Low-Cost Handheld Spectrometry for Detecting Flavescence Dorée in Vineyards
Abstract
:1. Introduction
- To compare the performances of the two portable mini spectrometers to detect the FD in vineyards.
- To compare the performance of different machine learning methods for the classification of healthy and infected plants with FD at different stages of disease development.
- To compare the best wavelengths selected by different feature selection methods.
2. Materials and Methods
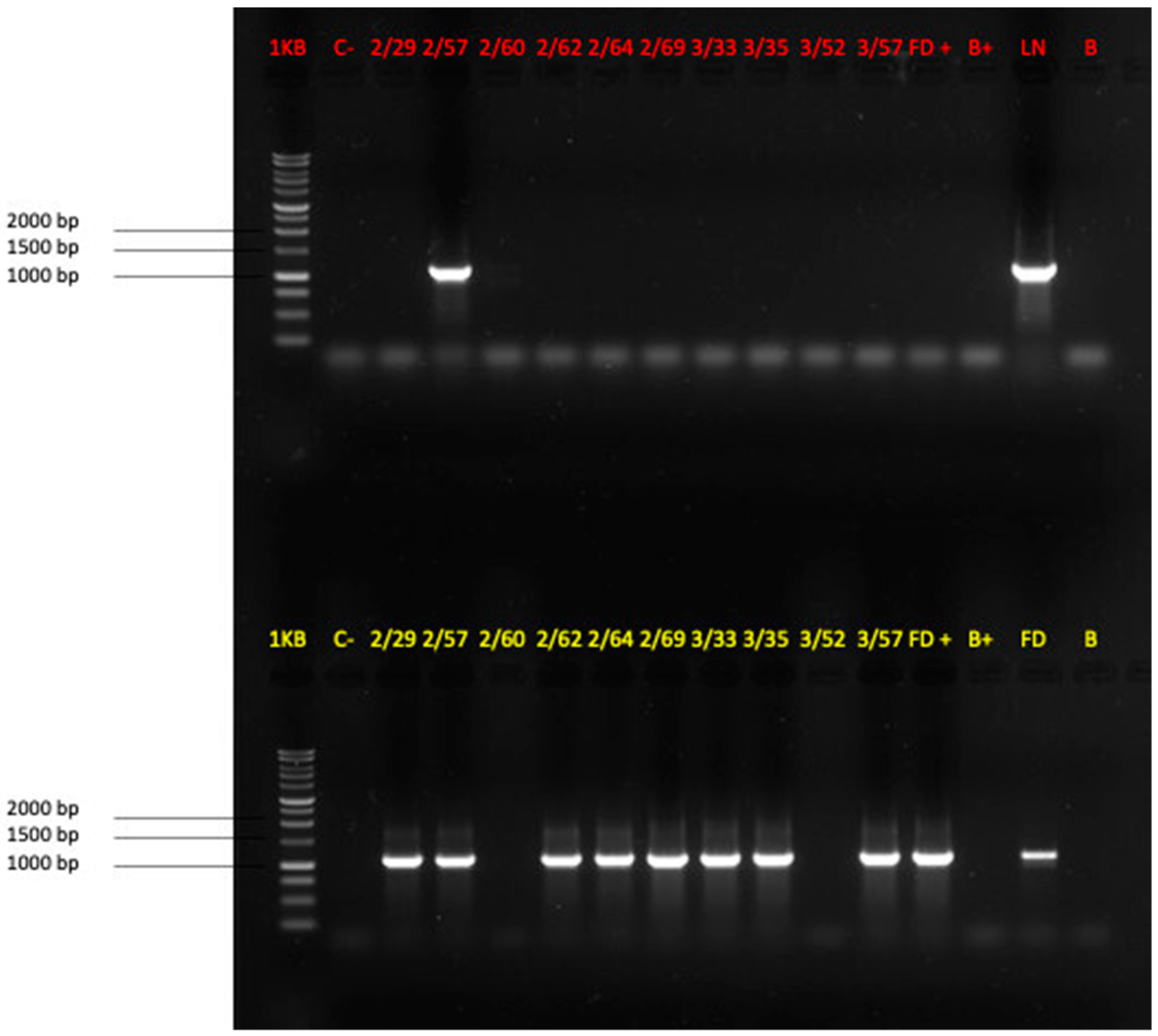
2.1. Leaf Sampling and Molecular Detection of Phytoplasmas
2.2. Spectral Data Collection
2.3. Machine Learning Pipeline for Classification
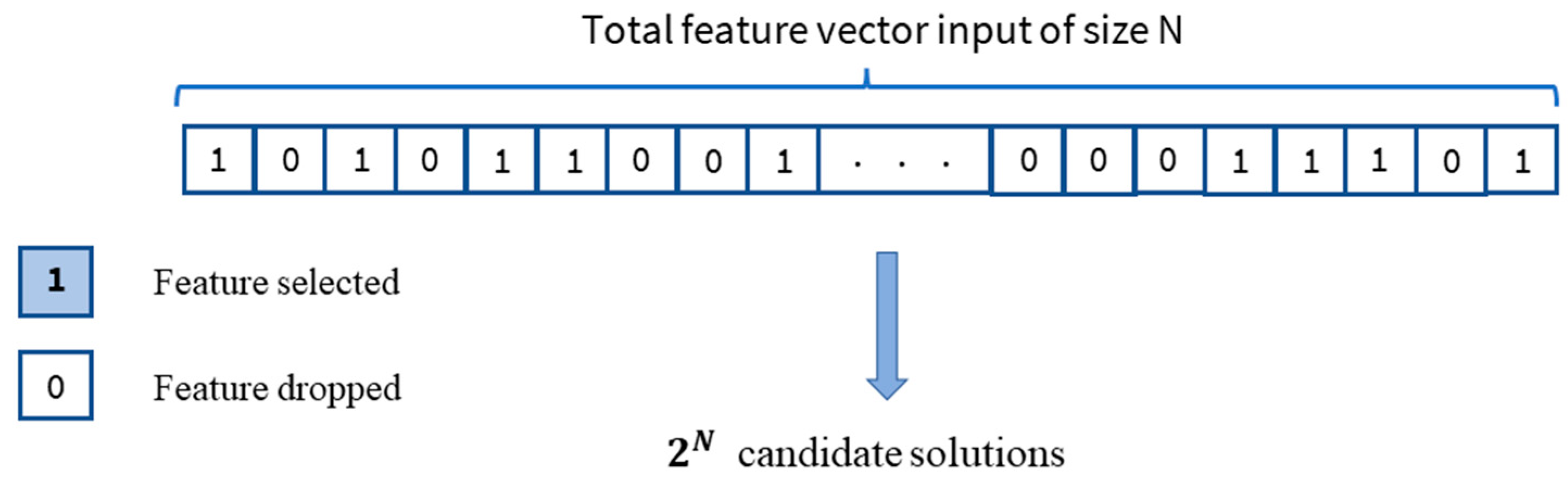
2.3.1. Feature Selection Algorithms
Ensemble-Based Feature Selection
Genetic Algorithms (GA)
2.3.2. Classification Models
- Logistic regression (LR): The LR classifier is a simple yet effective technique that takes advantage of logistic function to model dependent variables that are linearly related to the log odds for binary classification problems. After ranking the features in terms of relevance, the model can be employed to forecast the probability of the classes based on its input features [35]. For the purpose of describing the phenomenon of classification, it is useful to tie the likelihood of a class to a group of explanatory features. In order to avoid overfit, the “L2” penalty is exploited in addition to the “C” value hyperparameter governing the degree of regularization. LR is widely employed for binary classification tasks in the remote sensing community due to its simplicity, parallelizability, and high interpretability producing extremely competitive results [36].
- Support vector machines (SVMs): SVMs are known to be one of the most promising classification tools as they are widely used within the remote sensing community different classification and regression tasks [37,38,39]. Its main objective is to find an optimal hyperplane that separates a given dataset. Moreover, to cope with nonlinear patterns, an SVM kernel function aims at mapping the original data into a higher new dimensional space in which finding an optimal separation hyperplane between data becomes linearly feasible in the new transformed space. In our case, the radial basis function (RBF) has been exploited as an SVMs kernel. RBF function is widely used within the machine learning community, due to its flexibility, and similarity to the Gaussian distribution, it can overcome the feature space complexity. We put forth the SVMs classifier in this work motivated by their highly efficient performance in hyperdimensional classification tasks.
- Random forest classifier (RF): A decision tree is a supervised learning method based on a set of rules arranged in the shape of a tree for solving both classification and regression problems [31]. RF, as its name suggests, is an extension of the single decision tree yet better suited for a complex dataset. In order to cope with decision tree overfitting limitations, it takes advantage of exploiting averaging on a bunch of decision trees during the training phase. In doing so, it is possible to solve the problem with improved generalization.
- Gradient boosting classifier (XGBoost): Due to its high effectiveness and accuracy, extreme gradient boosting is one of the most used machine learning techniques. Similar to RF, it combines different machine learning algorithms to create a strong predictive classifier [40]. In contrast to gradient boosting decision trees, which is based on the gradient-powered decision trees exploiting linear combination of an ensemble of weak learning models where the trees are produced sequentially, correcting the error of prior weak learners, XGBoost builds its trees in parallel elevating its performance considerably with high computational speed. Chen and Guestrin [41], for this latter reason, chose XGBoost as the GA iterative optimization fitness assessment classifier.
- Cubist regression models (CB): Another model based on decision trees is the Cubist rule-based model [cubist]. A potent algorithm developing sequentially a series of trees. It induces accurate yet simple rules built and selected repeatedly at each iteration. Unlike RF, CB returns a set of rules linked to sets of multivariate models rather than a single final model. Then, depending on the rule that best matches the predictors, a particular collection of predictor variables will choose an actual prediction model [42,43]. To some extent, in order to interpret the complicated links between the influential features the Cubist model was seen to be very promising compared to tree methods counterparts [44].
- Trees-based models can produce a more thorough predictive accuracy when compared to traditional linear machine learning techniques. Their main benefits for classification tasks include (a) a reduced risk of overfitting, (b) a small number of effective tuning hyperparameters, (c) the ability to automatically determine variable importance in order to interpret the variable contribution mechanism in the final prediction model. To recapitulate, in order to capture the linear/non-linear relationships between the VIS-NIR hyperspectral bands, various predictors, random forests, Cubist and XGBoost-based tree ensemble models in addition to LR and SVMs algorithms are used in this context. A 3-fold cross-validation set has been exploited during the training process searching for the best hyper-parameters values of our respective supervised learning models. We use Savitzky–Golay filter (SG), a polynomial interpolation, for smoothing the input data as a pre-processing step with a polynomial order 2, and a passing window equal to 13. The K-fold cross-validation resulting scores in the training stage use the forward selection as in Kim et al. [29].
2.4. Classification Accuracy Assessment
3. Results
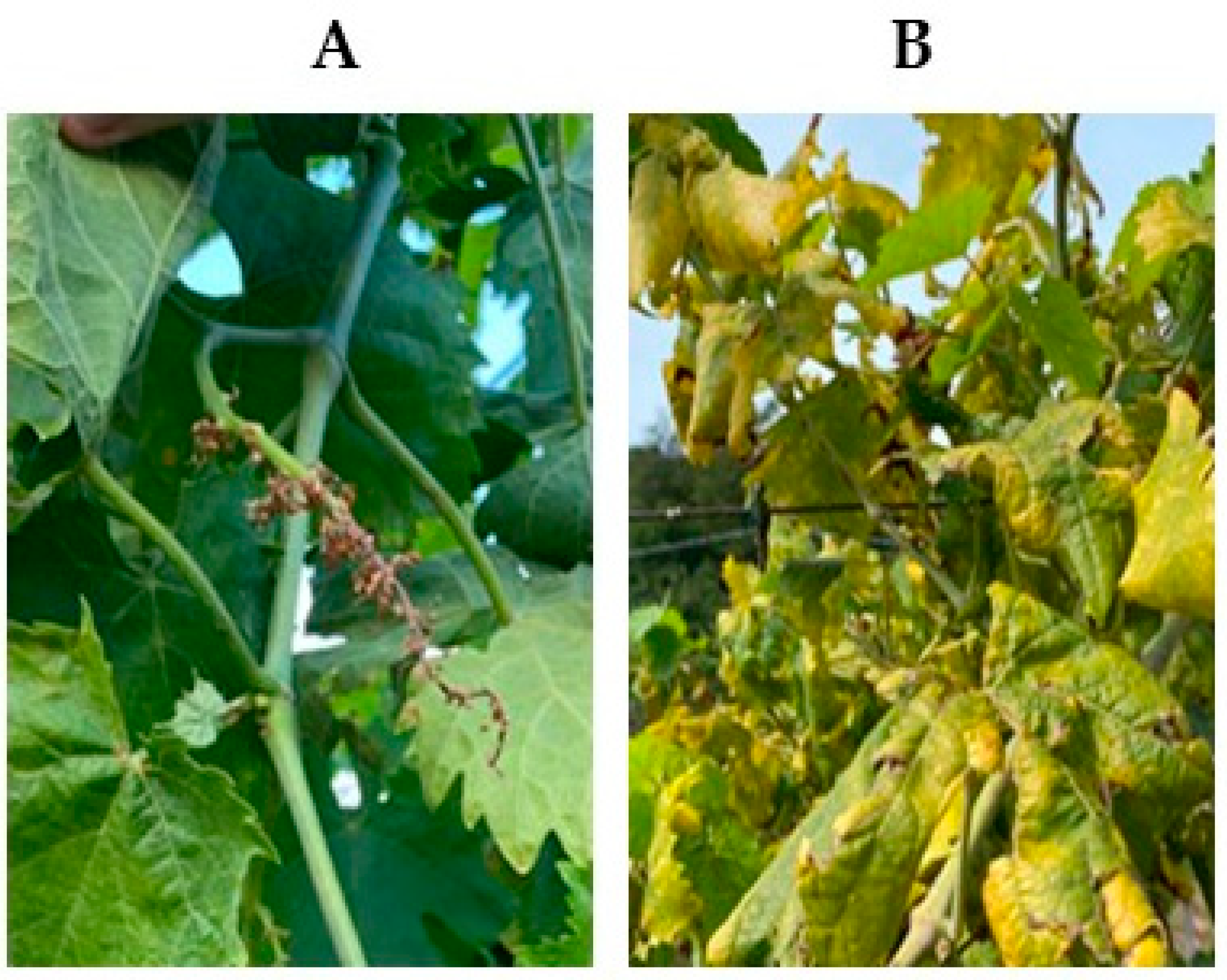
3.1. Visual Inspection of the Vineyard and Molecular Detection of FD and BN Phytoplasmas
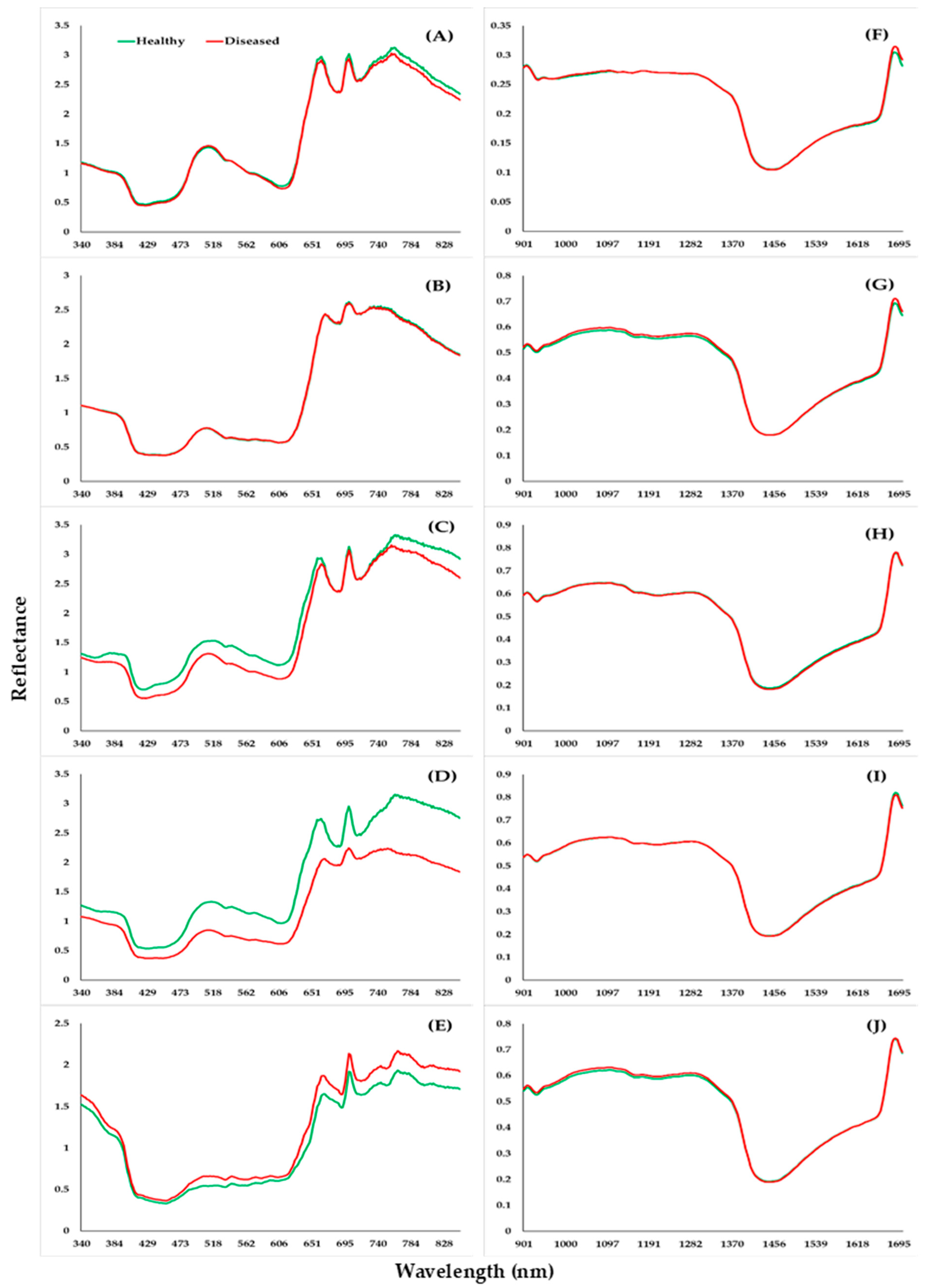
3.2. Healthy and Diseased Plants to Spectra
3.3. Classification Performance
3.4. Confusion MATRIX
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jeger, M.; Bragard, C.; Caffier, D.; Candresse, T.; Chatzivassiliou, E.; Dehnen-Schmutz, K.; Gilioli, G.; Jaques Miret, J.A.; MacLeod, A.; Navajas Navarro, M.; et al. Risk to Plant Health of Flavescence Dorée for the EU Territory. EFSA J. 2016, 14, e04603. [Google Scholar] [CrossRef]
- Ripamonti, M.; Pegoraro, M.; Rossi, M.; Bodino, N.; Beal, D.; Panero, L.; Marzachì, C.; Bosco, D. Prevalence of Flavescence Dorée Phytoplasma-Infected Scaphoideus Titanus in Different Vineyard Agroecosystems of Northwestern Italy. Insects 2020, 11, 301. [Google Scholar] [CrossRef] [PubMed]
- Martini, M.; Pavan, F.; Bianchi, G.L.; Loi, N.; Ermacora, P. Recent Spread of the “Flavescence Dorée” Disease in North-Eastern Italy. Phyt. Moll. 2019, 9, 207. [Google Scholar] [CrossRef]
- Al-Saddik, H.; Simon, J.C.; Cointault, F. Assessment of the Optimal Spectral Bands for Designing a Sensor for Vineyard Disease Detection: The Case of ‘Flavescence Dorée. Precis. Agric. 2019, 20, 398–422. [Google Scholar] [CrossRef]
- Sinha, R.; Khot, L.R.; Rathnayake, A.P.; Gao, Z.; Naidu, R.A. Visible-near Infrared Spectroradiometry-Based Detection of Grapevine Leafroll-Associated Virus 3 in a Red-Fruited Wine Grape Cultivar. Comput. Electron. Agric. 2019, 162, 165–173. [Google Scholar] [CrossRef]
- Musci, M.A.; Persello, C.; Lingua, A.M. Uav Images and Deep-Learning Algorithms for Detecting Flavescence Doree Disease in Grapevine Orchards. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2020, 43, 1483–1489. [Google Scholar] [CrossRef]
- Tessitori, M.; La Rosa, R.; Marzachì, C. Flavescence Dorée and Bois Noir Diseases of Grapevine Are Evolving Pathosystems. Plant Health Prog. 2018, 19, 136–138. [Google Scholar] [CrossRef]
- Wei, X.; Johnson, M.A.; Langston, D.B.; Mehl, H.L.; Li, S. Identifying Optimal Wavelengths as Disease Signatures Using Hyperspectral Sensor and Machine Learning. Remote Sens. 2021, 13, 2833. [Google Scholar] [CrossRef]
- Calamita, F.; Imran, H.A.; Vescovo, L.; Mekhalfi, M.L.; La Porta, N. Early Identification of Root Rot Disease by Using Hyperspectral Reflectance: The Case of Pathosystem Grapevine/Armillaria. Remote Sens. 2021, 13, 2436. [Google Scholar] [CrossRef]
- Mahlein, A.-K.; Kuska, M.T.; Behmann, J.; Polder, G.; Walter, A. Hyperspectral Sensors and Imaging Technologies in Phytopathology: State of the Art. Annu. Rev. Phytopathol. 2018, 56, 535–558. [Google Scholar] [CrossRef]
- Naidu, R.A.; Perry, E.M.; Pierce, F.J.; Mekuria, T. The Potential of Spectral Reflectance Technique for the Detection of Grapevine Leafroll-Associated Virus-3 in Two Red-Berried Wine Grape Cultivars. Comput. Electron. Agric. 2009, 66, 38–45. [Google Scholar] [CrossRef]
- Gao, L.; Wang, X.; Johnson, B.A.; Tian, Q.; Wang, Y.; Verrelst, J.; Mu, X.; Gu, X. Remote Sensing Algorithms for Estimation of Fractional Vegetation Cover Using Pure Vegetation Index Values: A Review. ISPRS J. Photogramm. Remote Sens. 2020, 159, 364–377. [Google Scholar] [CrossRef] [PubMed]
- Carter, G.A. Responses of Leaf Spectral Reflectance to Plant Stress. Am. J. Bot. 1993, 80, 239–243. [Google Scholar] [CrossRef]
- Jacquemoud, S.; Ustin, S.L. Leaf Optical Properties: A State of the Art. In Proceedings of the 8th International Symposium of Physical Measurements & Signatures in Remote Sensing, Aussois, France, 8–12 January 2001. [Google Scholar]
- Imran, H.A.; Gianelle, D.; Scotton, M.; Rocchini, D.; Dalponte, M.; Macolino, S.; Sakowska, K.; Pornaro, C.; Vescovo, L. Potential and Limitations of Grasslands α-Diversity Prediction Using Fine-Scale Hyperspectral Imagery. Remote Sens. 2021, 13, 2649. [Google Scholar] [CrossRef]
- Pagliarani, C.; Gambino, G.; Ferrandino, A.; Chitarra, W.; Vrhovsek, U.; Cantu, D.; Palmano, S.; Marzachì, C.; Schubert, A. Molecular Memory of Flavescence Dorée Phytoplasma in Recovering Grapevines. Hortic. Res. 2020, 7, 126. [Google Scholar] [CrossRef]
- Sinha, R.; Khot, L.R.; Gao, Z.; Chandel, A.K. Sensors III: Spectral Sensing and Data Analysis. In Fundamentals of Agricultural and Field Robotics; Agriculture Automation and Control; Karkee, M., Zhang, Q., Eds.; Springer International Publishing: Cham, Switzerland, 2021; pp. 79–110. ISBN 978-3-030-70400-1. [Google Scholar]
- Hira, Z.M.; Gillies, D.F. A Review of Feature Selection and Feature Extraction Methods Applied on Microarray Data. Adv. Bioinform. 2015, 2015, 198363. [Google Scholar] [CrossRef]
- AL-Saddik, H.; Simon, J.-C.; Cointault, F. Development of Spectral Disease Indices for ‘Flavescence Dorée’ Grapevine Disease Identification. Sensors 2017, 17, 2772. [Google Scholar] [CrossRef]
- Barjaktarović, M.; Faralli, M.; Bertamini, M.; Bruzzone, L. A Multispectral Acquisition System for Potential Detection of Flavescence Dorée. In Proceedings of the 2022 30th Telecommunications Forum (TELFOR), Belgrade, Serbia, 15–16 November 2022; pp. 1–4. [Google Scholar]
- Albetis, J.; Jacquin, A.; Goulard, M.; Poilvé, H.; Rousseau, J.; Clenet, H.; Dedieu, G.; Duthoit, S. On the Potentiality of UAV Multispectral Imagery to Detect Flavescence Dorée and Grapevine Trunk Diseases. Remote Sens. 2018, 11, 23. [Google Scholar] [CrossRef]
- Daglio, G.; Cesaro, P.; Todeschini, V.; Lingua, G.; Lazzari, M.; Berta, G.; Massa, N. Potential Field Detection of Flavescence Dorée and Esca Diseases Using a Ground Sensing Optical System. Biosyst. Eng. 2022, 215, 203–214. [Google Scholar] [CrossRef]
- Aitkenhead, M.; Gaskin, G.; Lafouge, N.; Hawes, C. PHYLIS: A Low-Cost Portable Visible Range Spectrometer for Soil and Plants. Sensors 2017, 17, 99. [Google Scholar] [CrossRef] [Green Version]
- Boudon-Padieu, E.; Béjat, A.; Clair, D.; Larrue, J.; Borgo, M.; Bertotto, L.; Angelini, E. Grapevine Yellows: Comparison of Different Procedures for DNA Extraction and Amplification with PCR for Routine Diagnosis of Phytoplasmas in Grapevine. VITIS–J. Grapevine Res. 2015, 42, 141–149. [Google Scholar] [CrossRef]
- Nees, P. Microstegium vimineum (Trin.) A. Camus. EPPO Bull. 2016, 46, 14–19. [Google Scholar]
- Doyle, J.J.; Doyle, J.L. A Rapid DNA Isolation Procedure for Small Quantities of Fresh Leaf Tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Smart, C.D.; Schneider, B.; Blomquist, C.L.; Guerra, L.J.; Harrison, N.A.; Ahrens, U.; Lorenz, K.H.; Seemüller, E.; Kirkpatrick, B.C. Phytoplasma-Specific PCR Primers Based on Sequences of the 16S-23S RRNA Spacer Region. Appl. Environ. Microbiol. 1996, 62, 2988–2993. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.M.; Gundersen, D.E.; Hammond, R.W.; Davis, R.E. Use of Mycoplasmalike Organism (MLO) Group-Specific Oligonucleotide Primers for Nested-PCR Assays to Detect Mixed-MLO Infections in a Single Host Plant. Phytopathology 1994, 84, 559–566. [Google Scholar] [CrossRef]
- Kim, Y.-E.; Kim, Y.-S.; Kim, H. Effective Feature Selection Methods to Detect IoT DDoS Attack in 5G Core Network. Sensors 2022, 22, 3819. [Google Scholar] [CrossRef] [PubMed]
- Babatunde, O.H.; Armstrong, L.; Leng, J.; Diepeveen, D. A Genetic Algorithm-Based Feature Selection. Int. J. Electron. Commun. Comput. Eng. 2014, 5, 2278–4209. [Google Scholar]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Zheng, A.; Casari, A. Feature Engineering for Machine Learning: Principles and Techniques for Data Scientists; O’Reilly Media, Inc.: Newton, MA, USA, 2018; ISBN 978-1-4919-5319-8. [Google Scholar]
- Lal, T.N.; Chapelle, O.; Weston, J.; Elisseeff, A. Embedded Methods. In Feature Extraction: Foundations and Applications; Studies in Fuzziness and Soft Computing; Guyon, I., Nikravesh, M., Gunn, S., Zadeh, L.A., Eds.; Springer: Berlin/Heidelberg, Germany, 2006; pp. 137–165. ISBN 978-3-540-35488-8. [Google Scholar]
- Alba, E.; Garcia-Nieto, J.; Jourdan, L.; Talbi, E.-G. Gene Selection in Cancer Classification Using PSO/SVM and GA/SVM Hybrid Algorithms. In Proceedings of the 2007 IEEE Congress on Evolutionary Computation, Singapore, 25–28 September 2007; pp. 284–290. [Google Scholar]
- Cramer, J.S. The Origins of Logistic Regression; Tinbergen Institute: Amsterdam, The Netherlands, 2002; pp. 167–178. [Google Scholar]
- Ling, X.; Zhu, Y.; Ming, D.; Chen, Y.; Zhang, L.; Du, T. Feature Engineering of Geohazard Susceptibility Analysis Based on the Random Forest Algorithm: Taking Tianshui City, Gansu Province, as an Example. Remote Sens. 2022, 14, 5658. [Google Scholar] [CrossRef]
- Cortes, C.; Vapnik, V. Support-Vector Networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Douha, L.; Benoudjit, N.; Douak, F.; Melgani, F. Support Vector Regression in Spectrophotometry: An Experimental Study. Crit. Rev. Anal. Chem. 2012, 42, 214–219. [Google Scholar] [CrossRef]
- Koda, S.; Zeggada, A.; Melgani, F.; Nishii, R. Spatial and Structured SVM for Multilabel Image Classification. IEEE Trans. Geosci. Remote Sens. 2018, 56, 5948–5960. [Google Scholar] [CrossRef]
- Friedman, J.H. Greedy Function Approximation: A Gradient Boosting Machine. Ann. Stat. 2001, 29, 1189–1232. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. XGBoost: A Scalable Tree Boosting System. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; Association for Computing Machinery: New York, NY, USA, 2016; pp. 785–794. [Google Scholar]
- Appelhans, T.; Mwangomo, E.; Hardy, D.R.; Hemp, A.; Nauss, T. Evaluating Machine Learning Approaches for the Interpolation of Monthly Air Temperature at Mt. Kilimanjaro, Tanzania. Spat. Stat. 2015, 14, 91–113. [Google Scholar] [CrossRef]
- Walton, J.T. Subpixel Urban Land Cover Estimation. Photogramm. Eng. Remote Sens. 2008, 74, 1213–1222. [Google Scholar] [CrossRef]
- Holmes, G.; Hall, M.; Prank, E. Generating Rule Sets from Model Trees. In Advanced Topics in Artificial Intelligence; Foo, N., Ed.; Springer: Berlin/Heidelberg, Germany, 1999; pp. 1–12. [Google Scholar]
- Tomkins, M.; Kliot, A.; Marée, A.F.; Hogenhout, S.A. A Multi-Layered Mechanistic Modelling Approach to Understand How Effector Genes Extend beyond Phytoplasma to Modulate Plant Hosts, Insect Vectors and the Environment. Curr. Opin. Plant Biol. 2018, 44, 39–48. [Google Scholar] [CrossRef]
- Jollard, C.; Foissac, X.; Desqué, D.; Razan, F.; Garcion, C.; Beven, L.; Eveillard, S. Flavescence Dorée Phytoplasma Has Multiple FtsH Genes That Are Differentially Expressed in Plants and Insects. Int. J. Mol. Sci. 2020, 21, 150. [Google Scholar] [CrossRef]
- Dermastia, M.; Škrlj, B.; Strah, R.; Anžič, B.; Tomaž, Š.; Križnik, M.; Schönhuber, C.; Riedle-Bauer, M.; Ramšak, Ž.; Petek, M.; et al. Differential Response of Grapevine to Infection with ‘Candidatus Phytoplasma solani’ in Early and Late Growing Season through Complex Regulation of MRNA and Small RNA Transcriptomes. Int. J. Mol. Sci. 2021, 22, 3531. [Google Scholar] [CrossRef]
- Dermastia, M. Interactions Between Grapevines and Grapevine Yellows Phytoplasmas BN and FD. In Grapevine Yellows Diseases and Their Phytoplasma Agents: Biology and Detection; Springer Briefs in Agriculture; Dermastia, M., Bertaccini, A., Constable, F., Mehle, N., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 47–67. ISBN 978-3-319-50648-7. [Google Scholar]
- Sims, D.A.; Gamon, J.A. Relationships between Leaf Pigment Content and Spectral Reflectance across a Wide Range of Species, Leaf Structures and Developmental Stages. Remote Sens. Environ. 2002, 81, 337–354. [Google Scholar] [CrossRef]
- Thenkabail, P.; Gumma, M.K.; Teluguntla, P.; Irshad Ahmed, M. Hyperspectral Remote Sensing of Vegetation and Agricultural Crops. Photogramm. Eng. Remote Sens. 2017, 80, 695–723. [Google Scholar]
- Nguyen, C.; Sagan, V.; Maimaitiyiming, M.; Maimaitijiang, M.; Bhadra, S.; Kwasniewski, M.T. Early Detection of Plant Viral Disease Using Hyperspectral Imaging and Deep Learning. Sensors 2021, 21, 742. [Google Scholar] [CrossRef]
- Cawley, G.C.; Talbot, N.L.C. Efficient Leave-One-out Cross-Validation of Kernel Fisher Discriminant Classifiers. Pattern Recognit. 2003, 36, 2585–2592. [Google Scholar] [CrossRef]
- Jackson, R.D. Remote Sensing of Biotic and Abiotic Plant Stress. Annu. Rev. Phytopathol. 1986, 24, 265–287. [Google Scholar] [CrossRef]
- Montero, R.; Pérez-Bueno, M.L.; Barón, M.; Florez-Sarasa, I.; Tohge, T.; Fernie, A.R.; Ouad, H.E.A.; Flexas, J.; Bota, J. Alterations in Primary and Secondary Metabolism in Vitis Vinifera ‘Malvasía de Banyalbufar’ upon Infection with Grapevine Leafroll-Associated Virus 3. Physiol. Plant. 2016, 157, 442–452. [Google Scholar] [CrossRef]
- Song, Y.; Hanner, R.H.; Meng, B. Probing into the Effects of Grapevine Leafroll-Associated Viruses on the Physiology, Fruit Quality and Gene Expression of Grapes. Viruses 2021, 13, 593. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, A.; Martins, V.; Frusciante, S.; Cruz, T.; Noronha, H.; Diretto, G.; Gerós, H. Flavescence Dorée-Derived Leaf Yellowing in Grapevine (Vitis vinifera L.) Is Associated to a General Repression of Isoprenoid Biosynthetic Pathways. Front. Plant Sci. 2020, 11, 896. [Google Scholar] [CrossRef] [PubMed]
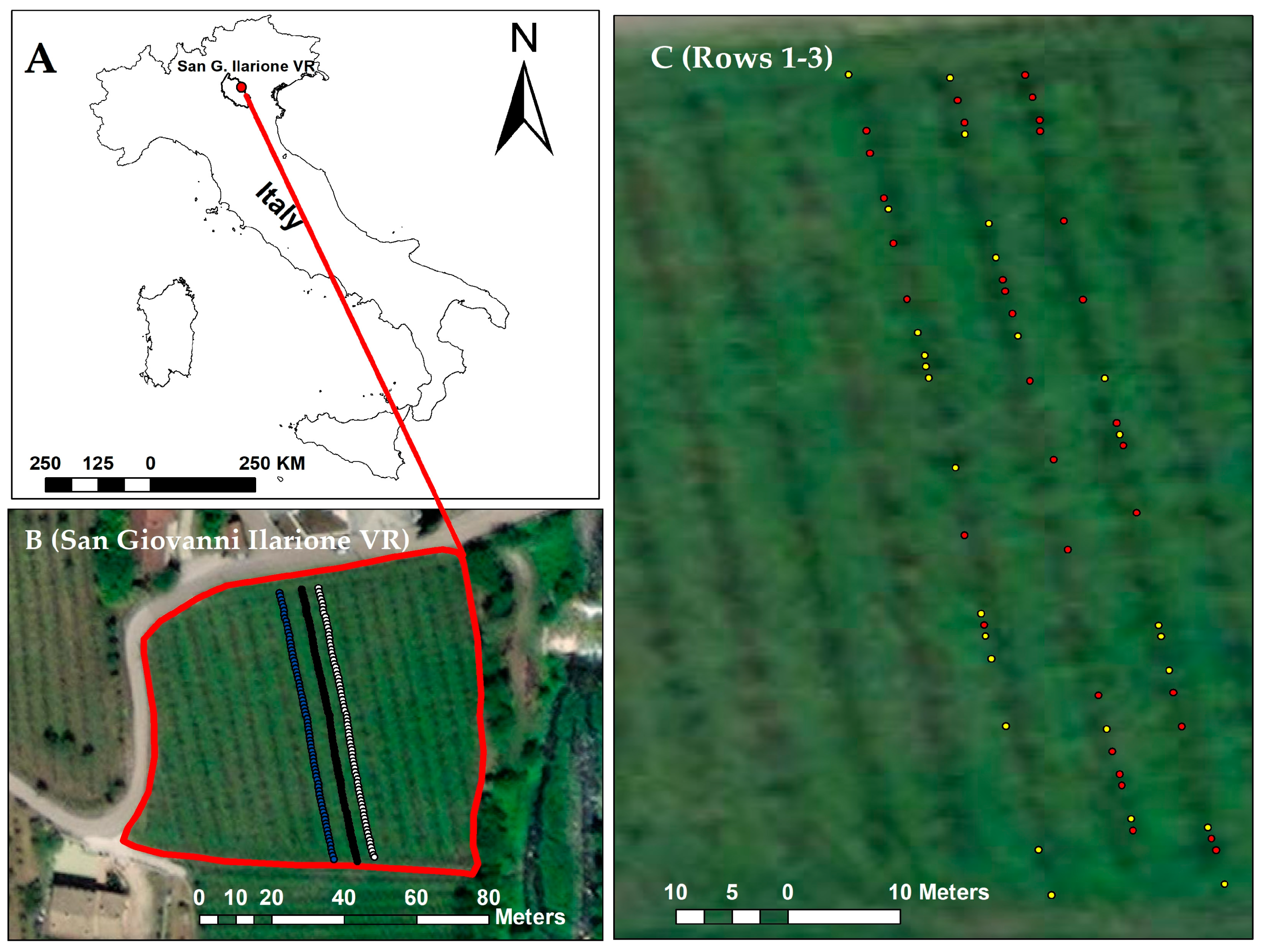




| Dates for Spectral Data Acquisition | Sensors | Vineyard Rows | No. of Samples | FD Positive Samples | FD Negative Samples |
|---|---|---|---|---|---|
| 8, 15, 21 June, 1 and 6 July 2022 | Hamamatsu (340–850 nm, 288 bands) and NIRScan sensor (900–1700 nm, 228 bands) | Row 1 | 20 | 07 | 13 |
| Row 2 | 20 | 13 | 07 | ||
| Row 3 | 20 | 13 | 07 | ||
| Total | 60 | 33 | 27 |
| Genetic Algorithm for Feature Selction |
|---|
| //Initialize variables , NbrTotalFeatures = 100, stop_criterion = False, α= 0.1, MaxNbrGeneration = 4000, N = TotalNumberOfGenes, = NbrOfPopulation // Initialize a number of binary chromosomes with random binary values of genes 1.Generate initial population While (stop_criterion ≠ True) { //compute the score of each chromosome in the population using fitness function 2.Evaluation //Evaluate the binary population chromosomes chosen genes For ∈ given that {1,.., } // For each binary chromosome in Population { // transoform the binary chosen chromoroses to the classification feature space =Train_features* // train classification model using 3-fold cross-validation = Train_model() //compute score function .nbrFeatures = Sum() = f (, .nbrFeatures, α, N) Equation (3) } end For 3. generate the progeny generation { //select the chromosomes allowed to reproduce the next generation based on the fitness function scores I. Selection //recombine chromosomes to choose which genes are transferred from parents to new progeny II. Crossover //random inversion in the selection progeny binary genes III. Mutation } 4. update with the new progeny generation k=k+1 // increment the generation tournament If (k >= MaxNbrGeneration) then { stop criterion ==True} } end While |
| Dates | Hamamatsu | NIRScan | ||||
|---|---|---|---|---|---|---|
| All Bands (288) | Features (100) Selected by Ensemble Method | Features Selected by GA Method | All Bands (228) | Features (100) Selected by Ensemble Method | Features Selected by GA Method | |
| 8 June 2022 | 67 | 72 | 61 (6) | 61 | 61 | 61 (10) |
| 15 June 2022 | 67 | 72 | 78 (9) | 61 | 67 | 61 (8) |
| 21 June 2022 | 72 | 78 | 61 (13) | 72 | 67 | 72 (6) |
| 1 July 2022 | 89 | 78 | 89 (13) | 61 | 61 | 61 (10) |
| 6 July 2022 | 61 | 61 | 61 (12) | 61 | 67 | 72 (7) |
| Dates | Hamamatsu | NIRScan | ||||
|---|---|---|---|---|---|---|
| All Bands (288) | Features (100) Selected by Ensemble Method | Features Selected by GA Method | All Bands (228) | Features (100) Selected by Ensemble Method | Features Selected by GA Method | |
| 8 June 2022 | 62 | 63 | 55 (10) | 58 | 57 | 55 (8) |
| 15 June 2022 | 62 | 85 | 63 (7) | 43 | 45 | 48 (18) |
| 21 June 2022 | 58 | 58 | 62 (8) | 60 | 62 | 63 (11) |
| 1 July 2022 | 72 | 77 | 72 (8) | 60 | 57 | 68 (3) |
| 6 July 2022 | 55 | 55 | 60 (7) | 55 | 55 | 63 (12) |
| Dates | Test Samples Accuracy | Test Samples Accuracy | ||||
|---|---|---|---|---|---|---|
| All Bands (516) | Features (100) Selected by Ensemble Method | Features Selected by GA Method | All Bands(516) | Features (100) Selected by Ensemble Method | Features Selected by GA Method | |
| 8 June 2022 | 61 | 67 | 67 (9) | 58 | 55 | 60 (14) |
| 15 June 2022 | 61 | 61 | 61 (8) | 55 | 58 | 60 (12) |
| 21 June 2022 | 67 | 61 | 72 (8) | 55 | 57 | 58 (15) |
| 1 July 2022 | 72 | 67 | 83 (7) | 72 | 77 | 72 (20) |
| 6 July 2022 | 67 | 67 | 72 (8) | 57 | 58 | 62 (20) |
| Dates | Sensors | Features | Classifiers | Accuracy (%) | Recall (%) | Precision (%) | F1 Score (%) |
|---|---|---|---|---|---|---|---|
| 1 July 2022 | Hamamatsu | GA (13) | LR | 89 | 100 | 85 | 92 |
| 21 June 2022 | NIRScan | GA (7) | RF | 72 | 91 | 71 | 80 |
| 1 July 2022 | Joined spectra | GA (7) | SVM, LR | 83 | 91 | 83 | 87 |
| Dates | Sensors | Features | Classifiers | Accuracy (%) | Recall (%) | Precision (%) | F1 Score (%) |
|---|---|---|---|---|---|---|---|
| 1 July 2022 | Hamamatsu | Ens method (100) | SVM | 85 | 88 | 85 | 87 |
| 21 June 2022 | NIRScan | GA (3) | GB | 68 | 88 | 66 | 75 |
| 1 July 2022 | Joined spectra | Ens method (100) | RF | 77 | 74 | 88 | 81 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Imran, H.A.; Zeggada, A.; Ianniello, I.; Melgani, F.; Polverari, A.; Baroni, A.; Danzi, D.; Goller, R. Low-Cost Handheld Spectrometry for Detecting Flavescence Dorée in Vineyards. Appl. Sci. 2023, 13, 2388. https://doi.org/10.3390/app13042388
Imran HA, Zeggada A, Ianniello I, Melgani F, Polverari A, Baroni A, Danzi D, Goller R. Low-Cost Handheld Spectrometry for Detecting Flavescence Dorée in Vineyards. Applied Sciences. 2023; 13(4):2388. https://doi.org/10.3390/app13042388
Chicago/Turabian StyleImran, Hafiz Ali, Abdallah Zeggada, Ivan Ianniello, Farid Melgani, Annalisa Polverari, Alice Baroni, Davide Danzi, and Rino Goller. 2023. "Low-Cost Handheld Spectrometry for Detecting Flavescence Dorée in Vineyards" Applied Sciences 13, no. 4: 2388. https://doi.org/10.3390/app13042388






