Model-Based Approach for the Semi-Automatic Analysis of Collagen Birefringence in Polarized Light Microscopy
Abstract
Featured Application
Abstract
1. Introduction
2. Materials and Methods
2.1. Birefringence Physical Model
2.2. Data Preprocessing

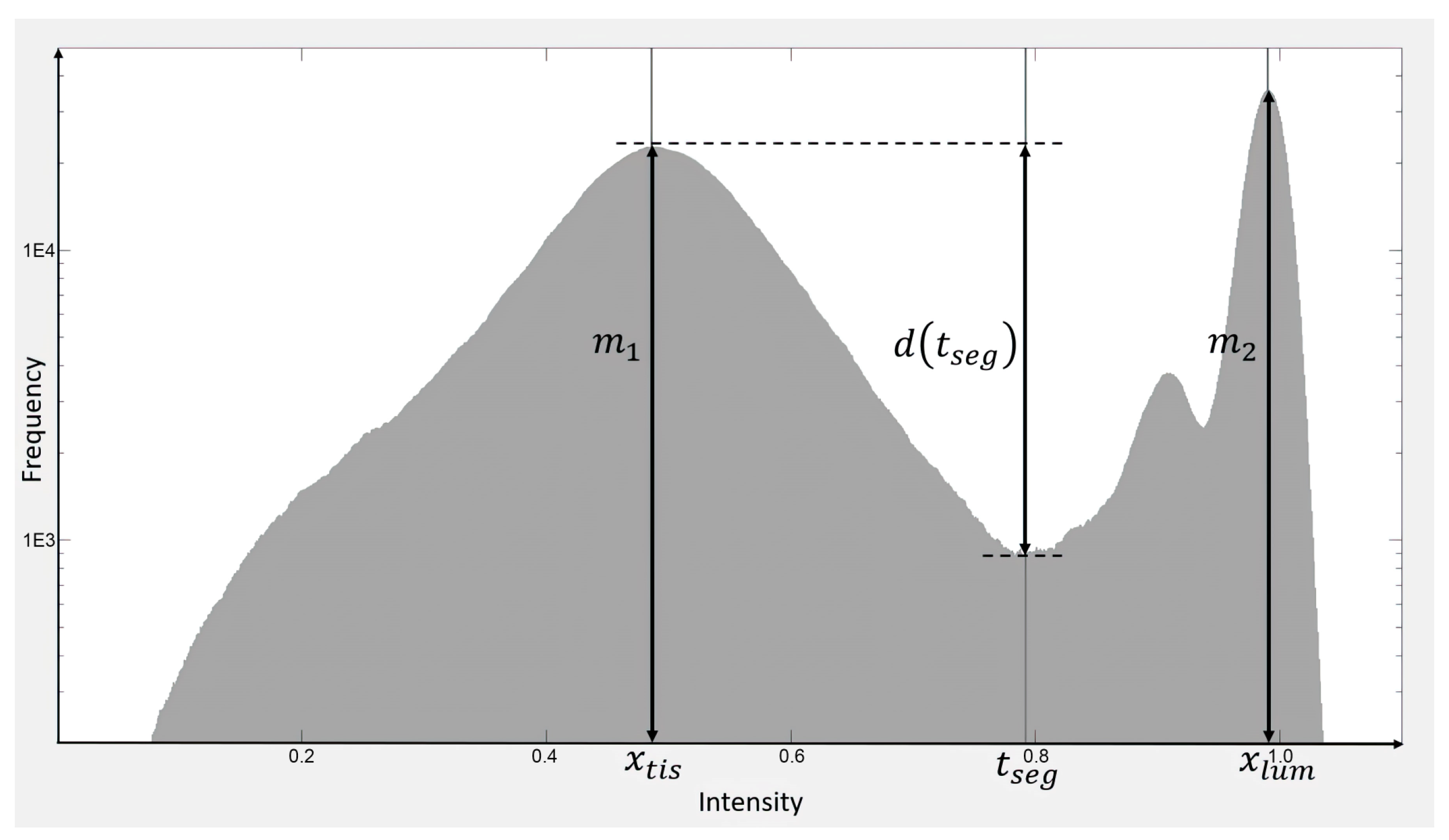
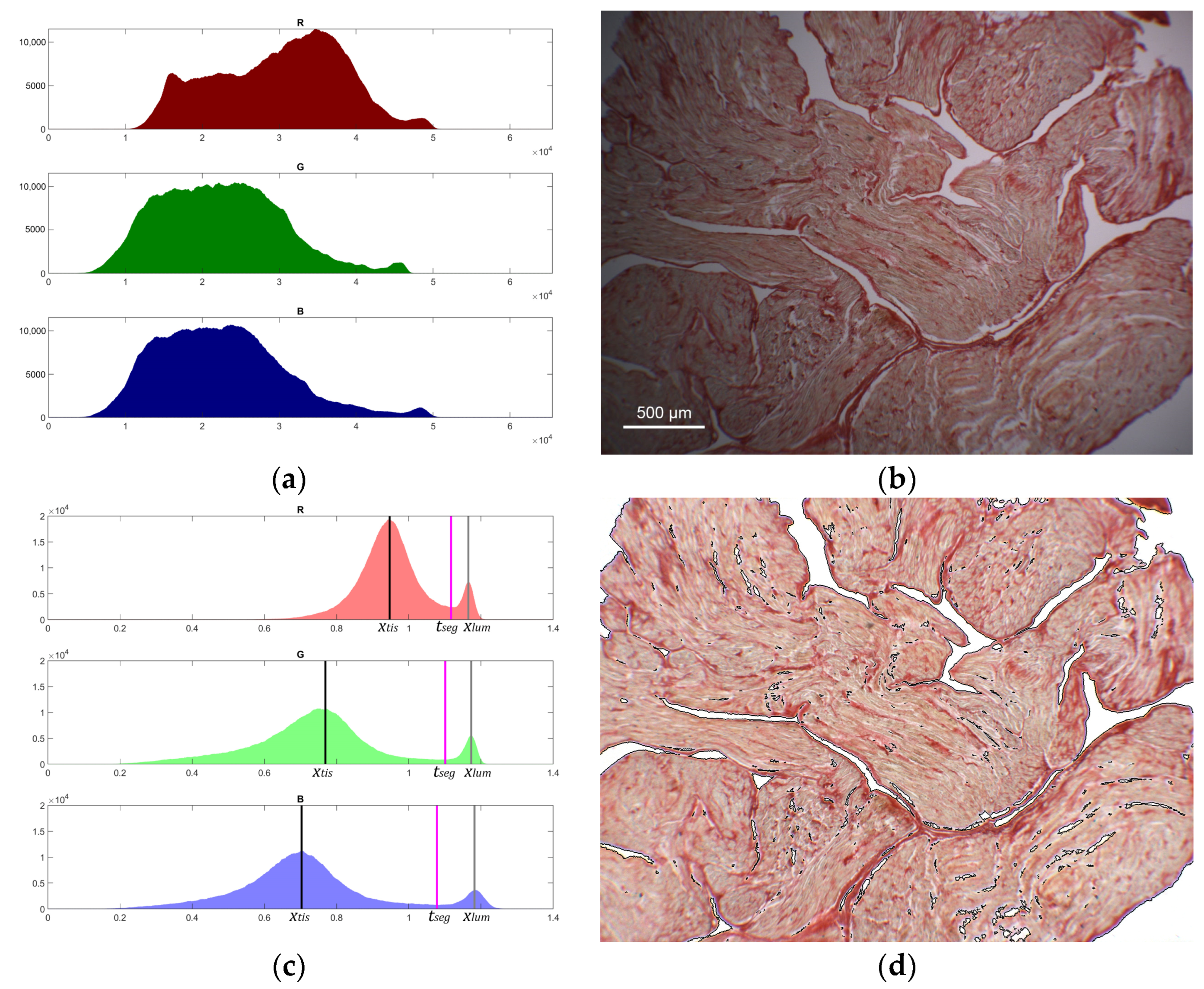
2.3. Tissue Segmentation, Transmittance, and Optical Thickness Estimation
2.4. Tissue Birefringence Quantification
2.5. Collagen Content Quantification
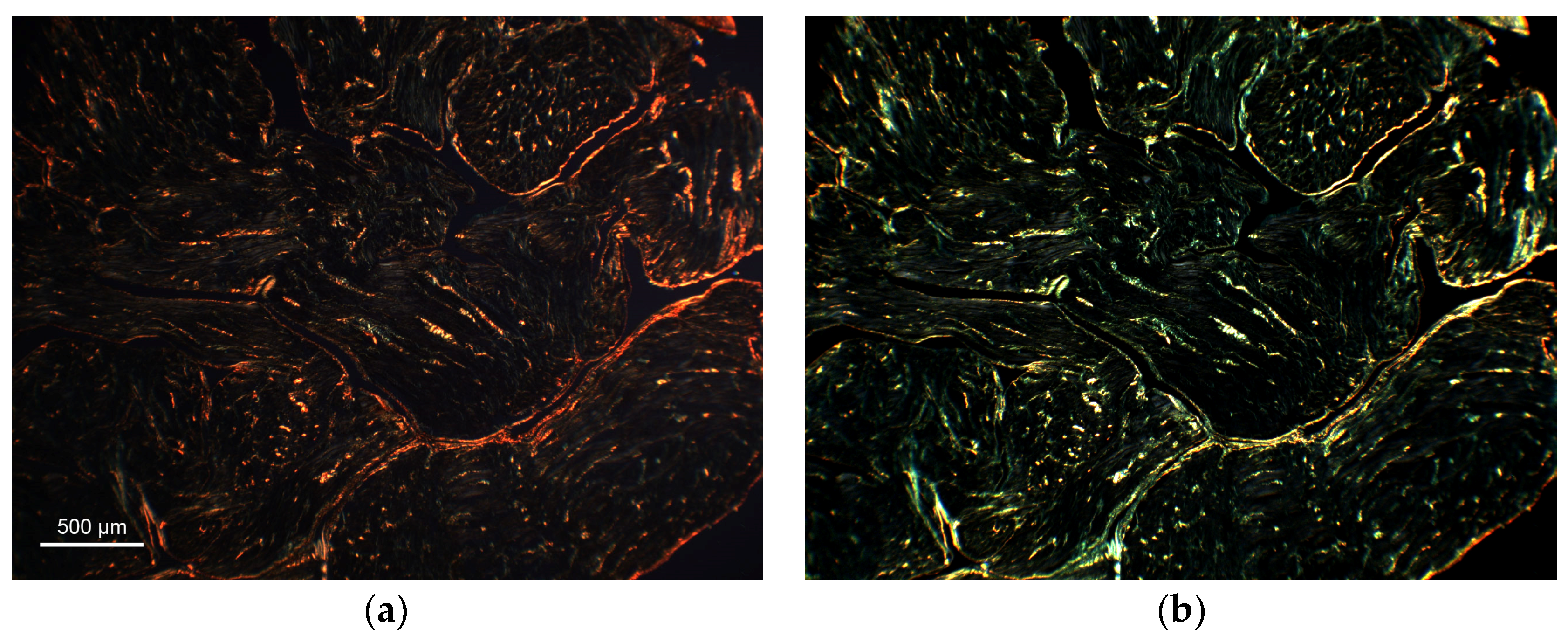
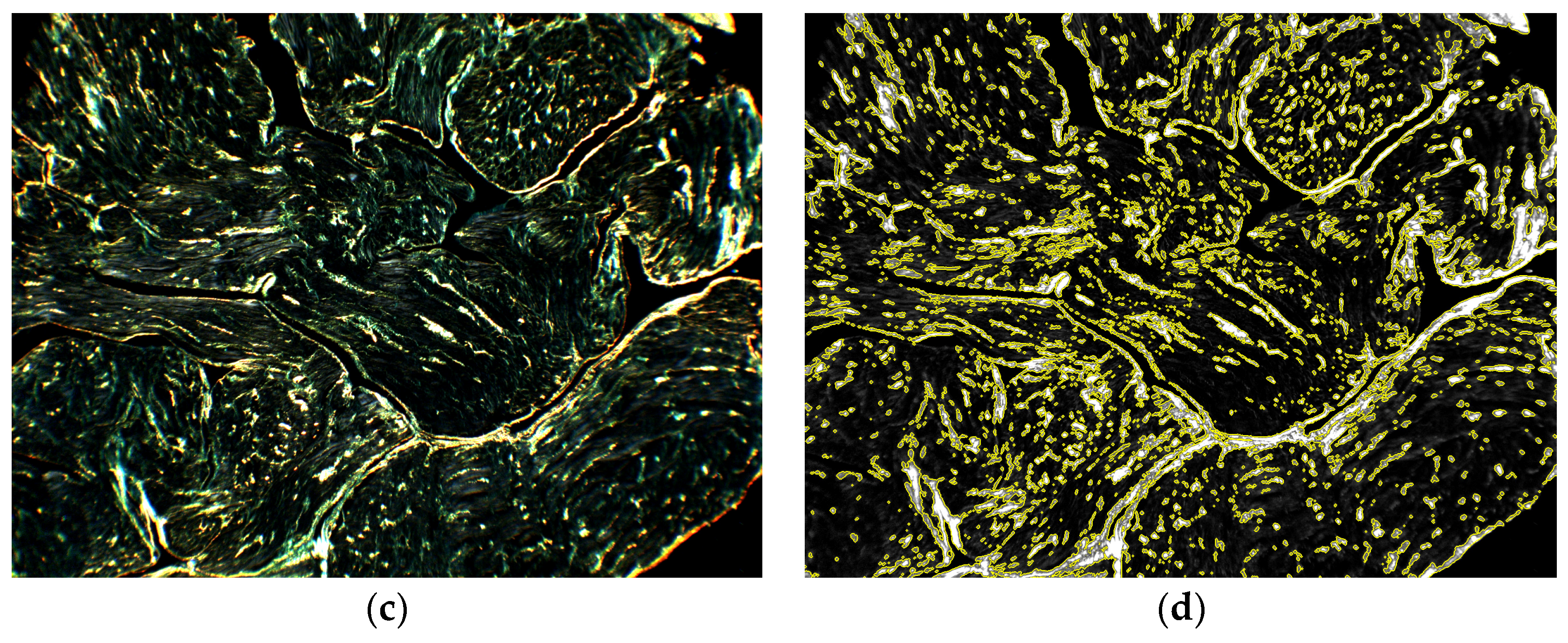
2.6. Proof of Concept Application of the Framework to Cardiac Histological Sections
2.7. Statistical Analysis
3. Results
3.1. Illumination Profile, Tissue Segmentation, and Transmittance Computation
3.2. Birefringence Analysis and Collagen Fraction Quantification
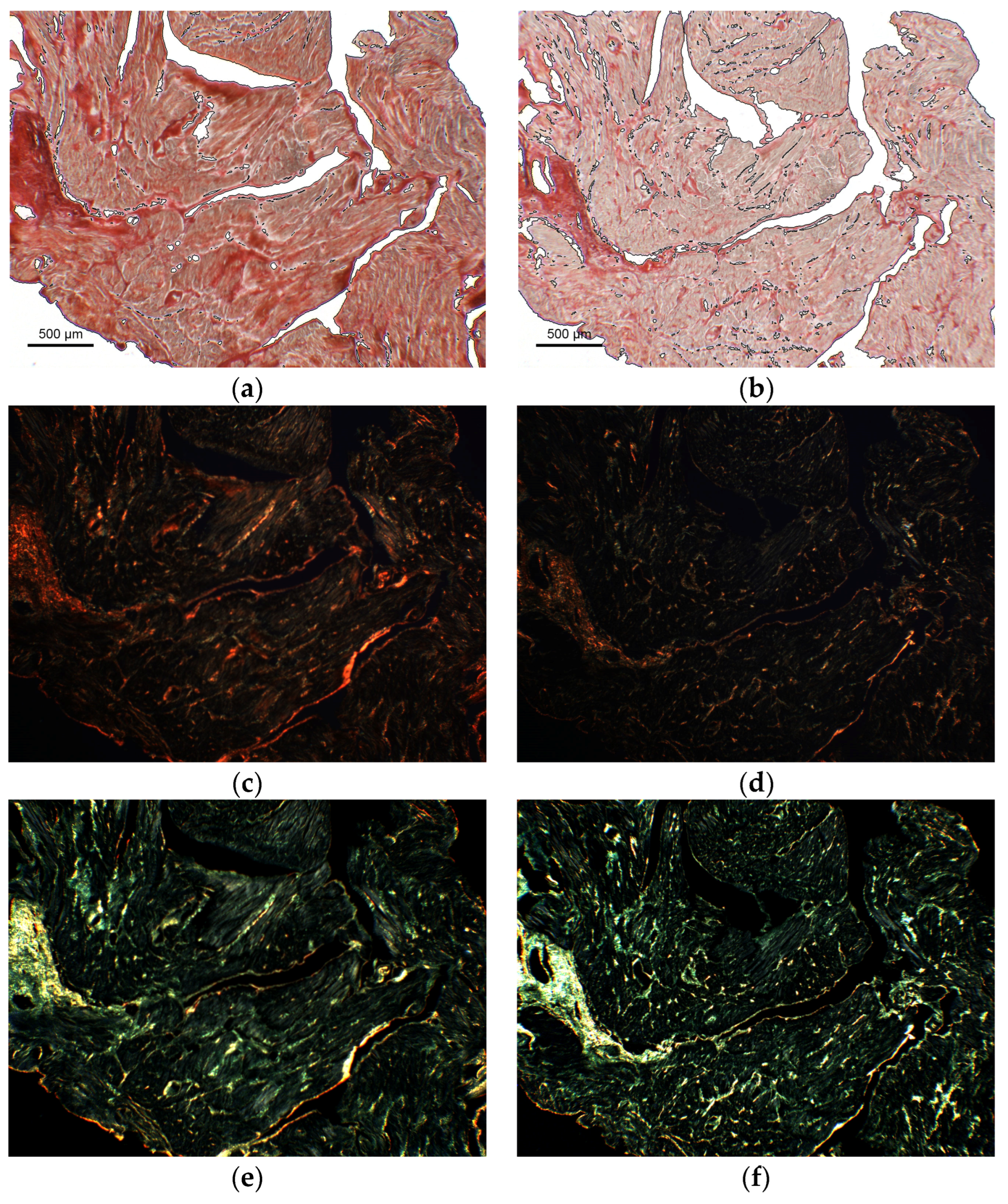
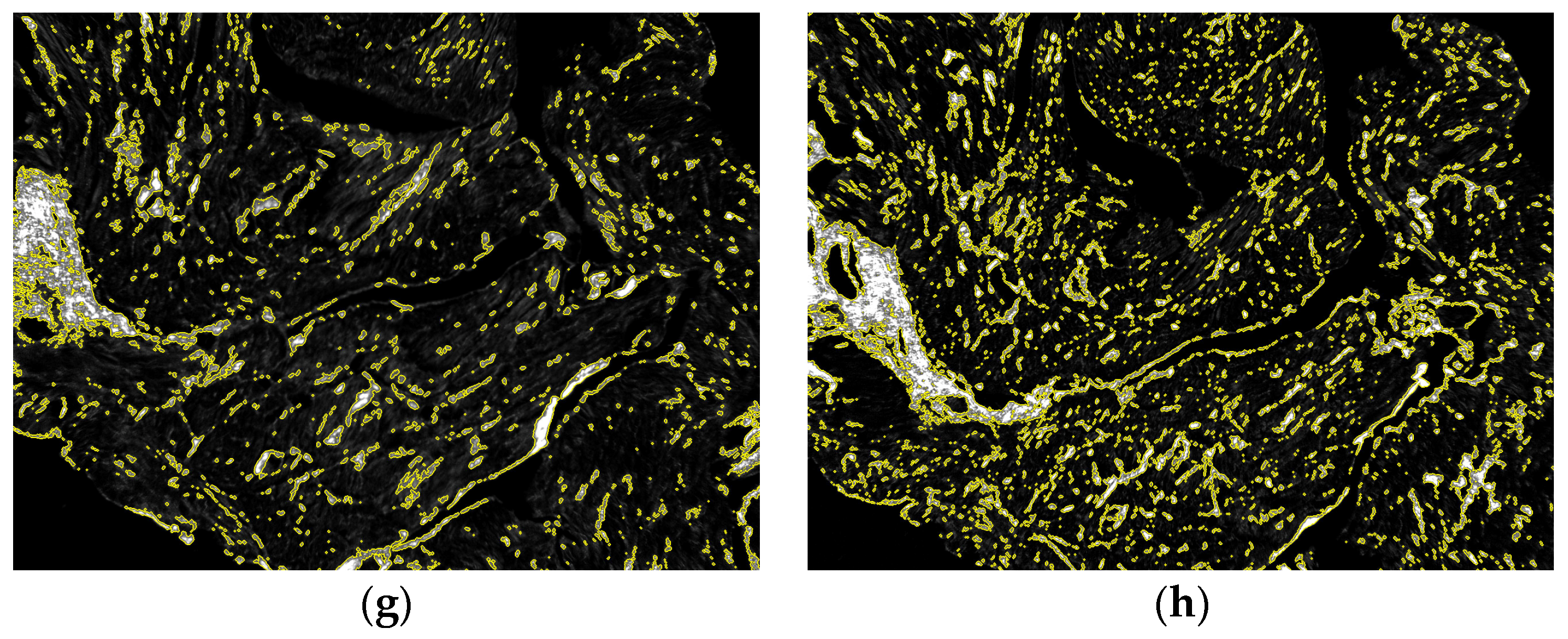
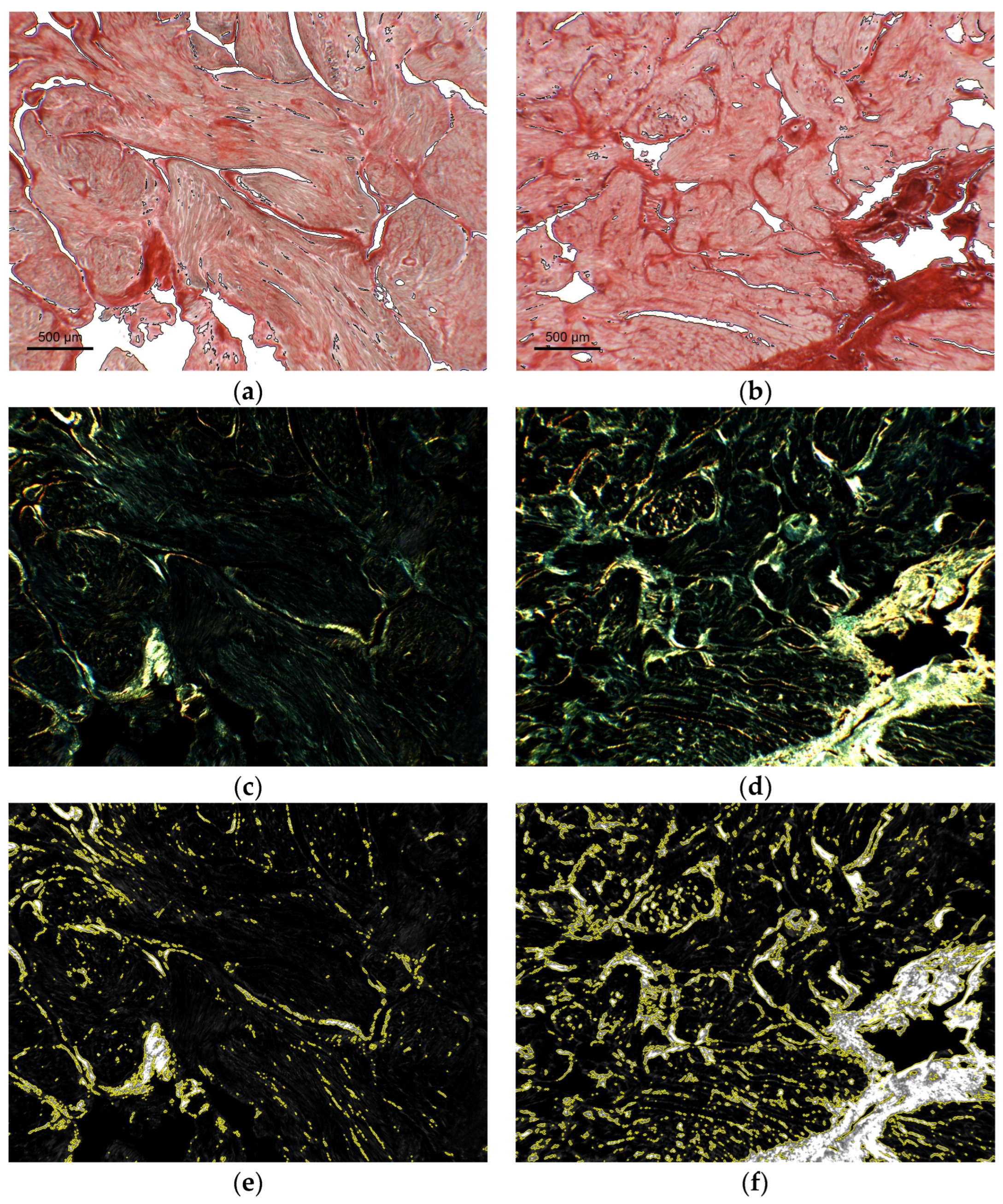
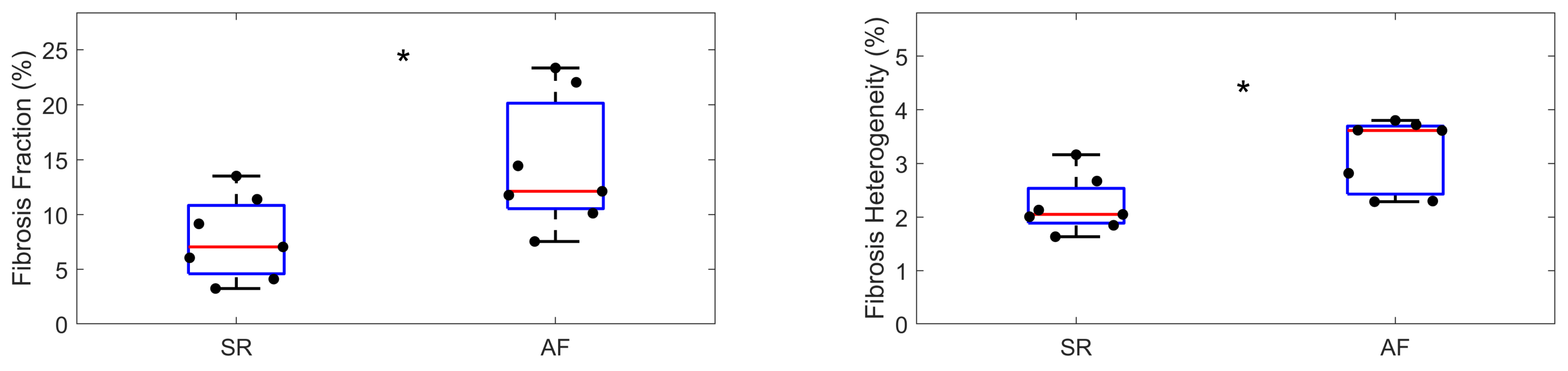
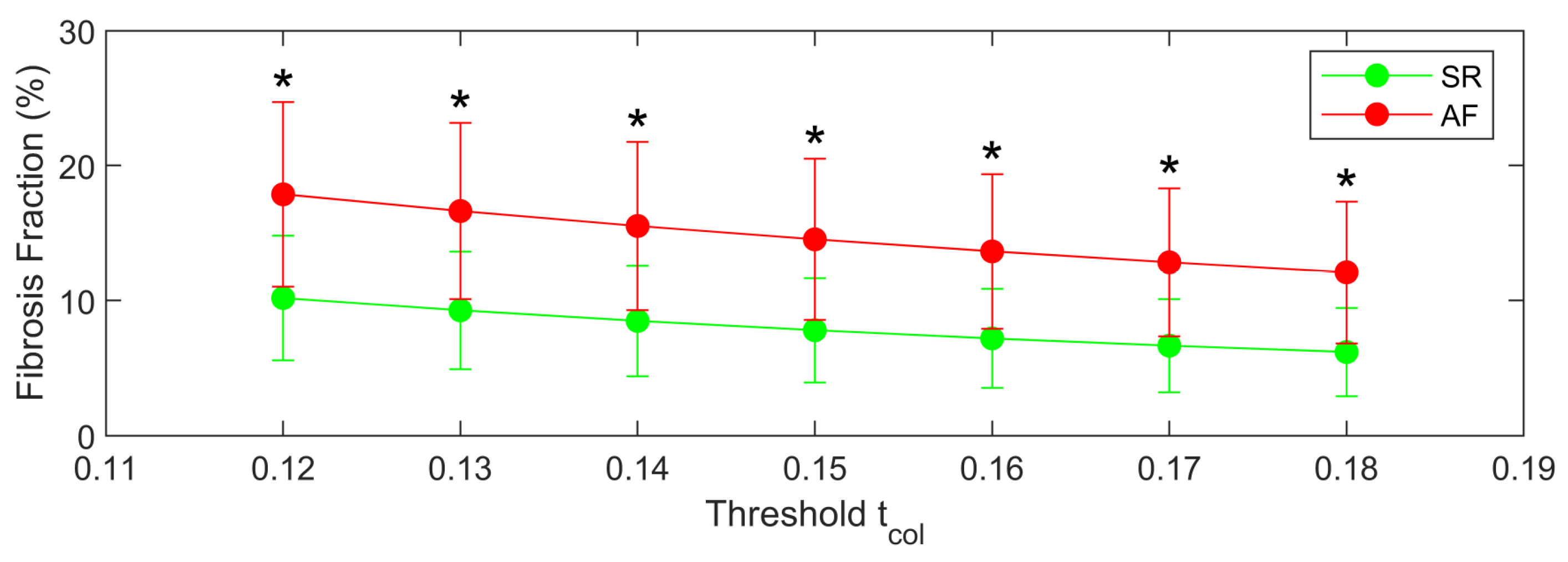
3.3. Proof of Concept Application of the Framework for the Quantification of Cardiac Fibrosis
4. Discussion
4.1. Technical Features of the Method
4.2. Applications of the Method
5. Limitations
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ricard-Blum, S. The Collagen Family. Cold Spring Harb. Perspect. Biol. 2011, 3, a004978. [Google Scholar] [CrossRef] [PubMed]
- Wynn, T.A. Cellular and Molecular Mechanisms of Fibrosis. J. Pathol. 2008, 214, 199–210. [Google Scholar] [CrossRef] [PubMed]
- Henderson, N.C.; Rieder, F.; Wynn, T.A. Fibrosis: From Mechanisms to Medicines. Nature 2020, 587, 555–566. [Google Scholar] [CrossRef] [PubMed]
- Li, A.-H.; Liu, P.P.; Villarreal, F.J.; Garcia, R.A. Dynamic Changes in Myocardial Matrix and Relevance to Disease: Translational Perspectives. Circ. Res. 2014, 114, 916–927. [Google Scholar] [CrossRef] [PubMed]
- Disertori, M.; Masè, M.; Ravelli, F. Myocardial Fibrosis Predicts Ventricular Tachyarrhythmias. Trends Cardiovasc. Med. 2017, 27, 363–372. [Google Scholar] [CrossRef] [PubMed]
- Hansen, B.J.; Zhao, J.; Csepe, T.A.; Moore, B.T.; Li, N.; Jayne, L.A.; Kalyanasundaram, A.; Lim, P.; Bratasz, A.; Powell, K.A.; et al. Atrial Fibrillation Driven by Micro-Anatomic Intramural Re-Entry Revealed by Simultaneous Sub-Epicardial and Sub-Endocardial Optical Mapping in Explanted Human Hearts. Eur. Heart J. 2015, 36, 2390–2401. [Google Scholar] [CrossRef]
- Ravelli, F.; Masè, M.; Cristoforetti, A.; Avogaro, L.; D’Amato, E.; Tessarolo, F.; Piccoli, F.; Graffigna, A. Quantitative Assessment of Transmural Fibrosis Profile in the Human Atrium: Evidence for a Three-Dimensional Arrhythmic Substrate by Slice-to-Slice Histology. Europace 2022, 25, euac187. [Google Scholar] [CrossRef]
- Wolman, M.; Kasten, F.H. Polarized Light Microscopy in the Study of the Molecular Structure of Collagen and Reticulin. Histochemistry 1986, 85, 41–49. [Google Scholar] [CrossRef]
- Whittaker, P.; Kloner, R.A.; Boughner, D.R.; Pickering, J.G. Quantitative Assessment of Myocardial Collagen with Picrosirius Red Staining and Circularly Polarized Light. Basic Res. Cardiol. 1994, 89, 397–410. [Google Scholar] [CrossRef]
- Rich, L.; Whittaker, P. Collagen and Picrosirius Red Staining: A Polarized Light Assessment of Fibrillar Hue and Spatial Distribution. J. Morphol. Sci. 2005, 22, 97–104. [Google Scholar]
- Lattouf, R.; Younes, R.; Lutomski, D.; Naaman, N.; Godeau, G.; Senni, K.; Changotade, S. Picrosirius Red Staining: A Useful Tool to Appraise Collagen Networks in Normal and Pathological Tissues. J. Histochem. Cytochem. 2014, 62, 751–758. [Google Scholar] [CrossRef] [PubMed]
- Rittié, L. Method for Picrosirius Red-Polarization Detection of Collagen Fibers in Tissue Sections. Methods Mol. Biol. 2017, 1627, 395–407. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Xu, M.-Y.; Wu, J.; Zhang, H.; Yang, L.; Lun, D.-X.; Hu, Y.-C.; Liu, B. Picrosirius-Polarization Method for Collagen Fiber Detection in Tendons: A Mini-Review. Orthop. Surg. 2021, 13, 701–707. [Google Scholar] [CrossRef] [PubMed]
- Daunoravicius, D.; Besusparis, J.; Zurauskas, E.; Laurinaviciene, A.; Bironaite, D.; Pankuweit, S.; Plancoulaine, B.; Herlin, P.; Bogomolovas, J.; Grabauskiene, V.; et al. Quantification of Myocardial Fibrosis by Digital Image Analysis and Interactive Stereology. Diagn. Pathol. 2014, 9, 114. [Google Scholar] [CrossRef]
- Hadi, A.M.; Mouchaers, K.T.B.; Schalij, I.; Grunberg, K.; Meijer, G.A.; Vonk-Noordegraaf, A.; van der Laarse, W.J.; Beliën, J.A.M. Rapid Quantification of Myocardial Fibrosis: A New Macro-Based Automated Analysis. Cell Oncol. 2011, 34, 343–354. [Google Scholar] [CrossRef]
- Gho, J.M.I.H.; van Es, R.; Stathonikos, N.; Harakalova, M.; Te Rijdt, W.P.; Suurmeijer, A.J.H.; van der Heijden, J.F.; de Jonge, N.; Chamuleau, S.A.J.; de Weger, R.A.; et al. High Resolution Systematic Digital Histological Quantification of Cardiac Fibrosis and Adipose Tissue in Phospholamban p.Arg14del Mutation Associated Cardiomyopathy. PLoS ONE 2014, 9, e94820. [Google Scholar] [CrossRef]
- Schipke, J.; Brandenberger, C.; Rajces, A.; Manninger, M.; Alogna, A.; Post, H.; Mühlfeld, C. Assessment of Cardiac Fibrosis: A Morphometric Method Comparison for Collagen Quantification. J. Appl. Physiol. 2017, 122, 1019–1030. [Google Scholar] [CrossRef]
- Gaspard, G.J.; Pasumarthi, K.B.S. Quantification of Cardiac Fibrosis by Colour-Subtractive Computer-Assisted Image Analysis. Clin. Exp. Pharmacol. Physiol. 2008, 35, 679–686. [Google Scholar] [CrossRef]
- Wülfers, E.M.; Greiner, J.; Giese, M.; Madl, J.; Kroll, J.; Stiller, B.; Kohl, P.; Rog-Zielinska, E.A.; Fürniss, H.E. Quantitative Collagen Assessment in Right Ventricular Myectomies from Patients with Tetralogy of Fallot. Europace 2021, 23, i38–i47. [Google Scholar] [CrossRef]
- Born, M.; Wolf, E. Principles of Optics, 6th ed.; Pergamon Press Ltd.: Oxford, UK, 1986. [Google Scholar]
- Bass, M. Handbook of Optics: Geometrical and Physical Optics, Polarized Light, Components and Instruments, 3rd ed.; The McGraw-Hill Companies, Inc.: London, UK, 2010. [Google Scholar]
- Moon, J.C.C.; Reed, E.; Sheppard, M.N.; Elkington, A.G.; Ho, S.Y.; Burke, M.; Petrou, M.; Pennell, D.J. The Histologic Basis of Late Gadolinium Enhancement Cardiovascular Magnetic Resonance in Hypertrophic Cardiomyopathy. J. Am. Coll. Cardiol. 2004, 43, 2260–2264. [Google Scholar] [CrossRef]
- Nart, P.; Williams, A.; Thompson, H.; Innocent, G.T. Morphometry of Bovine Dilated Cardiomyopathy. J. Comp. Pathol. 2004, 130, 235–245. [Google Scholar] [CrossRef] [PubMed]
- Ophof, R.; Maltha, J.C.; Von den Hoff, J.W.; Kuijpers-Jagtman, A.-M. Histologic Evaluation of Skin-Derived and Collagen-Based Substrates Implanted in Palatal Wounds. Wound Repair Regen. 2004, 12, 528–538. [Google Scholar] [CrossRef] [PubMed]
- Disertori, M.; Masè, M.; Marini, M.; Mazzola, S.; Cristoforetti, A.; Del Greco, M.; Kottkamp, H.; Arbustini, E.; Ravelli, F. Electroanatomic Mapping and Late Gadolinium Enhancement MRI in a Genetic Model of Arrhythmogenic Atrial Cardiomyopathy. J. Cardiovasc. Electrophysiol. 2014, 25, 964–970. [Google Scholar] [CrossRef] [PubMed]
- Goette, A.; Juenemann, G.; Peters, B.; Klein, H.U.; Roessner, A.; Huth, C.; Röcken, C. Determinants and Consequences of Atrial Fibrosis in Patients Undergoing Open Heart Surgery. Cardiovasc. Res. 2002, 54, 390–396. [Google Scholar] [CrossRef]
- Swartz, M.F.; Fink, G.W.; Lutz, C.J.; Taffet, S.M.; Berenfeld, O.; Vikstrom, K.L.; Kasprowicz, K.; Bhatta, L.; Puskas, F.; Kalifa, J.; et al. Left versus Right Atrial Difference in Dominant Frequency, K(+) Channel Transcripts, and Fibrosis in Patients Developing Atrial Fibrillation after Cardiac Surgery. Heart Rhythm 2009, 6, 1415–1422. [Google Scholar] [CrossRef]
- Nguyen, B.L.; Fishbein, M.C.; Chen, L.S.; Chen, P.-S.; Masroor, S. Histopathological Substrate for Chronic Atrial Fibrillation in Humans. Heart Rhythm 2009, 6, 454–460. [Google Scholar] [CrossRef]
- Platonov, P.G.; Mitrofanova, L.B.; Orshanskaya, V.; Ho, S.Y. Structural Abnormalities in Atrial Walls Are Associated with Presence and Persistency of Atrial Fibrillation but Not with Age. J. Am. Coll. Cardiol. 2011, 58, 2225–2232. [Google Scholar] [CrossRef]
- Eckstein, J.; Zeemering, S.; Linz, D.; Maesen, B.; Verheule, S.; van Hunnik, A.; Crijns, H.; Allessie, M.A.; Schotten, U. Transmural Conduction Is the Predominant Mechanism of Breakthrough during Atrial Fibrillation: Evidence from Simultaneous Endo-Epicardial High-Density Activation Mapping. Circ. Arrhythm. Electrophysiol. 2013, 6, 334–341. [Google Scholar] [CrossRef]
- Burton, R.A.B.; Lee, P.; Casero, R.; Garny, A.; Siedlecka, U.; Schneider, J.E.; Kohl, P.; Grau, V. Three-Dimensional Histology: Tools and Application to Quantitative Assessment of Cell-Type Distribution in Rabbit Heart. Europace 2014, 16 (Suppl. 4), iv86–iv95. [Google Scholar] [CrossRef]
- Camelliti, P.; Al-Saud, S.A.; Smolenski, R.T.; Al-Ayoubi, S.; Bussek, A.; Wettwer, E.; Banner, N.R.; Bowles, C.T.; Yacoub, M.H.; Terracciano, C.M. Adult Human Heart Slices Are a Multicellular System Suitable for Electrophysiological and Pharmacological Studies. J. Mol. Cell. Cardiol. 2011, 51, 390–398. [Google Scholar] [CrossRef]
- Schalla, S.; Bekkers, S.C.; Dennert, R.; van Suylen, R.J.; Waltenberger, J.; Leiner, T.; Wildberger, J.; Crijns, H.J.; Heymans, S. Replacement and Reactive Myocardial Fibrosis in Idiopathic Dilated Cardiomyopathy: Comparison of Magnetic Resonance Imaging with Right Ventricular Biopsy. Eur. J. Heart Fail. 2010, 12, 227–231. [Google Scholar] [CrossRef] [PubMed]
- Nogueira, D.M.B.; Figadoli, A.L.d.F.; Alcantara, P.L.; Pomini, K.T.; German, I.J.S.; Reis, C.H.B.; Júnior, G.M.R.; Rosso, M.P.d.O.; Santos, P.S.d.S.; Zangrando, M.S.R.; et al. Biological Behavior of Xenogenic Scaffolds in Alcohol-Induced Rats: Histomorphometric and Picrosirius Red Staining Analysis. Polymers 2022, 14, 584. [Google Scholar] [CrossRef] [PubMed]
- Grimm, P.C.; Nickerson, P.; Gough, J.; McKenna, R.; Stern, E.; Jeffery, J.; Rush, D.N. Computerized Image Analysis of Sirius Red-Stained Renal Allograft Biopsies as a Surrogate Marker to Predict Long-Term Allograft Function. J. Am. Soc. Nephrol. 2003, 14, 1662–1668. [Google Scholar] [CrossRef] [PubMed]
- Greiner, C.; Grainger, S.; Farrow, S.; Davis, A.; Su, J.L.; Saybolt, M.D.; Wilensky, R.; Madden, S.; Sum, S.T. Robust Quantitative Assessment of Collagen Fibers with Picrosirius Red Stain and Linearly Polarized Light as Demonstrated on Atherosclerotic Plaque Samples. PLoS ONE 2021, 16, e0248068. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cristoforetti, A.; Masè, M.; Ravelli, F. Model-Based Approach for the Semi-Automatic Analysis of Collagen Birefringence in Polarized Light Microscopy. Appl. Sci. 2023, 13, 2916. https://doi.org/10.3390/app13052916
Cristoforetti A, Masè M, Ravelli F. Model-Based Approach for the Semi-Automatic Analysis of Collagen Birefringence in Polarized Light Microscopy. Applied Sciences. 2023; 13(5):2916. https://doi.org/10.3390/app13052916
Chicago/Turabian StyleCristoforetti, Alessandro, Michela Masè, and Flavia Ravelli. 2023. "Model-Based Approach for the Semi-Automatic Analysis of Collagen Birefringence in Polarized Light Microscopy" Applied Sciences 13, no. 5: 2916. https://doi.org/10.3390/app13052916
APA StyleCristoforetti, A., Masè, M., & Ravelli, F. (2023). Model-Based Approach for the Semi-Automatic Analysis of Collagen Birefringence in Polarized Light Microscopy. Applied Sciences, 13(5), 2916. https://doi.org/10.3390/app13052916





