidiazuron: A Potent Regulator of in Vitro Plant Morphogenesis. In Vitro Cell. Dev. Biol.-Plant 1998, 34, 267–275. [Google Scholar] [CrossRef]
- Aziz, S.A.; Putri, A.A.; Sukma, D.; Syukur, M. Enhanced Growth of in Vitro Plantlets by Cytokinin and Pre-Colchicine Application in Phalaenopsis amabilis Blume Protocorms. Acta Hortic. 2019, 1262, 205–212. [Google Scholar] [CrossRef]
- Hebert, C.J.; Touchell, D.H.; Ranney, T.G.; LeBude, A.V. In Vitro Shoot Regeneration and Polyploid Induction of Rhododendron ‘Fragrantissimum Improved’. HortScience 2010, 45, 801–804. [Google Scholar] [CrossRef]
- Huy, N.P.; Tam, D.T.T.; Luan, V.Q.; Tung, H.T.; Hien, V.T.; Ngan, H.T.M.; Duy, P.N.; Nhut, D.T. In Vitro Polyploid Induction of Paphiopedilum villosum Using Colchicine. Sci. Hortic. 2019, 252, 283–290. [Google Scholar] [CrossRef]
- Kaensaksiri, T.; Soontornchainaksaeng, P.; Soonthornchareonnon, N.; Prathanturarug, S. In Vitro Induction of Polyploidy in Centella asiatica (L.) Urban. Plant Cell Tissue Organ Cult. 2011, 107, 187–194. [Google Scholar] [CrossRef]
- Shi, Q.H.; Liu, P.; Liu, M.J.; Wang, J.R.; Xu, J. A Novel Method for Rapid in Vivo Induction of Homogeneous Polyploids via Calluses in a Woody Fruit Tree (Ziziphus jujuba Mill.). Plant Cell Tissue Organ Cult. 2015, 121, 423–433. [Google Scholar] [CrossRef]
- Fritsche, Y.; Ornellas, T.S.; Stefenon, V.M.; Guerra, M.P. Ploidy Mosaics: Does Endopolyploidy in Explants Affect the Cytogenetic Stability of Orchids Regenerated from PLBs? Plant Cell Tissue Organ Cult. 2022, 149, 697–713. [Google Scholar] [CrossRef]
- Armijos-González, R.; Espinosa-Delgado, L.; Cueva-Agila, A. Indirect Shoot Regeneration Using 2,4-D Induces Somaclonal Variations in Cinchona officinalis. Floresta Ambiente 2021, 28, e20210017. [Google Scholar] [CrossRef]
- Bairu, M.W.; Aremu, A.O.; Van Staden, J. Somaclonal Variation in Plants: Causes and Detection Methods. Plant Growth Regul. 2011, 63, 147–173. [Google Scholar] [CrossRef]
- Flaishman, M.A.; Shargal, A.; Shlizerman, L.; Stern, R.A.; Lev-Yadun, S.; Graft, G. The Synthetic Cytokinins CPPU and TDZ Prolong the Phase of Cell Division in Developing Pear (Pyrus communis L.) Fruit. Acta Hortic. 2005, 671, 151–157. [Google Scholar] [CrossRef]
- Çelikel, F.G.; Zhang, Q.; Zhang, Y.; Reid, M.S.; Jiang, C.Z. A Cytokinin Analog Thidiazuron Suppresses Shoot Growth in Potted Rose Plants via the Gibberellic Acid Pathway. Front. Plant Sci. 2021, 12, 639717. [Google Scholar] [CrossRef]
- Zhai, L.; Xie, L.; Xu, J.; Xu, B.; Dong, J.; Zhang, X. Study on Exogenous Application of Thidiazuron on Seed Size of Brassica napus L. Front. Plant Sci. 2022, 13, 998698. [Google Scholar] [CrossRef]
- Slazak, B.; Sliwinska, E.; Saługa, M.; Ronikier, M.; Bujak, J.; Słomka, A.; Göransson, U.; Kuta, E. Micropropagation of Viola uliginosa (Violaceae) for Endangered Species Conservation and for Somaclonal Variation-Enhanced Cyclotide Biosynthesis. Plant Cell Tissue Organ Cult. 2015, 120, 179–190. [Google Scholar] [CrossRef] [Green Version]
- Żabicki, P.; Sliwinska, E.; Mitka, J.; Sutkowska, A.; Tuleja, M.; Migdałek, G.; Żabicka, J.; Słomka, A.; Kwiatkowska, M.; Kuta, E. Does Somaclonal Variation Play Advantageous Role in Conservation Practice of Endangered Species?: Comprehensive Genetic Studies of in Vitro Propagated Plantlets of Viola stagnina Kit. (Violaceae). Plant Cell Tissue Organ Cult. 2019, 136, 339–352. [Google Scholar] [CrossRef]
- Manzoor, A.; Ahmad, T.; Bashir, M.; Hafiz, I.; Silvestri, C. Studies on Colchicine Induced Chromosome Doubling for Enhancement of Quality Traits in Ornamental Plants. Plants 2019, 8, 194. [Google Scholar] [CrossRef] [Green Version]
- Marangelli, F.; Pavese, V.; Vaia, G.; Lupo, M.; Bashir, M.A.; Cristofori, V.; Silvestri, C. In Vitro Polyploid Induction of Highbush Blueberry through De Novo Shoot Organogenesis. Plants 2022, 11, 2349. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Jha, A.K.; Chen, R.; Doonan, J.H.; Yang, M. Polyploidy-Associated Genomic Instability in Arabidopsis thaliana. Genesis 2010, 48, 254–263. [Google Scholar] [CrossRef]
- Pecinka, A.; Fang, W.; Rehmsmeier, M.; Levy, A.A.; Mittelsten Scheid, O. Polyploidization Increases Meiotic Recombination Frequency in Arabidopsis. BMC Biol. 2011, 9, 24. [Google Scholar] [CrossRef] [PubMed]
- Kwiatkowska, M.; Izmaiłow, R. Ovules, Female Gametophytes and Embryos Are More Sensitive to Heavy Metal Pollution than Anthers and Pollen of Cardaminopsis arenosa (L.) Hayek (Brassicaceae), A Member of Calamine Flora. Acta Biol. Cracov. Bot. 2014, 56, 128–137. [Google Scholar] [CrossRef]
- Cohen, H.; Fait, A.; Tel-Zur, N. Morphological, Cytological and Metabolic Consequences of Autopolyploidization in Hylocereus (Cactaceae) Species. BMC Plant Biol. 2013, 13, 173. [Google Scholar] [CrossRef] [Green Version]
- Ramírez-Godina, F.; Robledo-Torres, V.; Foroughbakhch-Pournavab, R.; Benavides-Mendoza, A.; Alvarado-Vázquez, M.A. Pollen Viability, Density and Size of Stomata in Autotetraploids and Diploids of Physalis ixocarpa. Bot. Sci. 2013, 91, 11–18. [Google Scholar] [CrossRef] [Green Version]
- Ryder, P.; McHale, M.; Fort, A.; Spillane, C. Generation of Stable Nulliplex Autopolyploid Lines of Arabidopsis thaliana Using CRISPR/Cas9 Genome Editing. Plant Cell Rep. 2017, 36, 1005–1008. [Google Scholar] [CrossRef]


| Callus Induction on the ½ MS Medium Supplemented with 2 mg L−1 2,4-D and 2 mg L−1 BAP (after 6–8 Weeks) | Organogenesis (after 8 Weeks) | Rooting (after 4–12 Weeks) | |||
|---|---|---|---|---|---|
| Explant Type | Productivity | Medium | Productivity | Medium | Frequency (%) |
| Leaves | ++ | ½ MS | + | 0.5 IAA | 4.88 |
| Cotyledons + hypocotyl | +++ | 1 TDZ | +++ | 1 IAA | 16.25 |
| Roots | ++ | 2 TDZ | ++ | 0.5 IBA | 22.50 |
| 1 IBA | 11.11 | ||||
| ½ MS | 36.36 | ||||
| Plant Material | 4x (%) | 8x (%) | 16x (%) |
|---|---|---|---|
| Initial plants | 13 (100) | 0 (0) | 0 (0) |
| Callus obtained on ½ MS supplemented with 2 mg L−1 2,4-D + 2 mg L−1 BAP (8–16 weeks after induction) * | 15 (100) | 0 (0) | 0 (0) |
| Adventitious shoots after 4–6 weeks on ½ MS supplemented with 1 mg L−1 TDZ | 11 (45.8) | 13 (54.2) | 0 (0) |
| Rooted plants after 5–12 weeks on rooting ½ MS medium | 14 (23.7) | 38 (64.4) | 7 (11.9) |
| Regenerated plants (from hydroponic culture and soil) | 11 (22) | 39 (78) | 0 (0) |
| Initial Plant (Tetraploid) | Tetraploid Regenerant | Octaploid Regenerant | |
|---|---|---|---|
| Fv/Fm value | 0.822 [±0.059] a | 0.806 [±0.101] a | 0.815 [±0.074] a |
| Stomata length [µm] | 21.4 [±1.74] b | 20.5 [±2.28] a | 27.2 [±3.85] c |
| Pollen diameter [µm] | 19.3 [ ±1.70] a | 20.0 [±1.47] a | 24.7 [±1.47] b |
| Flower diameter [mm] | 3.4 [±0.362] a | 3.39 [±0.329] a | 4.6 [±0.555] b |
| Flower area [mm2] | 37.0 [±8.11] a | 35.6 [±4.50] a | 45.9 [±8.99] b |
| Pollen viability [%] | 96.5 [±3.67] a | 96.0 [±8.75] a | 96.8 [±5.58] a |
| Seed length [mm] | 0.892 [±0.086] a | n.m. | 1.170 [±0.109] b |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kurdziel, A.; Sychta, K.; Sliwinska, E.; Miszczak, S.; Szarek-Łukaszewska, G.; Rostański, A.; Słomka, A. Stable Artificial Autopolyploids of the Zn/Cd Accumulator Arabidopsis arenosa—A Promising Genetic Resource for Phytoremediation. Appl. Sci. 2023, 13, 1617. https://doi.org/10.3390/app13031617
Kurdziel A, Sychta K, Sliwinska E, Miszczak S, Szarek-Łukaszewska G, Rostański A, Słomka A. Stable Artificial Autopolyploids of the Zn/Cd Accumulator Arabidopsis arenosa—A Promising Genetic Resource for Phytoremediation. Applied Sciences. 2023; 13(3):1617. https://doi.org/10.3390/app13031617
Chicago/Turabian StyleKurdziel, Agnieszka, Klaudia Sychta, Elwira Sliwinska, Szymon Miszczak, Grażyna Szarek-Łukaszewska, Adam Rostański, and Aneta Słomka. 2023. "Stable Artificial Autopolyploids of the Zn/Cd Accumulator Arabidopsis arenosa—A Promising Genetic Resource for Phytoremediation" Applied Sciences 13, no. 3: 1617. https://doi.org/10.3390/app13031617
APA StyleKurdziel, A., Sychta, K., Sliwinska, E., Miszczak, S., Szarek-Łukaszewska, G., Rostański, A., & Słomka, A. (2023). Stable Artificial Autopolyploids of the Zn/Cd Accumulator Arabidopsis arenosa—A Promising Genetic Resource for Phytoremediation. Applied Sciences, 13(3), 1617. https://doi.org/10.3390/app13031617







