Automated Assessment of Radiographic Bone Loss in the Posterior Maxilla Utilizing a Multi-Object Detection Artificial Intelligence Algorithm
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design
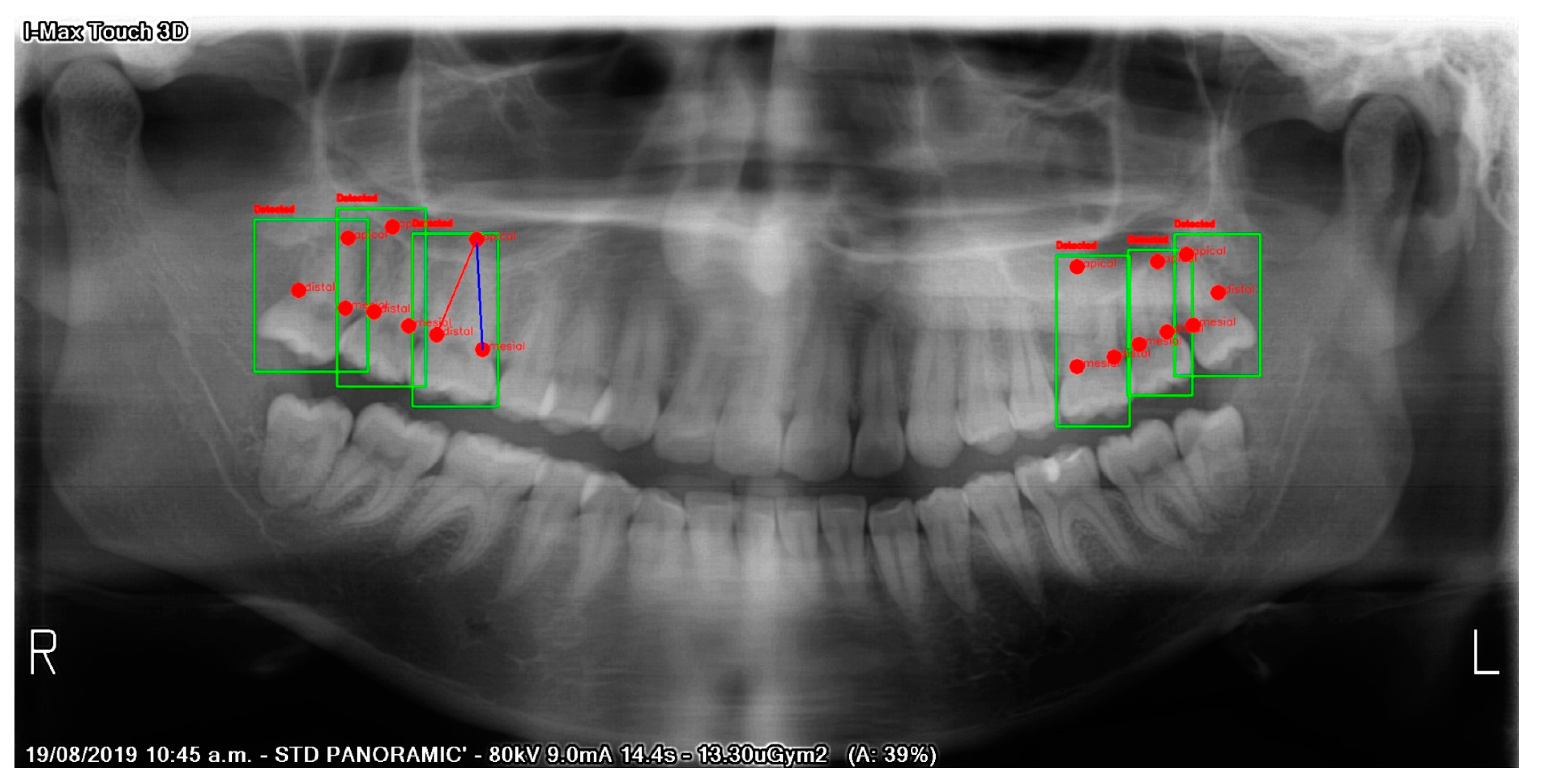
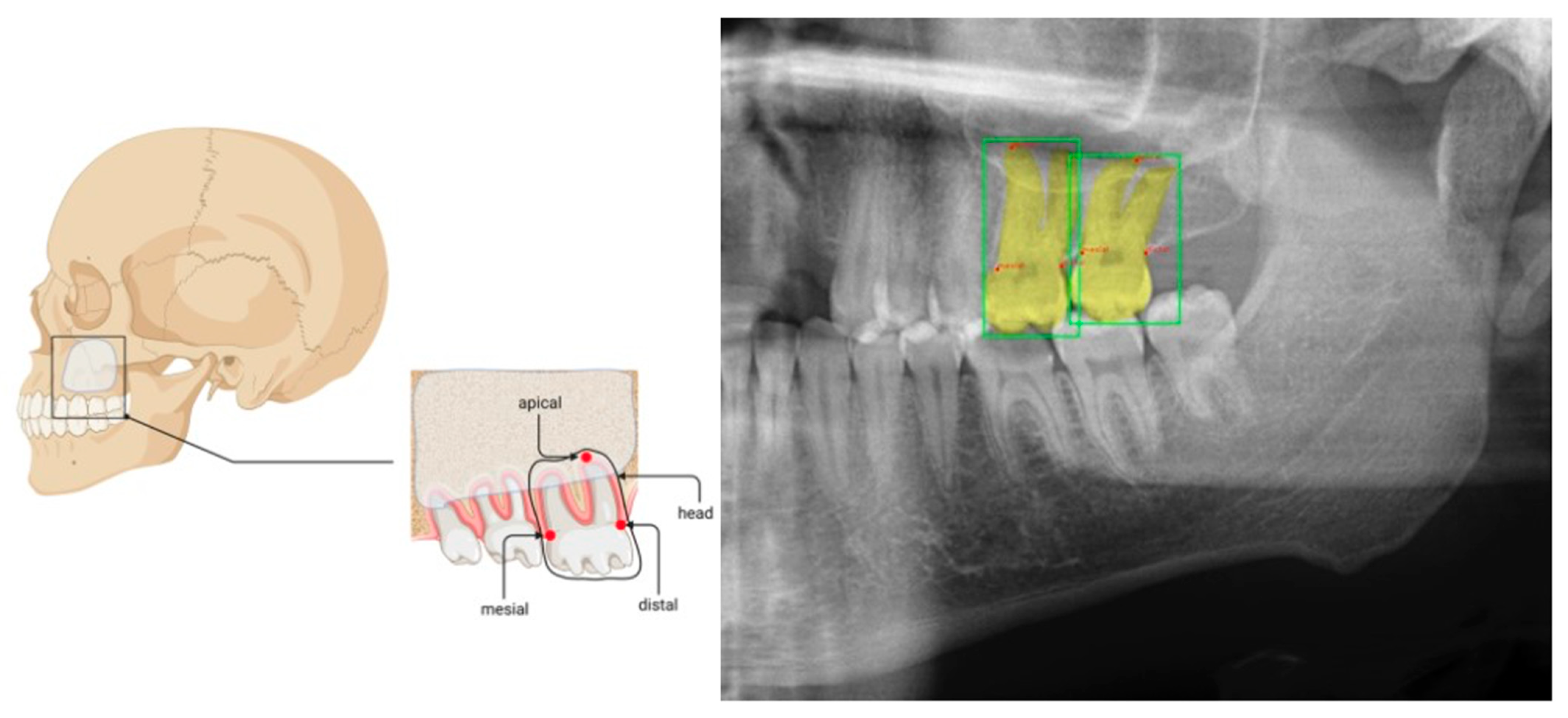
2.2. Image Processing
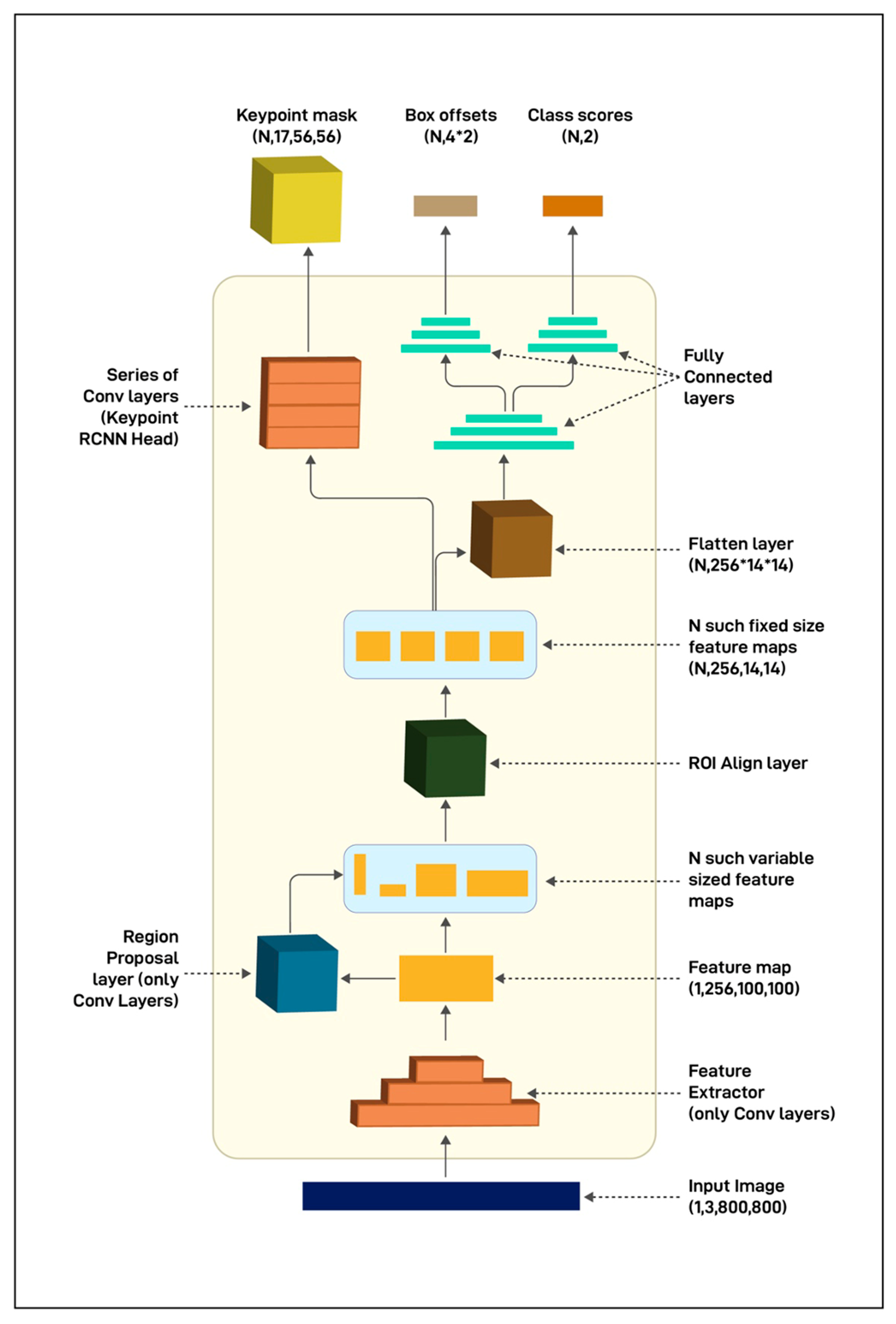
2.3. Model
2.4. Model Evaluation and Statistical Analysis
3. Results
3.1. Testing
3.2. Validation
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kinane, D.F.; Stathopoulou, P.G.; Papapanou, P.N. Periodontal Diseases. Nat. Rev. Dis. Primers 2017, 3, 17038. [Google Scholar] [CrossRef] [PubMed]
- Tonetti, M.S.; Greenwell, H.; Kornman, K.S. Staging and Grading of Periodontitis: Framework and Proposal of a New Classification and Case Definition. J. Periodontol. 2018, 89, S159–S172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nazir, M.; Al-Ansari, A.; Al-Khalifa, K.; Alhareky, M.; Gaffar, B.; Almas, K. Global Prevalence of Periodontal Disease and Lack of Its Surveillance. Sci. World J. 2020, 2020, 2146160. [Google Scholar] [CrossRef] [PubMed]
- Lang, N.P.; Bartold, P.M. Periodontal Health. J. Periodontol. 2018, 89, S9–S16. [Google Scholar] [CrossRef] [Green Version]
- Tonetti, M.S.; Jepsen, S.; Jin, L.; Otomo-Corgel, J. Impact of the Global Burden of Periodontal Diseases on Health, Nutrition and Wellbeing of Mankind: A Call for Global Action. J. Clin. Periodontol. 2017, 44, 456–462. [Google Scholar] [CrossRef] [Green Version]
- Trombelli, L.; Farina, R.; Silva, C.O.; Tatakis, D.N. Plaque-Induced Gingivitis: Case Definition and Diagnostic Considerations. J. Periodontol. 2018, 89 (Suppl. 1), S46–S73. [Google Scholar] [CrossRef] [Green Version]
- Garnick, J.J.; Silverstein, L. Periodontal Probing: Probe Tip Diameter. J. Periodontol. 2000, 71, 96–103. [Google Scholar] [CrossRef]
- Hefti, A.F. Periodontal Probing. Crit. Rev. Oral Biol. Med. 1997, 8, 336–356. [Google Scholar] [CrossRef] [Green Version]
- Machado, V.; Proença, L.; Morgado, M.; Mendes, J.J.; Botelho, J. Accuracy of Panoramic Radiograph for Diagnosing Periodontitis Comparing to Clinical Examination. JCM 2020, 9, 2313. [Google Scholar] [CrossRef]
- Chang, H.-J.; Lee, S.-J.; Yong, T.-H.; Shin, N.-Y.; Jang, B.-G.; Kim, J.-E.; Huh, K.-H.; Lee, S.-S.; Heo, M.-S.; Choi, S.-C.; et al. Deep Learning Hybrid Method to Automatically Diagnose Periodontal Bone Loss and Stage Periodontitis. Sci. Rep. 2020, 10, 7531. [Google Scholar] [CrossRef]
- Su, N.; van Wijk, A.; Berkhout, E.; Sanderink, G.; De Lange, J.; Wang, H.; van der Heijden, G.J.M.G. Predictive Value of Panoramic Radiography for Injury of Inferior Alveolar Nerve After Mandibular Third Molar Surgery. J. Oral Maxillofac. Surg. 2017, 75, 663–679. [Google Scholar] [CrossRef] [PubMed]
- Suomalainen, A.; Pakbaznejad Esmaeili, E.; Robinson, S. Dentomaxillofacial Imaging with Panoramic Views and Cone Beam CT. Insights Imaging 2015, 6, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corbet, E.; Ho, D.; Lai, S. Radiographs in Periodontal Disease Diagnosis and Management. Aust. Dent. J. 2009, 54, S27–S43. [Google Scholar] [CrossRef] [PubMed]
- Vigil, M.S.A.; Bharathi, V.S. Detection of Periodontal Bone Loss in Mandibular Area from Dental Panoramic Radiograph Using Image Processing Techniques. Concurr. Comput. Pract. Exp. 2021, 33, e6323. [Google Scholar] [CrossRef]
- Liu, M.; Wang, S.; Chen, H.; Liu, Y. A Pilot Study of a Deep Learning Approach to Detect Marginal Bone Loss around Implants. BMC Oral Health. 2022, 22, 11. [Google Scholar] [CrossRef]
- Albrektsson, T.; Dahlin, C.; Reinedahl, D.; Tengvall, P.; Trindade, R.; Wennerberg, A. An Imbalance of the Immune System Instead of a Disease Behind Marginal Bone Loss Around Oral Implants: Position Paper. Int. J. Oral Maxillofac. Implant. 2020, 35, 495–502. [Google Scholar] [CrossRef]
- Serino, G.; Sato, H.; Holmes, P.; Turri, A. Intra-Surgical vs. Radiographic Bone Level Assessments in Measuring Peri-Implant Bone Loss. Clin. Oral Implant. Res. 2017, 28, 1396–1400. [Google Scholar] [CrossRef]
- Al-Okshi, A.; Paulsson, L.; Rohlin, M.; Ebrahim, E.; Lindh, C. Measurability and Reliability of Assessments of Root Length and Marginal Bone Level in Cone Beam CT and Intraoral Radiography: A Study of Adolescents. Dentomaxillofac. Radiol. 2019, 48, 20180368. [Google Scholar] [CrossRef] [Green Version]
- Cassetta, M.; Altieri, F.; Giansanti, M.; Bellardini, M.; Brandetti, G.; Piccoli, L. Is There a Learning Curve in Static Computer-Assisted Implant Surgery? A Prospective Clinical Study. Int. J. Oral Maxillofac. Surg. 2020, 49, 1335–1342. [Google Scholar] [CrossRef]
- Khanagar, S.B.; Al-ehaideb, A.; Maganur, P.C.; Vishwanathaiah, S.; Patil, S.; Baeshen, H.A.; Sarode, S.C.; Bhandi, S. Developments, Application, and Performance of Artificial Intelligence in Dentistry—A Systematic Review. J. Dent. Sci. 2021, 16, 508–522. [Google Scholar] [CrossRef]
- Saravi, B.; Hassel, F.; Ülkümen, S.; Zink, A.; Shavlokhova, V.; Couillard-Despres, S.; Boeker, M.; Obid, P.; Lang, G.M. Artificial Intelligence-Driven Prediction Modeling and Decision Making in Spine Surgery Using Hybrid Machine Learning Models. J. Pers. Med. 2022, 12, 509. [Google Scholar] [CrossRef]
- Wong, S.H.; Al-Hasani, H.; Alam, Z.; Alam, A. Artificial Intelligence in Radiology: How Will We Be Affected? Eur. Radiol. 2019, 29, 141–143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bindushree Artificial Intelligence: In Modern Dentistry. Available online: https://www.jdrr.org/article.asp?issn=2348-2915;year=2020;volume=7;issue=1;spage=27;epage=31;aulast=Bindushree (accessed on 25 August 2022).
- Vollmer, A.; Saravi, B.; Vollmer, M.; Lang, G.M.; Straub, A.; Brands, R.C.; Kübler, A.; Gubik, S.; Hartmann, S. Artificial Intelligence-Based Prediction of Oroantral Communication after Tooth Extraction Utilizing Preoperative Panoramic Radiography. Diagnostics 2022, 12, 1406. [Google Scholar] [CrossRef] [PubMed]
- Vollmer, A.; Vollmer, M.; Lang, G.; Straub, A.; Kübler, A.; Gubik, S.; Brands, R.C.; Hartmann, S.; Saravi, B. Performance Analysis of Supervised Machine Learning Algorithms for Automatized Radiographical Classification of Maxillary Third Molar Impaction. Appl. Sci. 2022, 12, 6740. [Google Scholar] [CrossRef]
- Fazal, M.I.; Patel, M.E.; Tye, J.; Gupta, Y. The Past, Present and Future Role of Artificial Intelligence in Imaging. Eur. J. Radiol. 2018, 105, 246–250. [Google Scholar] [CrossRef]
- Artificial Intelligence in Healthcare: Past, Present and Future|Stroke and Vascular Neurology. Available online: https://svn.bmj.com/content/2/4/230.abstract (accessed on 25 August 2022).
- Sukegawa, S.; Yoshii, K.; Hara, T.; Yamashita, K.; Nakano, K.; Yamamoto, N.; Nagatsuka, H.; Furuki, Y. Deep Neural Networks for Dental Implant System Classification. Biomolecules 2020, 10, 984. [Google Scholar] [CrossRef] [PubMed]
- Cha, J.-Y.; Yoon, H.-I.; Yeo, I.-S.; Huh, K.-H.; Han, J.-S. Peri-Implant Bone Loss Measurement Using a Region-Based Convolutional Neural Network on Dental Periapical Radiographs. J. Clin. Med. 2021, 10, 1009. [Google Scholar] [CrossRef] [PubMed]
- Lerner, H.; Mouhyi, J.; Admakin, O.; Mangano, F. Artificial Intelligence in Fixed Implant Prosthodontics: A Retrospective Study of 106 Implant-Supported Monolithic Zirconia Crowns Inserted in the Posterior Jaws of 90 Patients. BMC Oral Health. 2020, 20, 80. [Google Scholar] [CrossRef] [Green Version]
- Schmidhuber, J. Deep Learning in Neural Networks: An Overview. Neural Netw. 2015, 61, 85–117. [Google Scholar] [CrossRef] [Green Version]
- Krois, J.; Ekert, T.; Meinhold, L.; Golla, T.; Kharbot, B.; Wittemeier, A.; Dörfer, C.; Schwendicke, F. Deep Learning for the Radiographic Detection of Periodontal Bone Loss. Sci. Rep. 2019, 9, 8495. [Google Scholar] [CrossRef]
- Kim, J.; Lee, H.-S.; Song, I.-S.; Jung, K.-H. DeNTNet: Deep Neural Transfer Network for the Detection of Periodontal Bone Loss Using Panoramic Dental Radiographs. Sci. Rep. 2019, 9, 17615. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, L.; Chen, D.; Cao, Z.; Wu, F.; Zhu, H.; Zhu, F. A Two-Stage Deep Learning Architecture for Radiographic Staging of Periodontal Bone Loss. BMC Oral Health. 2022, 22, 106. [Google Scholar] [CrossRef] [PubMed]
- Alotaibi, G.; Awawdeh, M.; Farook, F.F.; Aljohani, M.; Aldhafiri, R.M.; Aldhoayan, M. Artificial Intelligence (AI) Diagnostic Tools: Utilizing a Convolutional Neural Network (CNN) to Assess Periodontal Bone Level Radiographically—A Retrospective Study. BMC Oral Health. 2022, 22, 399. [Google Scholar] [CrossRef] [PubMed]
- Panetta, K.; Rajendran, R.; Ramesh, A.; Rao, S.; Agaian, S. Tufts Dental Database: A Multimodal Panoramic X-Ray Dataset for Benchmarking Diagnostic Systems. IEEE J. Biomed. Heal. Inf. 2022, 26, 1650–1659. [Google Scholar] [CrossRef] [PubMed]
- Román, J.C.M.; Fretes, V.R.; Adorno, C.G.; Silva, R.G.; Noguera, J.L.V.; Legal-Ayala, H.; Mello-Román, J.D.; Torres, R.D.E.; Facon, J. Panoramic Dental Radiography Image Enhancement Using Multiscale Mathematical Morphology. Sensors 2021, 21, 3110. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Phung, D.; Tran, T.; Gupta, S.; Rana, S.; Karmakar, C.; Shilton, A.; Yearwood, J.; Dimitrova, N.; Ho, T.B.; et al. Guidelines for Developing and Reporting Machine Learning Predictive Models in Biomedical Research: A Multidisciplinary View. J. Med. Internet Res. 2016, 18, e323. [Google Scholar] [CrossRef] [Green Version]
- Dutta, A.; Zisserman, A. The VIA Annotation Software for Images, Audio and Video. arXiv 2019, arXiv:1904.10699. [Google Scholar]
- He, K.; Gkioxari, G.; Dollar, P.; Girshick, R. Mask R-CNN. In Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 2980–2988. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Lin, T.-Y.; Dollár, P.; Girshick, R.; He, K.; Hariharan, B.; Belongie, S. Feature Pyramid Networks for Object Detection. arXiv 2016, arXiv:1612.03144. [Google Scholar]
- Lee, I.; Kim, D.; Wee, D.; Lee, S. An Efficient Human Instance-Guided Framework for Video Action Recognition. Sensors 2021, 21, 8309. [Google Scholar] [CrossRef]
- Xiao, B.; Wu, H.; Wei, Y. Simple Baselines for Human Pose Estimation and Tracking. arXiv 2018, arXiv:1804.06208v2. [Google Scholar] [CrossRef]
- Danks, R.P.; Bano, S.; Orishko, A.; Tan, H.J.; Moreno Sancho, F.; D’Aiuto, F.; Stoyanov, D. Automating Periodontal Bone Loss Measurement via Dental Landmark Localisation. Int. J. Comput. Assist. Radiol. Surg. 2021, 16, 1189–1199. [Google Scholar] [CrossRef] [PubMed]
- Persson, R.E.; Tzannetou, S.; Feloutzis, A.G.; Brägger, U.; Persson, G.R.; Lang, N.P. Comparison between Panoramic and Intra-Oral Radiographs for the Assessment of Alveolar Bone Levels in a Periodontal Maintenance Population. J. Clin. Periodontol. 2003, 30, 833–839. [Google Scholar] [CrossRef] [PubMed]
- Douglass, C.W.; Valachovic, R.W.; Wijesinha, A.; Chauncey, H.H.; Kapur, K.K.; McNeil, B.J. Clinical Efficacy of Dental Radiography in the Detection of Dental Caries and Periodontal Diseases. Oral Surg. Oral Med. Oral Pathol. 1986, 62, 330–339. [Google Scholar] [CrossRef] [PubMed]
- Tuzoff, D.V.; Tuzova, L.N.; Bornstein, M.M.; Krasnov, A.S.; Kharchenko, M.A.; Nikolenko, S.I.; Sveshnikov, M.M.; Bednenko, G.B. Tooth Detection and Numbering in Panoramic Radiographs Using Convolutional Neural Networks. Dentomaxillofac. Radiol. 2019, 48, 20180051. [Google Scholar] [CrossRef]
- Bouquet, A.; Coudert, J.-L.; Bourgeois, D.; Mazoyer, J.-F.; Bossard, D. Contributions of Reformatted Computed Tomography and Panoramic Radiography in the Localization of Third Molars Relative to the Maxillary Sinus. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endodontol. 2004, 98, 342–347. [Google Scholar] [CrossRef]
- Shan, T.; Tay, F.R.; Gu, L. Application of Artificial Intelligence in Dentistry. J. Dent. Res. 2021, 100, 232–244. [Google Scholar] [CrossRef]
- Gianfrancesco, M.A.; Tamang, S.; Yazdany, J.; Schmajuk, G. Potential Biases in Machine Learning Algorithms Using Electronic Health Record Data. JAMA Intern. Med. 2018, 178, 1544–1547. [Google Scholar] [CrossRef]
- England, J.R.; Cheng, P.M. Artificial Intelligence for Medical Image Analysis: A Guide for Authors and Reviewers. Am. J. Roentgenol. 2019, 212, 513–519. [Google Scholar] [CrossRef]
- Shorten, C.; Khoshgoftaar, T.M. A Survey on Image Data Augmentation for Deep Learning. J. Big Data 2019, 6, 60. [Google Scholar] [CrossRef]
- Waring, J.; Lindvall, C.; Umeton, R. Automated Machine Learning: Review of the State-of-the-Art and Opportunities for Healthcare. Artif. Intell. Med. 2020, 104, 101822. [Google Scholar] [CrossRef]
| Boxes | Keypoints | ||
|---|---|---|---|
| IoU = 0.50:0.95 | IoU = 0.50:0.95 | ||
| Fold 1 AP | 0.534 | Fold 1 AP | 0.639 |
| Fold 2 AP | 0.758 | Fold 2 AP | 0.622 |
| Fold 3 AP | 0.717 | Fold 3 AP | 0.726 |
| Fold 4 AP | 0.725 | Fold 4 AP | 0.643 |
| Fold 5 AP | 0.735 | Fold 5 AP | 0.531 |
| IoU = 0.50:0.95 | IoU = 0.50:0.95 | ||
| Fold 1 AR | 0.642 | Fold 1 AR | 0.616 |
| Fold 2 AR | 0.614 | Fold 2 AR | 0.513 |
| Fold 3 AR | 0.664 | Fold 3 AR | 0.556 |
| Fold 4 AR | 0.546 | Fold 4 AR | 0.633 |
| Fold 5 AR | 0.589 | Fold 5 AR | 0.579 |
| mAP | 0.6938 | mAP | 0.6322 |
| mAR | 0.611 | mAR | 0.5794 |
| Boxes | Keypoints | ||
|---|---|---|---|
| IoU = 0.50:0.95 | IoU = 0.50:0.95 | ||
| Fold 1 AP | 0.621 | Fold 1 AP | 0.544 |
| Fold 2 AP | 0.465 | Fold 2 AP | 0.358 |
| Fold 3 AP | 0.815 | Fold 3 AP | 0.671 |
| Fold 4 AP | 0.691 | Fold 4 AP | 0.604 |
| Fold 5 AP | 0.795 | Fold 5 AP | 0.599 |
| IoU = 0.50:0.95 | IoU = 0.50:0.95 | ||
| Fold 1 AR | 0.651 | Fold 1 AR | 0.512 |
| Fold 2 AR | 0.784 | Fold 2 AR | 0.655 |
| Fold 3 AR | 0.719 | Fold 3 AR | 0.498 |
| Fold 4 AR | 0.604 | Fold 4 AR | 0.577 |
| Fold 5 AR | 0.455 | Fold 5 AR | 0.681 |
| mAP | 0.6774 | mAP | 0.5552 |
| mAR | 0.6426 | mAR | 0.5846 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vollmer, A.; Vollmer, M.; Lang, G.; Straub, A.; Kübler, A.; Gubik, S.; Brands, R.C.; Hartmann, S.; Saravi, B. Automated Assessment of Radiographic Bone Loss in the Posterior Maxilla Utilizing a Multi-Object Detection Artificial Intelligence Algorithm. Appl. Sci. 2023, 13, 1858. https://doi.org/10.3390/app13031858
Vollmer A, Vollmer M, Lang G, Straub A, Kübler A, Gubik S, Brands RC, Hartmann S, Saravi B. Automated Assessment of Radiographic Bone Loss in the Posterior Maxilla Utilizing a Multi-Object Detection Artificial Intelligence Algorithm. Applied Sciences. 2023; 13(3):1858. https://doi.org/10.3390/app13031858
Chicago/Turabian StyleVollmer, Andreas, Michael Vollmer, Gernot Lang, Anton Straub, Alexander Kübler, Sebastian Gubik, Roman C. Brands, Stefan Hartmann, and Babak Saravi. 2023. "Automated Assessment of Radiographic Bone Loss in the Posterior Maxilla Utilizing a Multi-Object Detection Artificial Intelligence Algorithm" Applied Sciences 13, no. 3: 1858. https://doi.org/10.3390/app13031858
APA StyleVollmer, A., Vollmer, M., Lang, G., Straub, A., Kübler, A., Gubik, S., Brands, R. C., Hartmann, S., & Saravi, B. (2023). Automated Assessment of Radiographic Bone Loss in the Posterior Maxilla Utilizing a Multi-Object Detection Artificial Intelligence Algorithm. Applied Sciences, 13(3), 1858. https://doi.org/10.3390/app13031858









