Abstract
Liver diseases are among the most common diseases worldwide. Because of the high incidence and high mortality rate, these diseases diagnoses are vital. Several elements harm the liver. For instance, obesity, undiagnosed hepatitis infection, and alcohol abuse. This causes abnormal nerve function, bloody coughing or vomiting, insufficient kidney function, hepatic failure, jaundice, and liver encephalopathy.. The diagnosis of this disease is very expensive and complex. Therefore, this work aims to assess the performance of various machine learning algorithms at decreasing the cost of predictive diagnoses of chronic liver disease. In this study, five machine learning algorithms were employed: Logistic Regression, K-Nearest Neighbor, Decision Tree, Support Vector Machine, and Artificial Neural Network (ANN) algorithm. In this work, we examined the effects of the increased prediction accuracy of Generative Adversarial Networks (GANs) and the synthetic minority oversampling technique (SMOTE). Generative opponents’ networks (GANs) are a mechanism to produce artificial data with a distribution close to real data distribution. This is achieved by training two different networks: the generator, which seeks to produce new and real samples, and the discriminator, which classifies the augmented samples using supervised classifications. Statistics show that the use of increased data slightly improves the performance of the classifier.
1. Introduction
The liver is the largest organ in the body and is essential for food digestion and for processing the body’s toxic substances. Induced by viruses and alcohol, liver damage is life-threatening. Hepatitis, cirrhosis, liver tumors, liver cancer, and but a few common hepatitis diseases. The main cause of death is liver failure and cirrhosis [1]. Liver disease is, therefore, one of the world’s biggest health problems. Chronic liver disease is a significant contributor to mortality rates globally. A range of elements that harm the liver are responsible for this disease, such as, obesity, undiagnosed hepatitis infection and alcohol abuse. The causes of liver disease can include abnormal nerve function, coughing up or vomiting blood, insufficient kidney function, liver failure, jaundice, and liver encephalopathy. The diagnosis of this disease is very expensive and complex. About two million die of liver disease worldwide annually [2].
In 2010, one million people died from cirrhosis of the liver, with millions suffering from liver cancer, according to the Global Burden of Disease (G.B.D.) project published in B.M.C. Medicine [3]. For the prediction and diagnosis of liver diseases, machine learning has significantly affected the biomedical field [4,5,6]. Machine learning guarantees improved detection and prediction of diseases of biomedical interest and also enhances the objective nature of decision making [7]. Medical problems can be easily solved, and diagnosis costs can be reduced by using machine learning techniques. The main aspect of this study is to predict the results more effectively and to reduce diagnostic costs in the medical sector. Therefore, for the classification of patients with liver disease or not, we have used different classification techniques. There were five machine learning techniques including Logistic Regression, K-Nearest Neighbor, Decision Tree, Support Vector Machine, and Artificial Neural Network (ANN); and different perspectives such as accuracy, precision, remembering, and f-1 score were assessed on the performance of these techniques. Data augmentation has been developed over the last decade by treating each table column as a random variable, modeling a joint multivariate probability distribution, and then sampling from that distribution.
Due to the performance and flexibility provided in representing data, the creation of generative models using Variational Autoencoder (VAEs) and, later, GANs and their numerous extensions [8,9,10,11] has been very effective. GANs are also used to generate tabular data, especially in healthcare. For example, [12] uses GANs to generate continuous time-series medical records, and [13] proposes using GANs to generate discrete tabular data. To generate heterogeneous non-time-series continuous and/or binary data, GAN combines an auto-encoder and a GAN [14]. ehrGAN [15] is a software that produces augmented medical records.
Desk GAN [16] is a convolutional neural network that attempts to solve the problem of generating synthetic data. Recent attention has been paid to the Generative Adversarial Networks (GANs) because of their improved quality of data in a broad range of applications. Real-world data is used to produce artificial data that appears and acts like actual data, which significantly enhances machine learning algorithms. By using this augmented data in the training on larger data volumes, the efficiency of prediction algorithms can be greatly improved [17]. GANs are neural networks which train generators as well as discriminators. These networks are involved in a cooperative game which teaches generators to produce artificial data from real world data with the same distribution as real-world data. The resulting data is sent to the discriminator, together with real data, which uses a conventional supervised grade to distinguish between real and amplified data. The effect of vanilla GANs on improving the performance of liver disease prediction is also investigated in this paper. Two sets of different sample numbers will be expanded in order to investigate the impact of increased data volume on results. Liver disease is then predicted using algorithms for machine learning. To evaluate the performance of the model, classification accuracy is used. Liver diseases are among the most common diseases worldwide.
This work is therefore aimed at assessing the performance of various algorithms of machine learning in order to decrease the cost of predictive diagnoses of chronic liver disease. Since GAN’s results in computer vision, text and speech are remarkable, we will investigate it as a possible means of addressing liver disease classification. We generate artificial data to overcome the problem of the lack of data when using the predictive model. Building a high-quality classification model is considered a challenging task when small amounts of data are involved. In this paper, we address the training data augmentation issue, where a Generative Adversarial Networks GAN is used as an augmentation technique. We augmented the data by doubling and tripling the number of samples. The goal of this augmentation is to increase the generalization performance and stabilize the classifier to better fit the trained model as well as avoiding overfitting with a small size of labelled samples.
This study explores applying and evaluating five classification algorithms on the Indian Liver Patient Dataset (ILPD). The goal is to assist medical professionals in the early diagnosis and screening of liver diseases by identifying liver patients from healthy individuals. The algorithms are compared based on their performance factors, and the impact of data augmentation using GAN is also investigated. The main objective of this research is to develop a classification model with good generalizability, meaning the ability to perform well on new, unseen data. Previous research has shown that models with low generalizability tend to overfit the training data, which is a significant challenge in developing accurate and reliable models.
This paper is divided into seven sections. This section has introduced the paper’s problem and methodology. Section 2 presents some of the previous work done in the fields of liver disease and data augmentation. Section 3 explains the GANs framework. Section 4 provides an E.D.A. of the dataset used in this work. Section 5 presents the proposed approach. Section 6 shows the employed data augmentation techniques. Section 7 presents the experiments and results. Section 8 concludes the paper.
2. Related Works
2.1. Liver Disease Diagnosis
The liver is the main laboratory of the human body. About 20 million chemical reactions per minute occur in this organ. Here, blood proteins are synthesized (for example, immunoglobulins responsible for the so-called humoral immunity of the whole organism, albumin, which hold the required volume of fluid in the bloodstream, and others), the synpaper of bile acids, which are substances necessary for the digestion of food in the small intestine, and the accumulation and breakdown of glucose as the main source of energy for the body [18]. The liver metabolizes fats and detoxifies toxins. The slightest violation of at least one of the functions of the liver leads to serious disturbances in the work of the whole organism.
In mild cases, acute hepatitis is practically asymptomatic, being detected only during random or targeted examination (for example, in the workplace among persons in contact with hepatotropic poisons, or in the household with regard to poisoning with mushrooms, etc.). In more severe cases (for example, with toxic hepatitis), the clinical symptoms of the disease develop rapidly, often in combination with signs of general intoxication and toxic damage to other organs and systems. During the disease, icteric coloration of the skin and mucous membranes, whitish-clay-colored stools, saturated dark-colored (“beer-colored”) urine, and hemorrhagic phenomena are characteristic. The color of the skin is orange or saffron. However, in mild cases of jaundice, the yellowing of the skin and eyes may only be noticeable in daylight. The first sign of jaundice is typically the yellowing of the whites of the eyes and the soft palate’s mucous membrane. Other symptoms may include frequent nosebleeds and petechiae, itching, slow heartbeat, depression, irritability, insomnia, and other signs of damage to central nervous system.
The liver and spleen are slightly enlarged and slightly painful in liver disease. Its size may decrease with especially severe lesions and the predominance of necrotic changes in the liver (acute dystrophy).
Laboratory tests may show elevated bilirubin levels (100–300 μmol/L or higher), increased activity of certain serum enzymes such as aldolase, aspartate aminotransferase, and alanine aminotransferase (levels higher than 40 units), lactate dehydrogenase, decreased albumin levels, and increased globulin levels. Additionally, there may be indicators of protein deposits in sedimentary samples (such as thymol and sublimate).
The liver’s production of fibrinogen, prothrombin, and VII and V coagulation factors is impaired because of hemorrhagic phenomena, considering the epidemiological situation in identifying the nature and cause of the disease. In unclear cases, first, one should think about viral hepatitis. Detection of the so-called Australian antigen is characteristic of serum hepatitis B (it is also detected in virus carriers, but rarely in other diseases x). Mechanical (subhepatic) jaundice usually occurs acutely only when a stone in cholelithiasis blocks the common bile duct. However, in this case, jaundice is preceded by an attack of biliary colic; bilirubin in the blood is mostly straight, and the stool is discolored. With hemolytic adrenal jaundice, free (indirect) bilirubin is determined in the blood, the stool is intensely colored, and the osmotic resistance of erythrocytes is usually reduced. In the case of false jaundice (due to skin staining with carotene with prolonged and abundant consumption of oranges, carrots, and pumpkin), the sclera is usually not colored, and hyperbilirubinemia is absent. Scanning the liver allows for the determination of its size; with hepatitis, sometimes there is a reduced or uneven accumulation of a radioisotope drug in the liver tissue. In some cases, increased accumulation in the spleen occurs.
The research by Jeyalakshmi et al. [19] focuses on developing a prediction system for liver disease using a specific type of machine learning algorithm called Convolutional Neural Network (CNN). The aim is to improve the accuracy of liver disease diagnosis by utilizing the CNN algorithm. The performance of the proposed system is evaluated and compared to other traditional machine learning methods. The results show that using CNN improves accuracy in predicting liver disease compared to other methods.
The research paper by Islam et al. [20] focuses on machine learning and deep learning methods to analyze the essential factors affecting liver disease and make predictions about the presence of liver disease. The study applies various algorithms such as decision trees, random forests, and deep neural networks to the Indian Liver Patient Dataset (I.L.P.D.) and compares their performance. The results showed that the deep neural network achieved the highest accuracy in predicting liver disease, and essential factors affecting liver disease were identified through feature importance analysis.
Sravani et al. [21] focuses on using machine learning algorithms to predict liver diseases. The study evaluates the performance of various classification algorithms, such as k-nearest neighbors, decision trees, random forests, and support vector machines in detecting liver diseases. The performance of these algorithms is evaluated in terms of accuracy, sensitivity, specificity, and area under the receiver operating characteristic curve. The results show that the random forest algorithm best-predicted liver maladies.
Belavigi et al. [22] proposed a comparison of Rprop and SAG on text datasets and CNN on image datasets. This study uses deep learning for early liver disease prediction. All three methods use liver-related text and image datasets as inputs. Accuracy is determined by comparing the models’ respective outputs. Compared to results using word datasets, image datasets are more reliable.
Singh et al. [23] designed software based on classification algorithms (including logistic regression, random forest, and naive Bayes) to predict the risk of liver disease from a data set with liver function test results.
Differential diagnosis in cases with a vivid clinical picture of diffuse liver damage should be carried out with liver cirrhosis. With cirrhosis, the symptoms of the disease are more pronounced, and the liver is usually much denser than with hepatitis; it can be increased but often also decreased in size (atrophic phase of cirrhosis). As a rule, splenomegaly is observed, hepatic signs (vascular telangiectasias, hepatic tongue, hepatic palms) are often detected, and symptoms of portal hypertension may occur. Laboratory studies show significant deviations from the norm in the results of the so-called liver tests, with puncture biopsy—disorganization of the liver structure and the significant proliferation of connective tissue.
2.2. Data Augmentation
Increasing the variety of data samples is an excellent way to improve the performance of machine learning algorithms. This technology is predominantly utilized in computer vision applications. Simple algorithms can be used, such as transformations on existing images (flipping, cropping, and rotation) [18,24]. However, these methods only modify the existing samples and do not generate new ones. To address this issue, researchers have begun adapting algorithms for increasing data more effectively in tabular data.
Synthetic data is a type of data that is generated by combining real-world characteristics and sampling from a distribution. Each approach to generating synthetic data has its unique advantages and disadvantages. Historically, synthetic data has been used for data anonymization and software testing, an is sometimes even intertwined. With the rise of Big Data and deep learning, privacy has become an increasingly important concern, making creating realistic synthetic data a critical aspect. A study published in [16] implements an uncontrolled data enhancement method in a semi-supervised learning environment to make the model more consistent when training both real and unmarked data. This method replaces random noise with noise that is more like the real world. This makes it easier to predict what will happen based on real data and predictions based on data that has been improved. Another study [25] presents a data enhancement method for training a convolutional neural network (CNN) known as random erasing.
However, this approach faces the challenge of occlusion, which is prevalent in using CNNs. To address this challenge, the study [24] proposes AutoAugment. This process identifies the most suitable data enhancement policy using reinforcement learning to find the optimal combination of decisions and feature order for maximum accuracy. Additionally, the authors of [26] propose the Fast AutoAugment algorithm, motivated by Bayesian data enhancement. This algorithm optimizes the search for missing data points and results in faster search times and improved error rates compared to AutoAugment. Meanwhile, the population-based increase (P.B.A.) algorithm replaces the fixed increase policy during training periods, producing the optimal schedule for each training period of increased policies. This algorithm requires less computing power and training time, as demonstrated in [27].
The current research on liver disease diagnosis using machine learning primarily focuses on using existing datasets and applying different algorithms for prediction. However, there needs to be more knowledge about using newly generated tabular data to improve liver disease diagnosis. This type of research would involve creating new datasets from relevant sources and using these datasets to train machine learning models for improved predictions. Creating these new datasets could involve data pre-processing, feature selection, and data generation techniques such as synthetic data generation and oversampling to overcome class imbalance issues. The use of newly generated tabular data has the potential to provide more accurate and robust predictions in liver disease diagnosis as compared to the use of existing datasets. Further research in this area can help fill this knowledge gap and advance the field of liver disease diagnosis using machine learning.
3. I.L.P.D. Dataset: Exploratory Data Analysis
E.D.A. is an iterative brainstorming around the dataset and has no strict instructions. One question can be asked based on ideas already formed prior to analysis, but another can be asked after a pattern or outlier has been noticed. This means that E.D.A. does not provide an exhaustive list of approaches available for data collection considering the small number of processes that can be taken to understand the data being studied.
3.1. Dataset Description
In this paper, the databases of 583 entries are obtained from the I.L.P.D. (Indian Liver Patient Dataset). Using the U.C.I. Machine Learning Repository (https://archive.ics.uci.edu/ml/index.php (accessed on 1 January 2023)), the dataset was downloaded successfully. Inside the I.L.P.D. dataset is thorough information regarding 583 Indian liver patients. In particular, 416 of these are liver patients, and the rest of the 167 are non-liver patients. The dataset was collected from the northeast of Andhra Pradesh in India. A class label called selector is used to split patients into liver and non-liver categories. It should be noted that patients of 89 years and older are specified as the age of “90”. Some information on the attributes are shown in Table 1.

Table 1.
Description of Liver patient dataset.
3.2. Exploratory Data Analysis
Most of the results we derived from this analysis confirm the research and other resources available (Kaggle website Indian liver disease analysis); such patterns must be confirmed in several sets to be beneficial for data understanding. Furthermore, more data allows us to zoom in on multiple facets in the data. In the analysis, E.D.A. is only a tool that gives the analyst insight into the nature of the data since it includes the word «explorational». There are stronger statistical procedures such as modelling and inference, which can help us make better use of data and draw more solid conclusions.
We have found that the results of blood tests may vary between men and women with liver disease. Without solid evidence, the conclusion could be reached that men are more severely affected by liver disease than women or that there is a sexual confounder (e.g., physiological differences or risk factors of the behavior) which causes the difference. On the other hand, gender does not seem to influence the presence or lack of liver disease, i.e., every gender has “shared” relatively positive and negative cases. It is worth noting, however, that the average age diagnosed as positive is higher in both women and men than the average age diagnosed as negative.
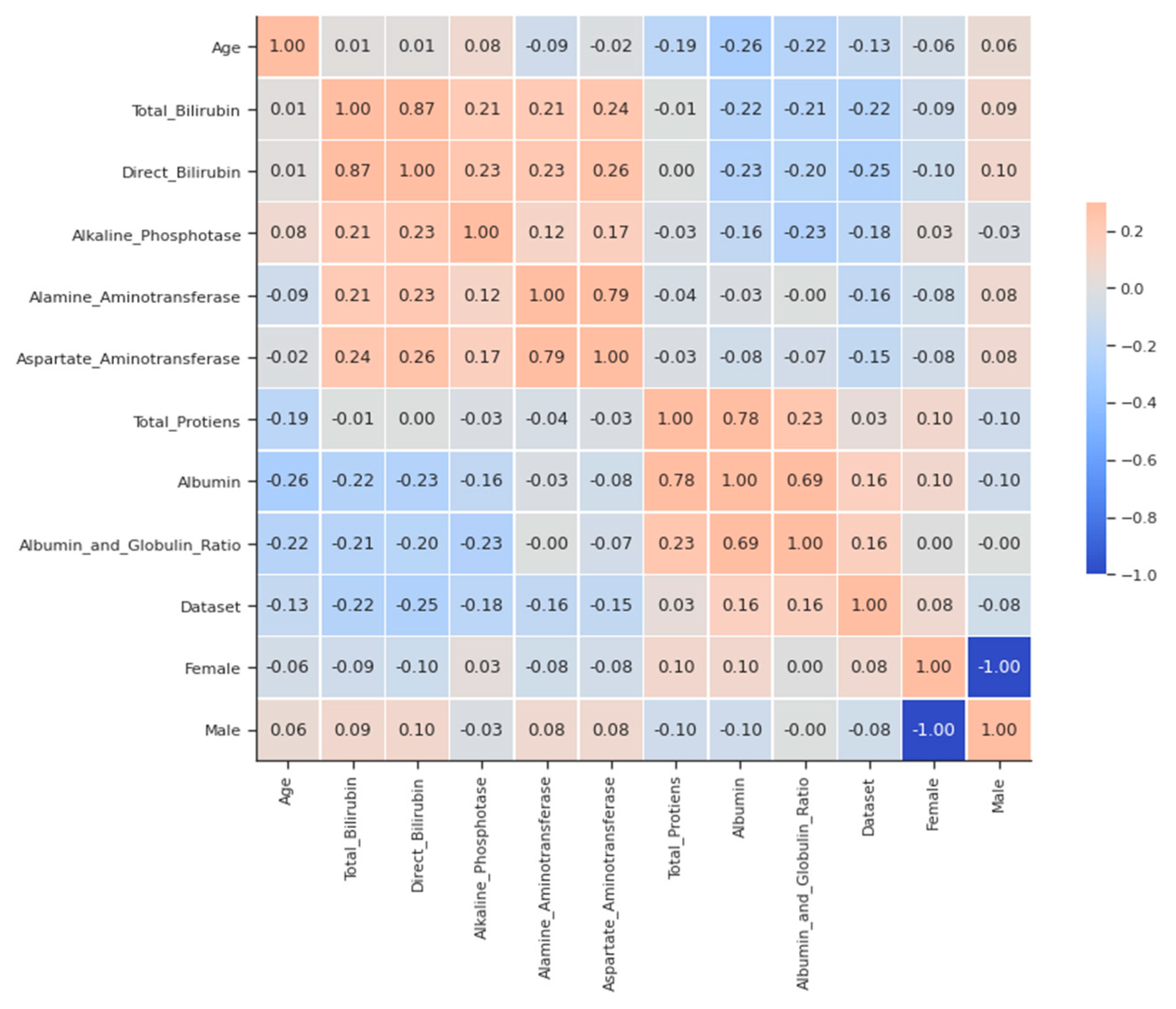
A matrix for correlation, as shown in Figure 1, can be an extremely concise way of examining links between numerical variables. Some of the surprisingly strong correlations between ALT (both liver enzymes), AST and total protein (the former is only the most plentiful), and albumin (being a protein in abundance) are direct bilirubins with total proteins. This is also high and not as large as other strong correlations because globulins introduce some variability into the ratio, which is not shared with albumin. The ratio is not as large.
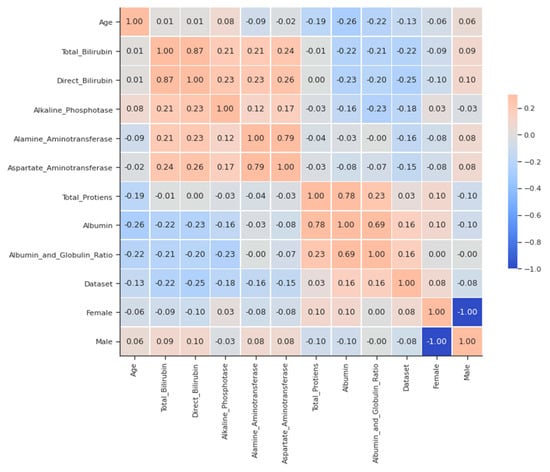
Figure 1.
Simple Correlation Plot- liver disease.
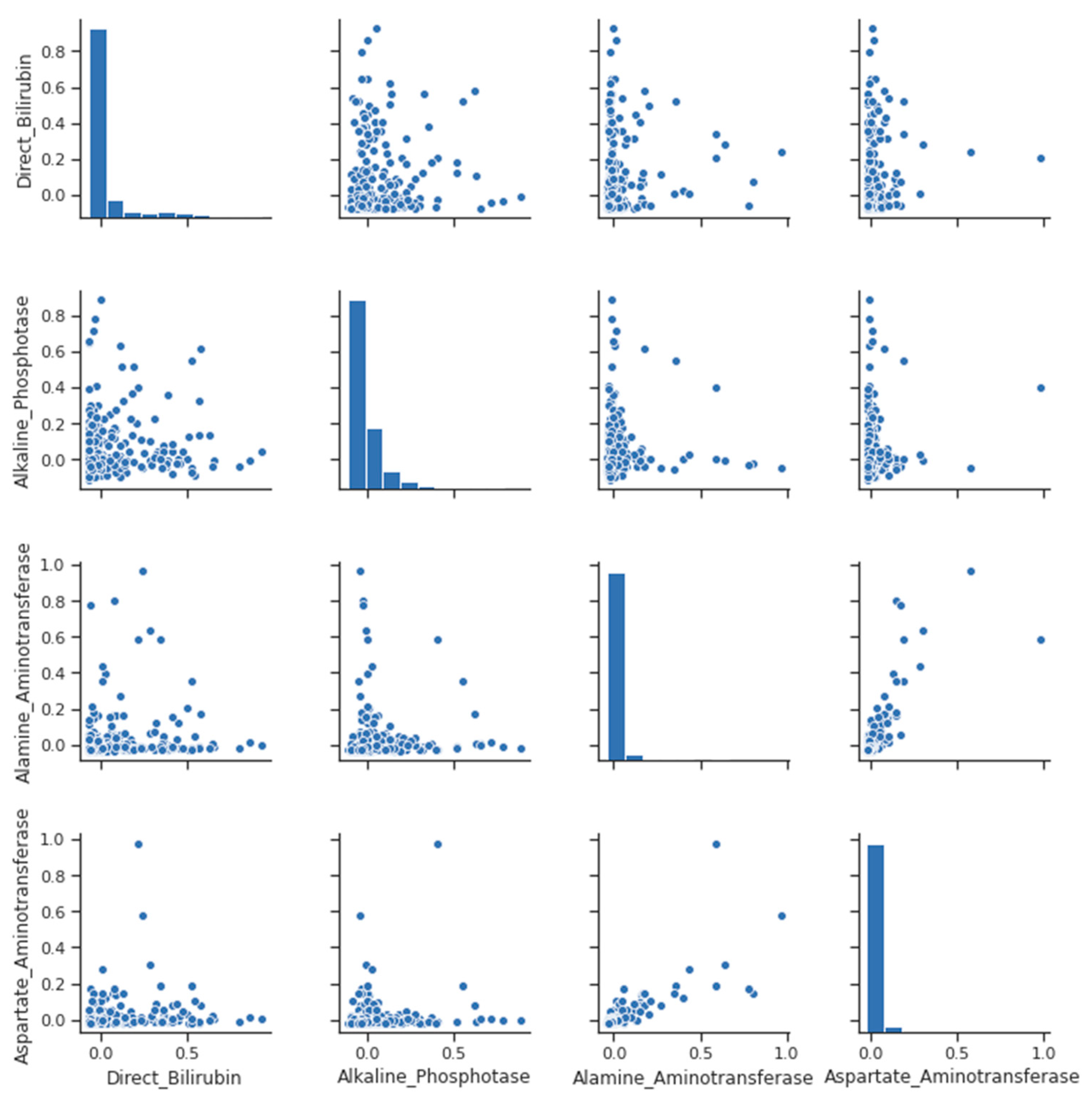
Other strong correlations exist between direct bilirubin and total bilirubin (as the former is only a component of the latter) and between AST and ALT (both liver enzymes) and albumin (which is the most abundant protein). This is also high and not as large as other strong correlations because globulins introduce some variability into the ratio that is not shared with albumin, as seen from the pairplot for clarification in Figure 2 and Figure 3.
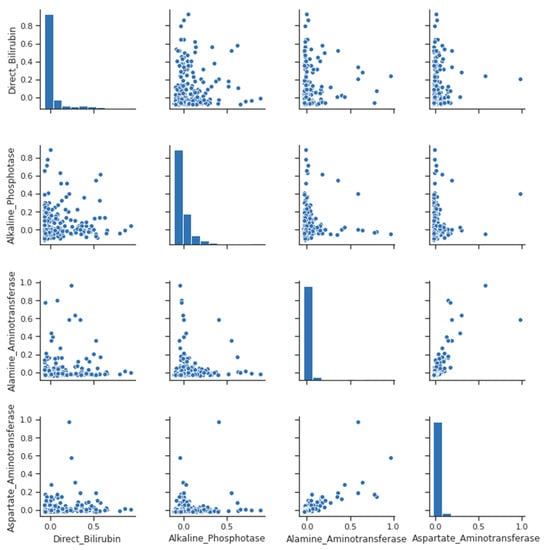
Figure 2.
Pair Plot- liver disease.
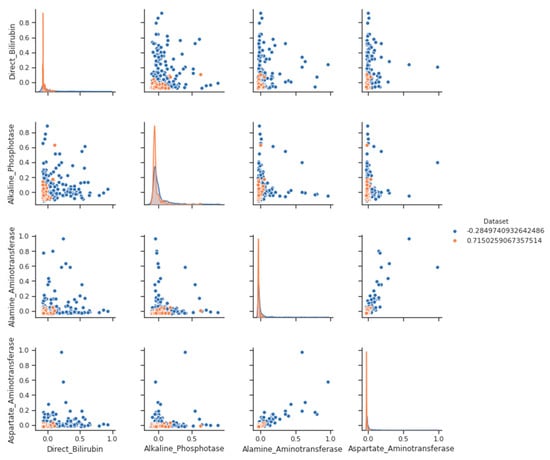
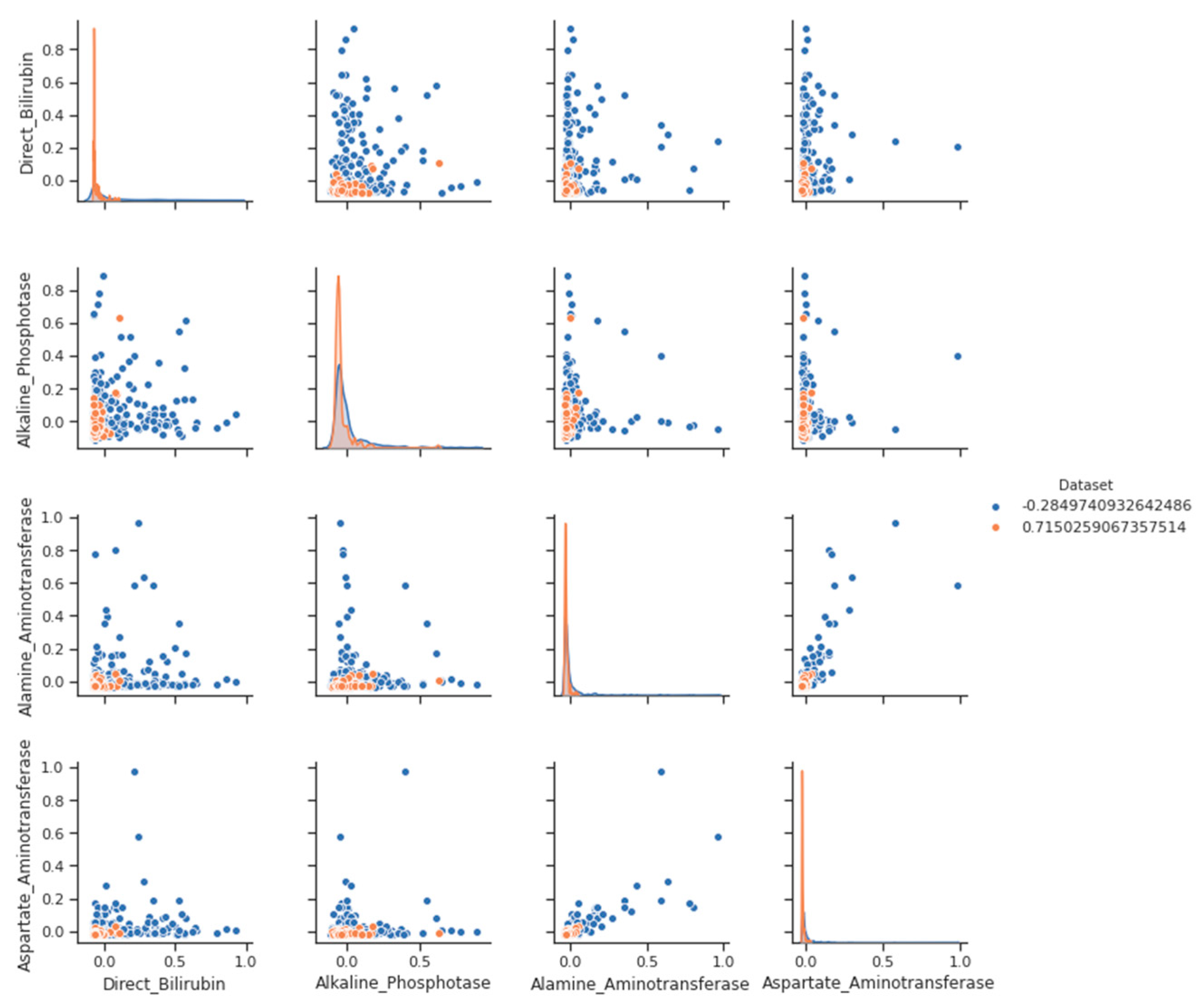
Figure 3.
Pair Plot- a liver disease with label categories.
A pair-plot is a graphical tool that allows visualizing the pairwise relationships between multiple variables in a dataset. Each variable is plotted against all the other variables in the dataset, resulting in a grid of scatterplots. A pair plot can help identify patterns and trends in the relationships between variables and is useful for exploring the structure of the data, as we can see in in Figure 2.
On the other hand, a pair plot with label categories Figure 3 is a variation of the basic pair plot that includes information about label categories. Label categories are categorical variables that provide additional information about each data point, such as group membership or a binary classification label.
We see a negative trend. Even weakly, higher values are associated with the lower A/G ratio of direct bilirubin. It makes sense because both the low A/G ratio and the high direct bilirubin indicate liver disease. After removing outliers, the coefficient of correlation between two variables may indicate a stronger correlation. It is important to note that outliers are not necessarily incorrect; they may represent a distinct group of observations that should be isolated rather than rejected.
4. Model Construction
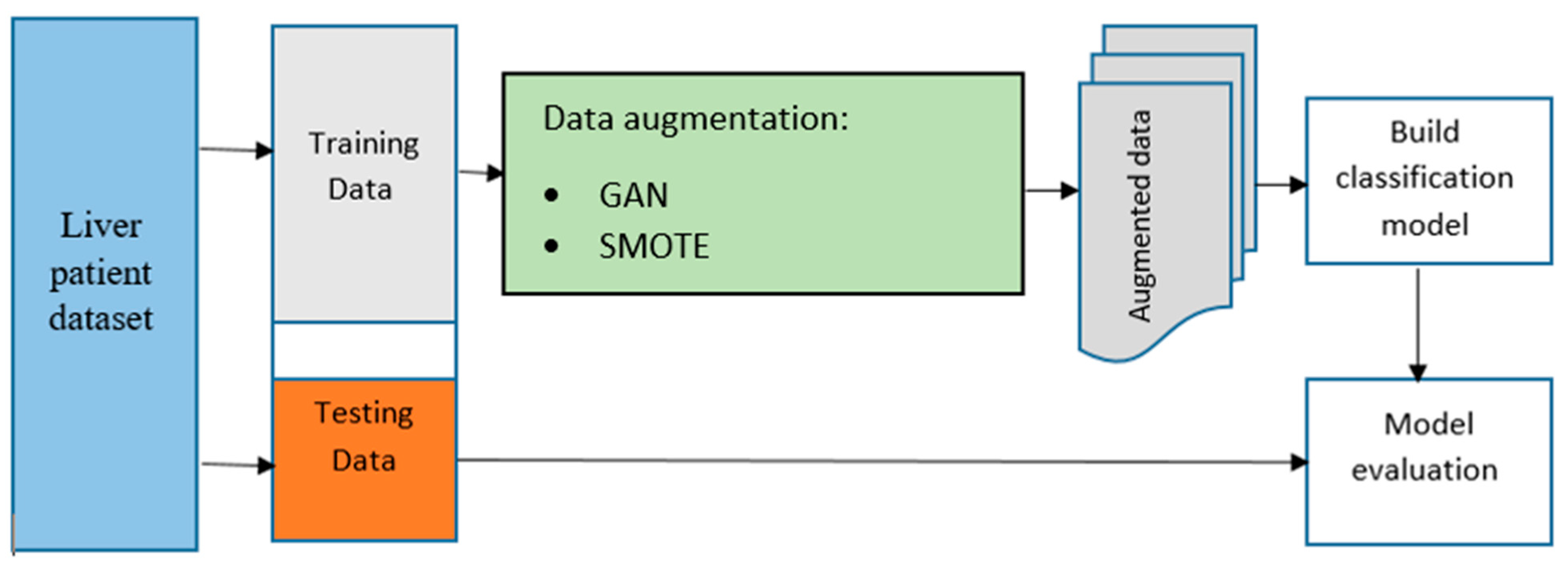
In Figure 4, we can see a flowchart that outlines the process for creating the model. This chart provides an overview of the system used to enhance the data and carry out classification. The augmented data generated from this system will be used to train and develop the final model using various machine-learning algorithms. To get started, the raw data is split into training and test data sets. The raw training data is then enhanced, and the raw and enhanced training data are combined to train the machine learning algorithms. The test data that was separated earlier is then used to evaluate the performance of the trained models. The GAN method was chosen in this research because it has shown high training stability and sample quality, as reported in previous studies. To make the predictive model better at identifying the decision boundaries between the two classes in the liver disease data set, the original sigmoid cross-entropy loss function for the discriminator was used during data augmentation. For comparison purposes, the SMOTE was also used in the study.
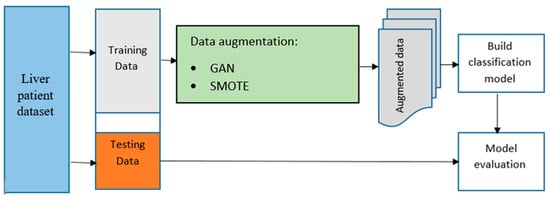
Figure 4.
The architecture of the proposed approach.
5. Classification Algorithms
5.1. Artificial Neural Networks (ANN)
Artificial Neural Networks (ANNs) are a class of machine learning algorithms inspired by the human brain’s structure and function [28,29]. They consist of interconnected nodes called artificial neurons, which process and transmit information. ANNs are highly flexible and can be used for many tasks, including image recognition, natural language processing, and prediction. They can also handle complex, non-linear relationships between inputs and outputs and learn from large amounts of data. However, ANNs can be prone to overfitting and require a significant amount of computational resources and careful tuning of their hyperparameters. Despite these limitations, ANNs have become a popular and powerful tool for many machine-learning applications due to their ability to model complex patterns in data.
5.2. Support Vector Machines (SVM)
Support Vector Machines (SVM) are a popular automated learning method used for classification and are linked to the latest algorithms [16,30,31]. This technique calculates the margin between classes and separates them with deliberate margins. The aim is to maximize the distance between classes, thereby reducing errors in the classification process. The SVM algorithm is highly effective in classifying non-linear data and is widely used in various domains such as text classification, image recognition, and bioinformatics [32].
5.3. Decision Trees (DT)
A decision tree predicts outcomes by dividing the input space into smaller subspaces through recursive partitioning. The instance tree method is a catalyst for the last node. It has its rules. It is referred to as the tree structure resemblance to the node number. In most cases, there are two options: a decision node or a leaf node. The data begins from the root and top of the node in the lower part of the data category. We adopt this classification type in this research, starting with the most common standard binary divisions. The C4.5 decision tree, distributed by Ross Quinlan in 1993, will also be used [33,34]. In addition to a rather traditional method, this method has the power and speed to express data structures. Moreover, the diversity of options has good effects on the grading process and the quality of its results.
5.4. K-Nearest Neighbours Algorithm (K.N.N.)
K-Nearest Neighbors (K.N.N.) is a widely used instance-based learning algorithm for classification and regression problems. It operates by finding the K closest points in the training set to a new data point, using a specified distance metric, and assigning the majority class label or the average target value of the nearest neighbors as the prediction for the new data point [30,35]. The strengths of K.N.N. include its simplicity, its ability to handle multi-class problems, and its suitability for large datasets. However, its performance can be affected by the choice of a distance metric, sensitivity to irrelevant features and the scale of the data, and the computational cost of finding the nearest neighbors. Despite its limitations, K.N.N. remains a popular and effective machine learning method.
5.5. Logistic Regression Classifier (L.R.)
The Logistic Regression Classifier (L.R.) [34,36] is a widely used supervised learning algorithm for binary and multi-class classification problems. It models the relationship between a set of input features and the probability of a binary outcome through a logistic function. L.R. optimizes its parameters by maximizing the likelihood of the observed class labels in the training data. The optimized model can then be used to make predictions on new data points by computing the estimated probability of each class and choosing the class with the highest probability as the prediction. L.R. has several advantages, including its simplicity, interpretability, and the availability of efficient algorithms for optimization. However, it can be sensitive to outliers and require the independence assumption between features. Despite these limitations, L.R. remains a widely used and effective method for binary and multi-class classification problems.
6. Data Augmentation Methods
6.1. Generative Adversarial Networks
In the area of data augmentation, generative models that include adversarial networks have recently come to be regarded as one of the most fascinating and potentially fruitful strategies. This is because the generation of images has significantly improved because of incorporating these models. GANs are superior to other algorithms in sample quality and usability across a broad variety of applications, including translating text to images. This is because GANs can learn from their missteps. It is important to train not just one but two deep neural networks in parallel for GANs to function properly. The first network, referred to as the generator, is in charge of producing synthetic data by imitating the training data distribution. The discriminator is the second component of a generative adversarial network (GAN) and its function is to differentiate between real and fake data by utilizing traditional supervised learning techniques [9]. The quality of the generated samples is evaluated by the discriminator to determine if they are sufficiently accurate to distinguish between genuine and false data. This determination is made by the network’s ability to determine whether the produced samples are of high enough quality. Algorithm 1 shows the GAN pseudocode.
| Algorithm 1: Generative Adversarial Networks (GANs) Algorithm |
| For number of training iterations do: |
| For K steps do: |
| Sample minibatch of m noise samples from noise prior |
| Sample minibatch of m examples from data-generating distribution |
| Update the discriminator by ascending it is stochastic gradient |
| End For |
| Sample minibatch of m noise samples from noise prior |
| Update the generator by descending it is stochastic gradient |
| End For |
6.2. Synthetic Minority Oversampling Technique
If the data are not evenly distributed, a problem known as unbalanced target classes may occur, in which classifiers used by machine learning systems may be biased in favor of one category over another. However, the distribution of classes that was supposed to occur differs significantly across datasets. As a result, we chose to make use of the Synthetic Minority Oversampling Method (SMOTE) to guarantee that all of the data was distributed fairly. SMOTE [37] produces synthetic data samples to increase the number of minority class data samples by first locating the K nearest neighbors, measuring the distance between them, and then increasing the distance by a random value between 0 and 1.
7. Experiments and Evaluation
The 10-fold cross-validation is widely used as the standard for conducting experiments. The data is divided into 10 equal parts, referred to as subsets, with one subset being used for testing while the rest are utilized for training. This process is repeated 10 times, with each subset being used for testing only once, ensuring that every instance in the feature matrix is included in both the testing and training phases. The results obtained from each iteration are then averaged to produce a single classification rate after being repeated 10 times.
7.1. Evaluation Performance Measures
The evaluation of the various classifiers on the original dataset before augmentation (No-AUG) after doubling the dataset (DD-AUG) and tripling the dataset (TD-AUG) was conducted using various measures. The Receiver Operating Characteristic (ROC) parameters, including True Positive (TP), False Negative (FN), False Positive (FP), and True Negative (TN), were utilized to evaluate and compare the performance. TP denotes the number of accurate positive diagnoses where the individual is classified as healthy and is indeed healthy; TN denotes the individual who is classified as infected and is indeed infected; and FP denotes the individual who is classified as infected but is healthy. FN denotes the individual who is classified as healthy but is infected. The ROC parameters demonstrate the consistency of the used classifier.
In addition to the ROC parameters, this study also employs the measures of Sensitivity, Precision, F1, and Specificity to evaluate the model. These performance metrics are derived from the confusion matrices and are calculated using the following equations
It is not easy to get all aspects of performance in a contingency table, because only half of its information is used to calculate PPV, N.P.V., Sensitivity and Specificity. The Matthews correlation coefficient (MMC) and the precision factor can benefit from four numbers (all existing numbers), providing a more comprehensive, more balanced and representative representation than if they are linear or vertical. In all of the measures that have been put forward here, we can generalize that a higher value is better, but the MCC method doesn’t follow this theory. If we take the values from 0 to 1 (0, −1, 1), 1 represents perfect correlation, 0 represents random distribution, and −1 represents perfect negative correlation. In risky cases, class imbalance affects the MCC and the level of accuracy. MCC has been developing slowly compared to others. Of 75% of cases predicted, MCC has achieved only 0.5. Based on random results, 50% of the cases that were predicted correctly (positive and negative) gave a value of 0, unlike other measures that provided 0.5, such as accuracy, N.P.V., Sensitivity, PPV and Specificity. It is reliable to examine performance in data that has been evenly distributed.
In this form, biases can be considered as deviations from the relationships. We must evaluate all measures to imagine an integrated form of how to perform a prediction. To obtain a clear picture of the performance of the prediction, we can use a receiver operating characteristics (R.O.C.) study. We can analyze this from finding the appropriate classifier. It also illustrates the tradeoffs between the Specificity on one hand and the Sensitivity on the other. There are special programs to draw R.O.C. curves, and we can rely on them when our predictor type is dependent on probabilities and provides a good degree of classification. However, we must be aware that this degree is not a real value for P, but it represents its reliable value in ranking for predictions.
Sensitivity or recall [38] is one of the reliable measures which are used to verify the ability of a model or system to retrieve the situation in a non-defect case. Depending on the formula’s calculations, the score for Sensitivity or recall is calculated between [0, 1] in an interval. However, for Specificity, it is one of the reliable measures to verify the ability of a model or system that cannot retrieve the condition in a non-defect case. Depending on the calculations of this formula, the score of Specificity is calculated between [0, 1] in an interval [39].
7.2. Experimental Results
In this work, various measures are applied to evaluate differential evolution’s performance and efficiency and to build upon different algorithms for supervised machine learning. This section focuses on how measures in different types and situations are implemented. The results of tests conducted to assess the proposed method’s effectiveness are presented, including a study of the impact of different distance methods on classification accuracy. Information about the performance of five different types of classifiers, including (ANN), (SVM), (L.R.), (D.T.), and K-nearest neighbor (K-NN) with data augmentation (NO-AUG), double data augmentation (DD-AUG), and triple data augmentation (TD-AUG), is provided in Table 2, Table 3, Table 4, Table 5, Table 6 and Table 7. All experimental results are based on the average of 10-fold cross-validation. The results of the SVM technique are presented in Table 2, and the experiments conducted with different data set increments showed little difference in results.

Table 2.
Evaluation of SVM Classifier for NO-AUG, DD-AUG and TD-AUG.

Table 3.
Evaluation of D.T. Classifier for NO-AUG, DD-AUG and TD-AUG.

Table 4.
Evaluation of kNN Classifier for NO-AUG, DD-AUG and TD-AUG.

Table 5.
Evaluation of L.R. Classifier for NO-AUG, DD-AUG and TD-AUG.

Table 6.
Evaluation of ANN Classifier for NO-AUG, DD-AUG and TD-AUG.

Table 7.
Comparison with other research work.
In Table 2, all cases that achieved approximately 0.71% accuracy are observed for all Class 2 cases, such as NO-AUG, DD-AUG, and TD-AUG. With regard to the F-measurement, the results are 0.83. The output ranged in precision from 0.71304 to 0.73041 for the TD-AUG sets. On the basis of these results, we can conclude that this classification does not have good results that make it an inappropriate way for people who are healthy to diagnose liver disease in terms of the different status of the patient. SMOTE, on the other hand, outperforms GAN in terms of accuracy, with an increase ranging from 12 to 22%.
Nonetheless, GAN outperformed SMOTE in terms of precision recall and F-measure. In light of this, GAN outperforms SMOTE in terms of model stability. Notably, the accuracy increase with data augmentation is quite stable. For clarity, accuracy improves somewhat as data creation progresses. Additionally, the average of evaluation metrics was calculated to emphasize the performance of data augmentation techniques. The findings reveal that SMOTE surpasses GAN with an average accuracy of 93% across all data augmentation cases.
In Table 3, all cases achieved around 0.60% of accuracy are observed, ad for all Class 2 cases NO-AUG, DD-AUG and TD-AUG we note the decrease in the accuracy of the samples. As regards F-measurement, the results are 0.74. The output ranged in precision from 0.73105 to 0.76553 for the TD-AUG sets. Based on these findings, we may deduce that this classification does not provide satisfactory outcomes and is hence unsuitable for use by healthy individuals in the diagnosis of liver disease in patients with varying degrees of illness. SMOTE, on the other hand, is superior to additional data augmentation in terms of its ability to enhance accuracy. Additionally, improvements may be noticed in terms of accuracy, recall, and F-measure. SMOTE’s average accuracy of 97% is a gain in accuracy of 38% when compared to GAN’s average accuracy of 59%.
In Table 4, all cases that achieved approximately 0.69% of accuracy are observed, ad for all Class 2 cases, such as NO-AUG, DD-AUG, and TD-AUG, we note the decrease in the accuracy of the samples. With regard to F-measurement, the results are 0.79. The output ranged in precision from 0.74889 to 0.77001 for the TD-AUG sets. Based on these data, it appears that the existing classification scheme is inappropriate for discriminating between the various phases of liver disease in otherwise healthy individuals. However, in basic data augmentation scenarios, SMOTE exhibits a greater improvement in outcomes than GAN, with an average accuracy of 99%.
Table 5 shows that all cases achieved around 0.75% of accuracy are observed, and for all Class 2 cases, including NO-AUG, DD-AUG, and TD-AUG we note the decrease in the accuracy of the samples. As regards F-measurement, results are 0.84. The output ranged in precision from 0.71306 to 0.71786 for the TD-AUG sets. Given these data, we can conclude that this classification is an inaccurate method for identifying liver sickness in individuals who are otherwise healthy. This is because it fails to account for individual variations in liver function. On the other hand, in comparison to GAN, SMOTE demonstrates better stability in terms of improvement in accuracy. Despite this, we found that employing L.R. with SMOTE performs much better than GAN, with an average accuracy of 98% across all cases.
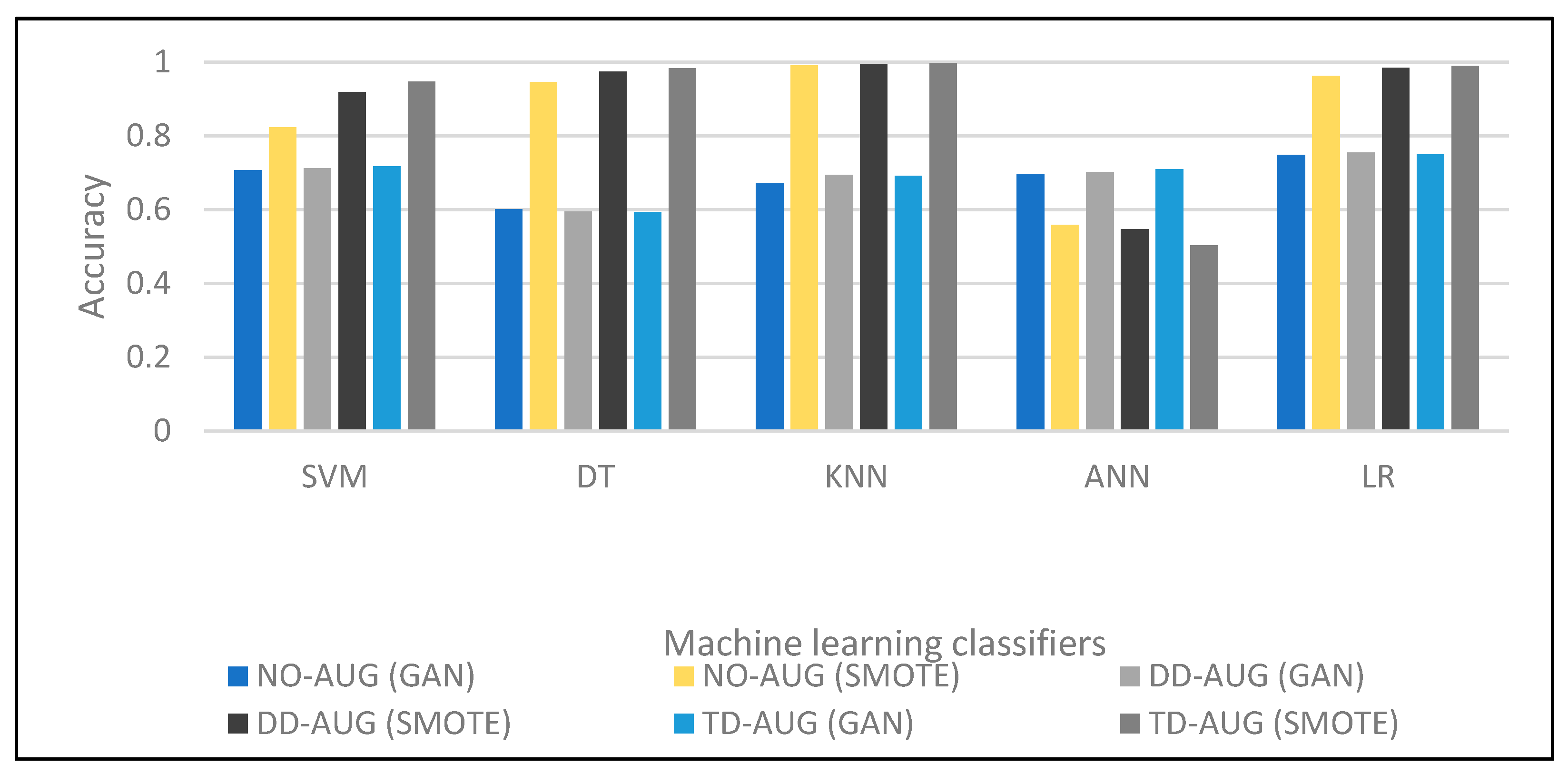
In Table 6, all cases that achieved around 0.71% of accuracy are observed, and for all Class 2 cases NO-AUG, DD-AUG, and TD-AUG, we note the increase in the accuracy of the samples. With regard to F-measurement, the results are 0.80. The output ranged in precision from 0.75004 to 0.75471 for the TD-AUG sets. Although GAN improves accuracy, SMOTE shows a loss in overall accuracy. To the best of our understanding, the higher the quantity of data, the larger the classifier size should be to anticipate better performance. In this study, we use ANN with 10 epochs. As a result, we concluded that ANN performs better with GAN overall data sizes, even with the same neural network size. ANN is also more stable when utilizing GAN. Based on the data, it can be determined that the classification does not yield satisfactory results, thus indicating that SMOTE is an inadequate approach for diagnosing liver disease in individuals considered healthy. The accuracy of the five machine classifiers is compared in Figure 5. The figure shows that most classifiers’ classification performance improves for double and triple data augmentation (DD-AUG, TD-AUG). In addition, it shows that SMOTE outperforms GAN in overall data augmentation cases.
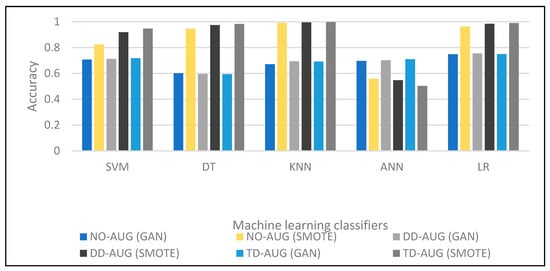
Figure 5.
Performance comparison of different machine learning techniques.
The comparison of the results in Table 7 shows that the proposed approach has the highest accuracy among all the other approaches for liver disease prediction. The proposed approach uses multiple classification algorithms such as ANN, SVM, L.R., D.T., and K-NN, and has improved accuracy by applying SMOTE on the dataset. The highest accuracy of 0.9872 is achieved using Logistic Regression with SMOTE. All of the other algorithms also show good accuracy with SMOTE. On the other hand, in the other studies, the highest accuracy achieved is approximately 90% using a Convolutional Neural Network (CNN), and the highest accuracy among the traditional machine learning algorithms is around 78% using K-NN. The results suggest that the proposed approach is better in terms of accuracy compared to other approaches.
8. Conclusions
A growing number of people are developing liver illness as a result of the use of excessive amounts of alcohol, the inhaling of gas, and the consumption of tainted food, pickles, and medicines. The earlier a liver diagnosis is made, the better the prognosis. Liver disease can be diagnosed based on blood enzyme levels. Diagnosing this condition is a difficult and time-consuming process that can be highly expensive. This work is consequently geared toward evaluating the performance of various machine learning algorithms to reduce the cost of prediction diagnostics of chronic liver disease. We used five different logistic regression techniques, as well as the K-Nearest neighbor algorithm, the Decision Tree methodology, the Support Vector Machine algorithm, and the ANN algorithm. In this study, we investigated how an improvement in the accuracy of predictions made by Generative Adversarial Networks (GANs) and a technique called synthetic minority oversampling impacted the results (SMOTE). The experimental results demonstrate that SMOTE surpasses GAN in its effectiveness when utilizing the proposed classifiers across all data augmentation scenarios (NO-AUG, DD-AUG, and TD-AUG). Furthermore, we found that K.N.N. outperforms others with an average accuracy of 99%. However, GAN results demonstrate better model stability when compared to SMOTE. As a future direction, we intend to experiment with other cost-sensitive data resampling techniques and compare their performances with GANs over larger data augmentation.
Author Contributions
Conceptualization, M.A. and Y.A.-S.; Methodology, A.A., A.A.-q. and M.A.; Resources, F.A.; Writing—original draft, A.A.-q. and Y.A.-S.; Writing—review & editing, A.M.A., B.A. and O.R.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The author would like to thank the Deanship of Scientific Research at Shaqra University for supporting this research.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Lin, R.-H. An intelligent model for liver disease diagnosis. Artif. Intell. Med. 2009, 47, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Maddrey, W.C.; Sorrell, M.F.; Schiff, E.R. Schiff’s Diseases of the Liver; John Wiley & Sons: Hoboken, NJ, USA, 2011. [Google Scholar]
- Oniśko, A.; Druzdzel, M.J.; Wasyluk, H. Learning Bayesian network parameters from small data sets: Application of Noisy-OR gates. Int. J. Approx. Reason. 2001, 27, 165–182. [Google Scholar] [CrossRef]
- Babu, M.S.P.; Ramana, B.V.; Kumar, B.R.S. New automatic diagnosis of liver status using bayesian classification. In Proceedings of the International Conference on Intelligent Network and Computing) ICINC, Kuala Lumpur, Malaysia, 26–28 November 2010. [Google Scholar]
- Domingos, P. Metacost: A general method for making classifiers cost-sensitive. In Proceedings of the Fifth ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Diego, CA USA, 15–18 August 1999; pp. 155–164. [Google Scholar]
- Ramana, B.V.; Babu, M.S.P.; Venkateswarlu, N. A critical study of selected classification algorithms for liver disease diagnosis. Int. J. Database Manag. Syst. 2011, 3, 101–114. [Google Scholar] [CrossRef]
- Kim, S.; Jung, S.; Park, Y.; Lee, J.; Park, J. Effective liver cancer diagnosis method based on machine learning algorithm. In Proceedings of the 2014 7th International Conference on Biomedical Engineering and Informatics, Dalian, China, 14–16 October 2014; pp. 714–718. [Google Scholar]
- Al-Qerem, A.; Alsalman, Y.S.; Mansour, K. Image Generation Using Different Models of Generative Adversarial Network. In Proceedings of the 2019 International Arab Conference on Information Technology (ACIT), Al Ain, United Arab Emirates, 3–5 December 2019; pp. 241–245. [Google Scholar]
- Al-Qerem, A.; Kharbat, F.; Nashwan, S.; Ashraf, S.; Blaou, K. General model for best feature extraction of EEG using discrete wavelet transform wavelet family and differential evolution. Int. J. Distrib. Sens. Netw. 2020, 16, 1550147720911009. [Google Scholar] [CrossRef]
- Al-Qerem, A. An efficient machine-learning model based on data augmentation for pain intensity recognition. Egypt. Inform. J. 2020, 21, 241–257. [Google Scholar] [CrossRef]
- Arjovsky, M.; Bottou, L. Towards principled methods for training generative adversarial networks. arXiv 2017, arXiv:1701.04862. [Google Scholar]
- Borji, A. Pros and cons of gan evaluation measures. Comput. Vis. Image Underst. 2019, 179, 41–65. [Google Scholar] [CrossRef]
- Ho, D.; Liang, E.; Chen, X.; Stoica, I.; Abbeel, P. Population based augmentation: Efficient learning of augmentation policy schedules. In Proceedings of the International Conference on Machine Learning, Long Beach, CA, USA, 9–15 June 2019; pp. 2731–2741. [Google Scholar]
- Perez, L.; Wang, J. The effectiveness of data augmentation in image classification using deep learning. arXiv 2017, arXiv:1712.04621. [Google Scholar]
- Che, Z.; Cheng, Y.; Zhai, S.; Sun, Z.; Liu, Y. Boosting deep learning risk prediction with generative adversarial networks for electronic health records. In Proceedings of the 2017 IEEE International Conference on Data Mining (ICDM), New Orleans, LA, USA, 18–21 November 2017; pp. 787–792. [Google Scholar]
- Pradhan, A. Support vector machine-a survey. Int. J. Emerg. Technol. Adv. Eng. 2012, 2, 82–85. [Google Scholar]
- Radford, A.; Metz, L.; Chintala, S. Unsupervised representation learning with deep convolutional generative adversarial networks. arXiv 2015, arXiv:1511.06434. [Google Scholar]
- Al-Qerem, A.; Salem, A.A.; Jebreen, I.; Nabot, A.; Samhan, A. Comparison between Transfer Learning and Data Augmentation on Medical Images Classification. In Proceedings of the 2021 22nd International Arab Conference on Information Technology (ACIT), Muscat, Oman, 21–23 December 2021; pp. 1–7. [Google Scholar]
- Jeyalakshmi, K.; Rangaraj, R. Accurate liver disease prediction system using convolutional neural network. Indian J. Sci. Technol. 2021, 14, 1406–1421. [Google Scholar] [CrossRef]
- Islam, M.K.; Alam, M.M.; Rony, M.R.A.H.; Mohiuddin, K. Statistical Analysis and Identification of Important Factors of Liver Disease using Machine Learning and Deep Learning Architecture. In Proceedings of the 2019 3rd International Conference on Innovation in Artificial Intelligence, Suzhou, China, 15–18 March 2019; pp. 131–137. [Google Scholar]
- Sravani, K.; Anushna, G.; Maithraye, I.; Chetan, P.; Yeruva, S. Prediction of Liver Malady Using Advanced Classification Algorithms. In Machine Learning Technologies and Applications: Proceedings of ICACECS 2020; Springer: Singapore, 2021; pp. 39–49. [Google Scholar]
- Belavigi, D.; Veena, G.; Harekal, D. Prediction of liver disease using Rprop, SAG and CNN. Int. J. Innov. Technol. Expl. Eng. IJITEE 2019, 8, 3290–3295. [Google Scholar]
- Singh, J.; Bagga, S.; Kaur, R. Software-based prediction of liver disease with feature selection and classification techniques. Procedia Comput. Sci. 2020, 167, 1970–1980. [Google Scholar] [CrossRef]
- Shorten, C.; Khoshgoftaar, T.M. A survey on image data augmentation for deep learning. J. Big Data 2019, 6, 1–48. [Google Scholar] [CrossRef]
- Salimans, T.; Goodfellow, I.; Zaremba, W.; Cheung, V.; Radford, A.; Chen, X. Improved techniques for training gans. Adv. Neural Inf. Process. Syst. 2016, 29, 2234–2242. [Google Scholar]
- Tran, T.; Pham, T.; Carneiro, G.; Palmer, L.; Reid, I. A bayesian data augmentation approach for learning deep models. Adv. Neural Inf. Process. Syst. 2017, 30, 2794–2803. [Google Scholar]
- Turhan, C.G.; Bilge, H.S. Recent trends in deep generative models: A review. In Proceedings of the 2018 3rd International Conference on Computer Science and Engineering (UBMK), Sarajevo, Bosnia and Herzegovina, 20–23 September 2018; pp. 574–579. [Google Scholar]
- Zou, J.; Han, Y.; So, S.-S. Overview of artificial neural networks. Artif. Neural Netw. 2008, 458, 14–22. [Google Scholar]
- Ecer, F.; Ardabili, S.; Band, S.S.; Mosavi, A. Training Multilayer Perceptron with Genetic Algorithms and Particle Swarm Optimization for Modeling Stock Price Index Prediction. Entropy 2020, 22, 1239. [Google Scholar] [CrossRef]
- Bansal, M.; Goyal, A.; Choudhary, A. A comparative analysis of K-Nearest Neighbor, Genetic, Support Vector Machine, Decision Tree, and Long Short Term Memory algorithms in machine learning. Decis. Anal. J. 2022, 3, 100071. [Google Scholar] [CrossRef]
- Xia, D.; Tang, H.; Sun, S.; Tang, C.; Zhang, B. Landslide Susceptibility Mapping Based on the Germinal Center Optimization Algorithm and Support Vector Classification. Remote Sens. 2022, 14, 2707. [Google Scholar] [CrossRef]
- Awad, M.; Khanna, R. Support vector machines for classification. In Efficient Learning Machines; Apress: Berkeley, CA, USA, 2015; pp. 39–66. [Google Scholar]
- Osei-Bryson, K.-M. Evaluation of decision trees: A multi-criteria approach. Comput. Oper. Res. 2004, 31, 1933–1945. [Google Scholar] [CrossRef]
- Saxena, R.; Sharma, S.K.; Gupta, M.; Sampada, G.C. A Novel Approach for Feature Selection and Classification of Diabetes Mellitus: Machine Learning Methods. Comput. Intell. Neurosci. 2022, 2022, 3820360. [Google Scholar] [CrossRef] [PubMed]
- Kataria, A.; Singh, M. A review of data classification using k-nearest neighbour algorithm. Int. J. Emerg. Technol. Adv. Eng. 2013, 3, 354–360. [Google Scholar]
- Lemon, S.C.; Roy, J.; Clark, M.A.; Friedmann, P.D.; Rakowski, W. Classification and regression tree analysis in public health: Methodological review and comparison with logistic regression. Ann. Behav. Med. 2003, 26, 172–181. [Google Scholar] [CrossRef]
- Fernández, A.; Garcia, S.; Herrera, F.; Chawla, N.V. SMOTE for learning from imbalanced data: Progress and challenges, marking the 15-year anniversary. J. Artif. Intell. Res. 2018, 61, 863–905. [Google Scholar] [CrossRef]
- Laakso, M.; Soininen, H.; Partanen, K.; Lehtovirta, M.; Hallikainen, M.; Hänninen, T.; Helkala, E.-L.; Vainio, P.; Riekkinen, P. MRI of the hippocampus in Alzheimer’s disease: Sensitivity, specificity, and analysis of the incorrectly classified subjects. Neurobiol. Aging 1998, 19, 23–31. [Google Scholar] [CrossRef]
- Sokolova, M.; Japkowicz, N.; Szpakowicz, S. Beyond accuracy, F-score and ROC: A family of discriminant measures for performance evaluation. In Proceedings of the Australasian joint conference on artificial intelligence, Hobart, Australia, 4–8 December 2006; pp. 1015–1021. [Google Scholar]
- Dritsas, E.; Trigka, M. Supervised Machine Learning Models for Liver Disease Risk Prediction. Computers 2023, 12, 19. [Google Scholar] [CrossRef]
- Behera, M.P.; Sarangi, A.; Mishra, D.; Sarangi, S.K. A Hybrid Machine Learning algorithm for Heart and Liver Disease Prediction Using Modified Particle Swarm Optimization with Support Vector Machine. Procedia Comput. Sci. 2023, 218, 818–827. [Google Scholar] [CrossRef]
- Mostafa, F.; Hasan, E.; Williamson, M.; Khan, H. Statistical Machine Learning Approaches to Liver Disease Prediction. Livers 2021, 1, 294–312. [Google Scholar] [CrossRef]
- Wu, C.-C.; Yeh, W.-C.; Hsu, W.-D.; Islam, M.M.; Nguyen, P.A.; Poly, T.N.; Wang, Y.-C.; Yang, H.-C.; Li, Y.-C. Prediction of fatty liver disease using machine learning algorithms. Comput. Methods Programs Biomed. 2019, 170, 23–29. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).