Differential Expression Analysis of Blood MicroRNA in Identifying Potential Genes Relevant to Alzheimer’s Disease Pathogenesis, Using an Integrated Bioinformatics and Machine Learning Approach
Abstract
:1. Introduction
2. Related Works
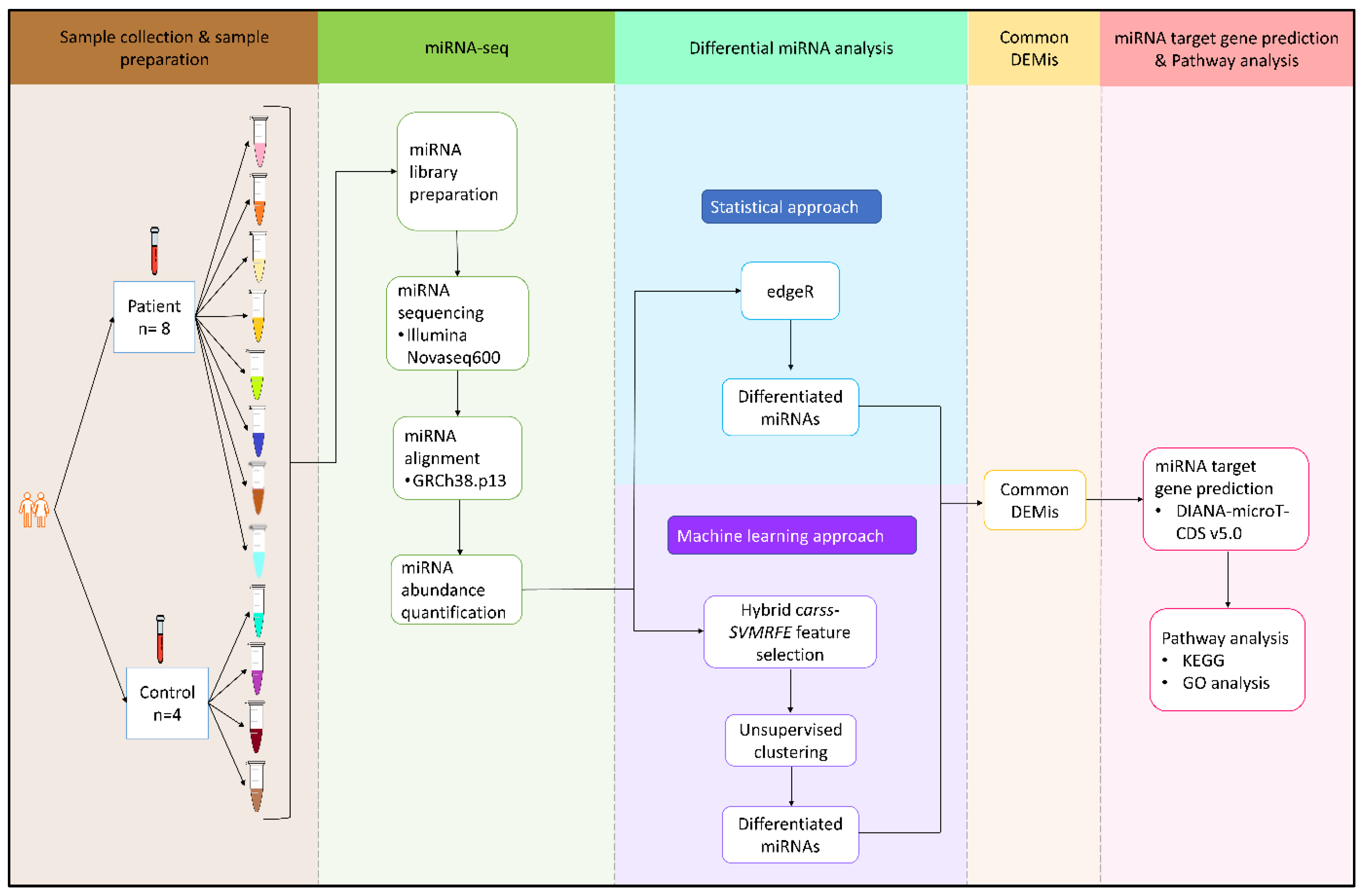
3. Methods and Materials
3.1. Subjects
3.2. Sample Preparation
3.3. Small RNA-Sequencing Analysis
3.4. Integrative Statistical and Machine Learning Analysis
3.4.1. Differential Expression Analysis
- (i)
- Statistical approach: EdgeR
- (ii)
- Machine learning approach
3.4.2. miRNA Target Gene Prediction
3.4.3. miRNA Pathway and Gene Ontology (GO) Analysis
4. Results
4.1. Statistical Approach: edgeR
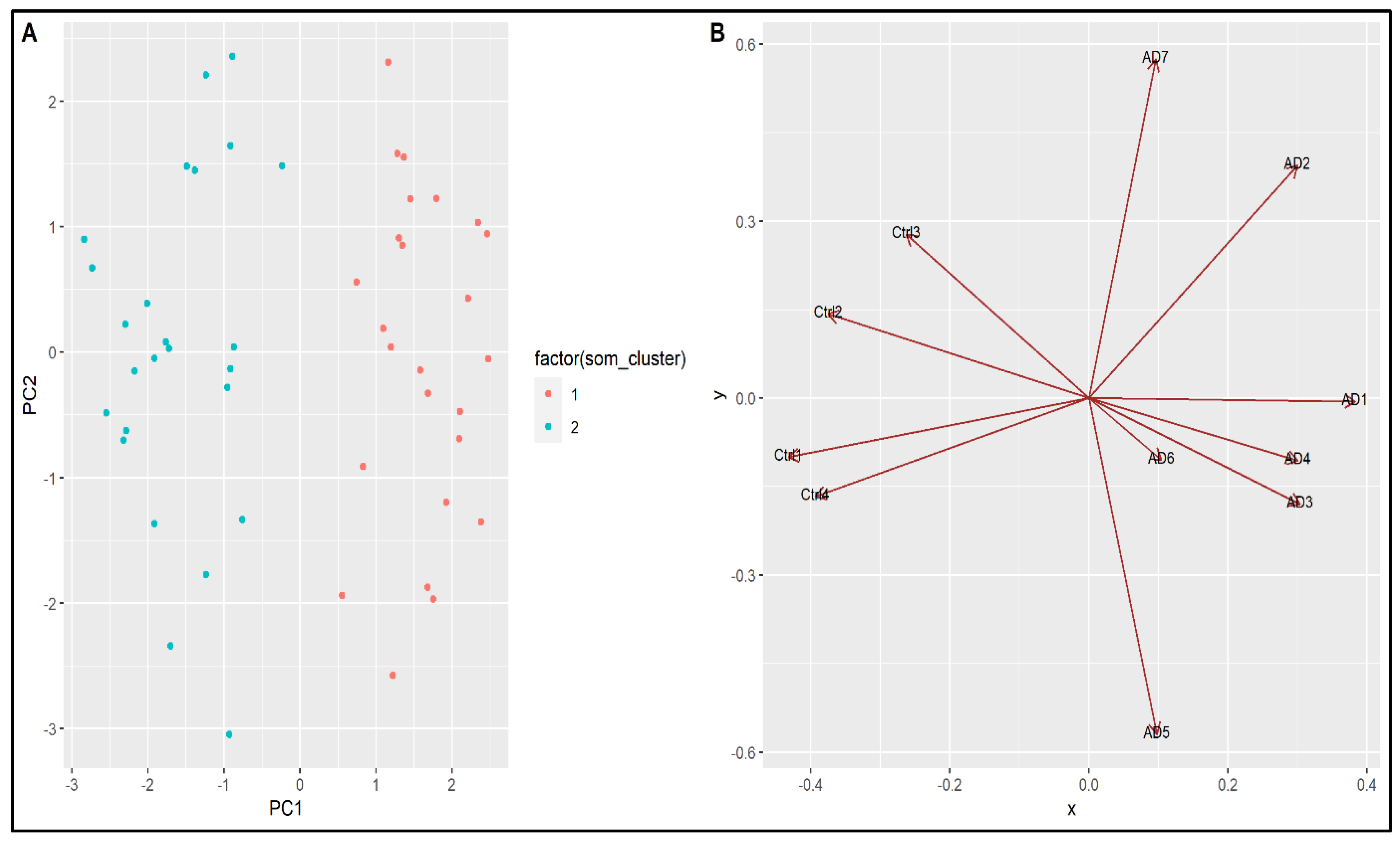
4.2. Machine Learning Approach
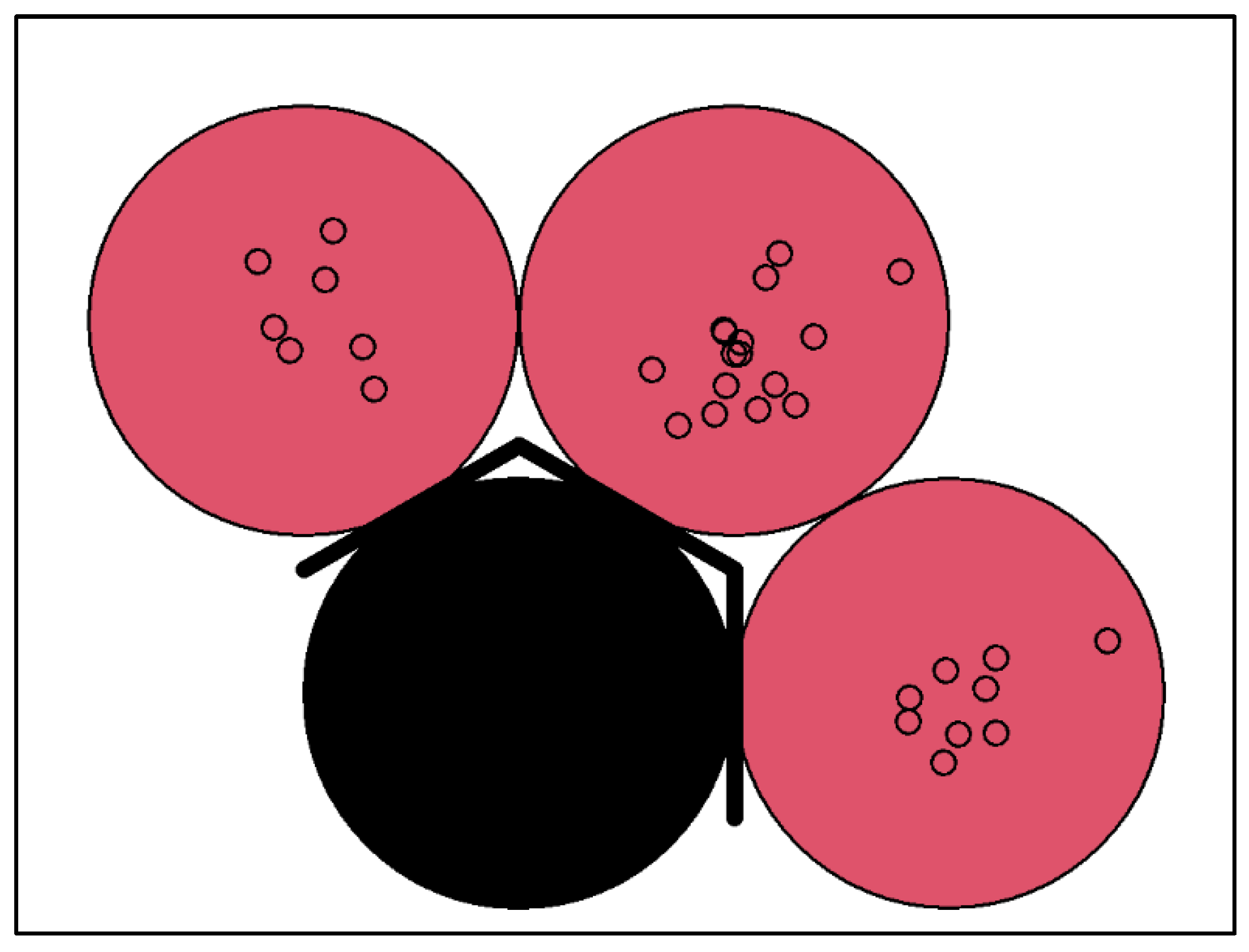
4.3. Integrated Bioinformatics and Machine Learning Approach
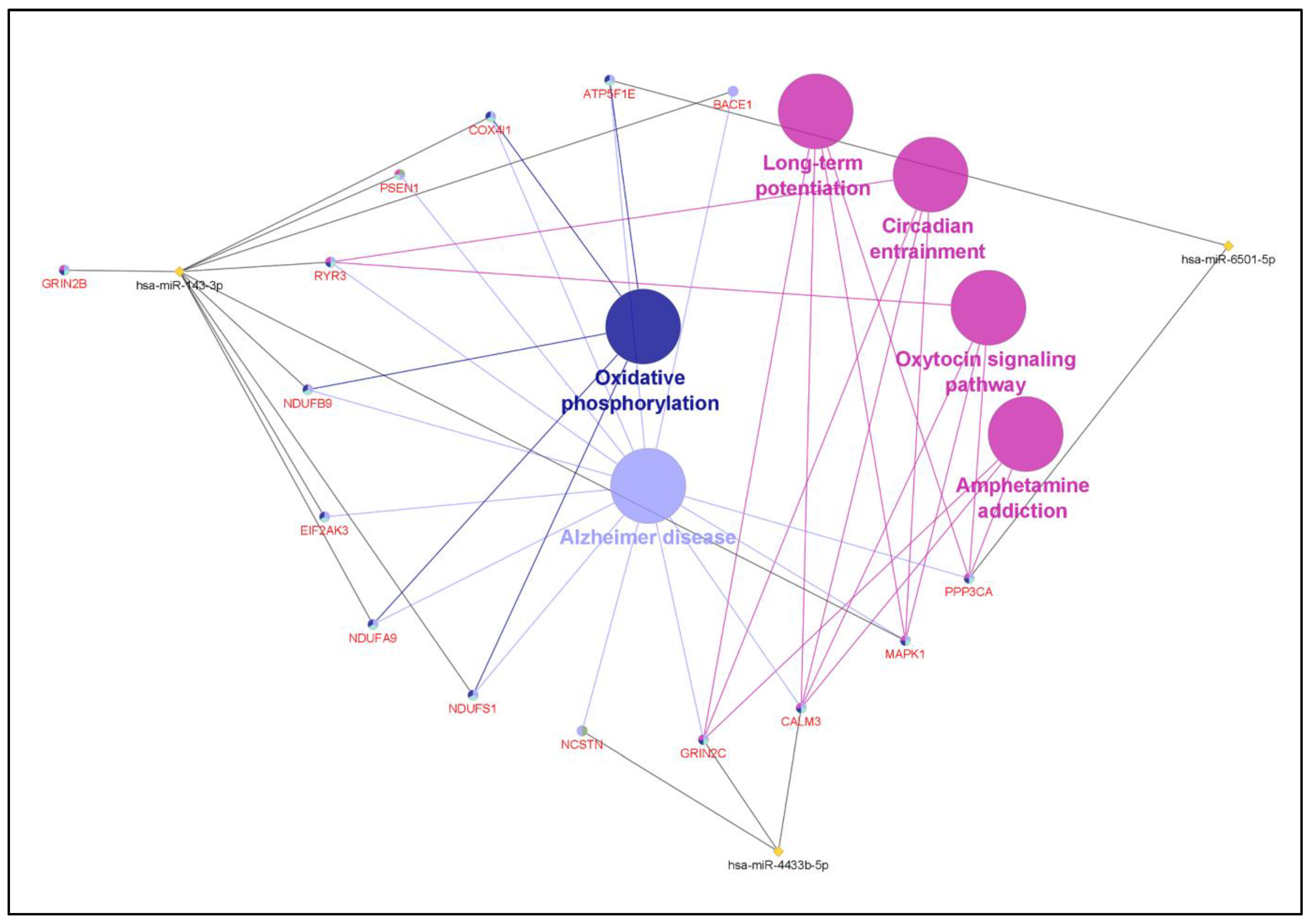
4.4. Target Gene Prediction
4.5. miRNA Pathway and Gene Ontology (GO) Analysis
5. Discussion
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wolk, D.A.; Dickerson, B.C. Clinical Features and Diagnosis of Alzheimer Disease; UpToDate: Waltham, MA, USA, 2016. [Google Scholar]
- Abuelezz, N.Z.; Nasr, F.E.; AbdulKader, M.A.; Bassiouny, A.R.; Zaky, A. MicroRNAs as Potential Orchestrators of Alzheimer’s Disease-Related Pathologies: Insights on Current Status and Future Possibilities. Front. Aging Neurosci. 2021, 653, 743573. [Google Scholar] [CrossRef] [PubMed]
- Miya Shaik, M.; Tamargo, I.A.; Abubakar, M.B.; Kamal, M.A.; Greig, N.H.; Gan, S.H. The role of microRNAs in Alzheimer’s disease and their therapeutic potentials. Genes 2018, 9, 174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Xu, Y.; Yu, M. MicroRNA-4722-5p and microRNA-615-3p serve as potential biomarkers for Alzheimer’s disease. Exp. Ther. Med. 2022, 23, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Guévremont, D.; Tsui, H.; Knight, R.; Fowler, C.J.; Masters, C.L.; Martins, R.N.; Abraham, W.C.; Tate, W.P.; Cutfield, N.J.; Williams, J.M. Plasma microRNA vary in association with the progression of Alzheimer’s disease. Alzheimer’s Dement. Diagn. Assess. Dis. Monit. 2022, 14, e12251. [Google Scholar]
- Zhang, W.; Zhang, G.; Gao, S. The potential diagnostic accuracy of circulating microRNAs for Alzheimer’s disease: A meta-analysis. Neurologia, 2021; in press. [Google Scholar] [CrossRef]
- Walgrave, H.; Zhou, L.; De Strooper, B.; Salta, E. The promise of microRNA-based therapies in Alzheimer’s disease: Challenges and perspectives. Mol. Neurodegener. 2021, 16, 1–16. [Google Scholar] [CrossRef]
- Angelucci, F.; Cechova, K.; Valis, M.; Kuca, K.; Zhang, B.; Hort, J. MicroRNAs in Alzheimer’s disease: Diagnostic markers or therapeutic agents? Front. Pharmacol. 2019, 10, 665. [Google Scholar] [CrossRef]
- Wu, H.Z.Y.; Thalamuthu, A.; Cheng, L.; Fowler, C.; Masters, C.L.; Sachdev, P.; Mather, K.A. Differential blood miRNA expression in brain amyloid imaging-defined Alzheimer’s disease and controls. Alzheimer’s Res. Ther. 2020, 12, 1–11. [Google Scholar] [CrossRef]
- Kumar, S.; Reddy, P.H. Are circulating microRNAs peripheral biomarkers for Alzheimer’s disease? Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2016, 1862, 1617–1627. [Google Scholar] [CrossRef]
- Liu, S.; Fan, M.; Zheng, Q.; Hao, S.; Yang, L.; Qi, C.; Ge, J.; Xia, Q. MicroRNAs in Alzheimer’s disease: Potential diagnostic markers and therapeutic targets. Biomed. Pharmacother. 2022, 148, 112681. [Google Scholar] [CrossRef]
- Fransquet, P.D.; Ryan, J. Micro RNA as a potential blood-based epigenetic biomarker for Alzheimer’s disease. Clin. Biochem. 2018, 58, 5–14. [Google Scholar] [CrossRef] [PubMed]
- Takousis, P.; Sadlon, A.; Schulz, J.; Wohlers, I.; Dobricic, V.; Middleton, L.; Lill, C.M.; Perneczky, R.; Bertram, L. Differential expression of microRNAs in Alzheimer’s disease brain, blood, and cerebrospinal fluid. Alzheimer’s Dement. 2019, 15, 1468–1477. [Google Scholar] [CrossRef] [PubMed]
- Mahendran, N.; Vincent, P.D.R.; Srinivasan, K.; Chang, C.-Y. Improving the classification of alzheimer’s disease using hybrid gene selection pipeline and deep learning. Front. Genet. 2021, 12. [Google Scholar] [CrossRef]
- Tan, M.S.; Cheah, P.-L.; Chin, A.-V.; Looi, L.-M.; Chang, S.-W. A review on omics-based biomarkers discovery for Alzheimer’s disease from the bioinformatics perspectives: Statistical approach vs machine learning approach. Comput. Biol. Med. 2021, 139, 104947. [Google Scholar] [CrossRef] [PubMed]
- Robles, J.A.; Qureshi, S.E.; Stephen, S.J.; Wilson, S.R.; Burden, C.J.; Taylor, J.M. Efficient experimental design and analysis strategies for the detection of differential expression using RNA-Sequencing. BMC Genom. 2012, 13, 484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ziats, M.N.; Rennert, O.M. Identification of differentially expressed microRNAs across the developing human brain. Mol. Psychiatry 2014, 19, 848–852. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takahashi, Y.; Ueki, M.; Tamiya, G.; Ogishima, S.; Kinoshita, K.; Hozawa, A.; Minegishi, N.; Nagami, F.; Fukumoto, K.; Otsuka, K. Machine learning for effectively avoiding overfitting is a crucial strategy for the genetic prediction of polygenic psychiatric phenotypes. Transl. Psychiatry 2020, 10, 294. [Google Scholar] [CrossRef]
- Bommert, A.; Welchowski, T.; Schmid, M.; Rahnenführer, J. Benchmark of filter methods for feature selection in high-dimensional gene expression survival data. Brief. Bioinform. 2022, 23, bbab354. [Google Scholar] [CrossRef]
- Yuen, S.C.; Liang, X.; Zhu, H.; Jia, Y.; Leung, S.-w. Prediction of differentially expressed microRNAs in blood as potential biomarkers for Alzheimer’s disease by meta-analysis and adaptive boosting ensemble learning. Alzheimer’s Res. Ther. 2021, 13, 126. [Google Scholar] [CrossRef]
- Zhao, Y.; Chen, X.; Yin, J. Adaptive boosting-based computational model for predicting potential miRNA-disease associations. Bioinformatics 2019, 35, 4730–4738. [Google Scholar] [CrossRef]
- Lugli, G.; Cohen, A.M.; Bennett, D.A.; Shah, R.C.; Fields, C.J.; Hernandez, A.G.; Smalheiser, N.R. Plasma exosomal miRNAs in persons with and without Alzheimer disease: Altered expression and prospects for biomarkers. PLoS ONE 2015, 10, e0139233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ludwig, N.; Fehlmann, T.; Kern, F.; Gogol, M.; Maetzler, W.; Deutscher, S.; Gurlit, S.; Schulte, C.; Von Thaler, A.-K.; Deuschle, C. Machine learning to detect Alzheimer’s disease from circulating non-coding RNAs. Genom. Proteom. Bioinform. 2019, 17, 430–440. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Metpally, R.P.R.; Nasser, S.; Malenica, I.; Courtright, A.; Carlson, E.; Ghaffari, L.; Villa, S.; Tembe, W.; Van Keuren-Jensen, K. Comparison of analysis tools for miRNA high throughput sequencing using nerve crush as a model. Front. Genet. 2013, 4, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [Green Version]
- Lang, M.; Binder, M.; Richter, J.; Schratz, P.; Pfisterer, F.; Coors, S.; Au, Q.; Casalicchio, G.; Kotthoff, L.; Bischl, B. mlr3: A modern object-oriented machine learning framework in R. J. Open Source Softw. 2019, 4, 1903. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X.; Tuck, D.P. MSVM-RFE: Extensions of SVM-RFE for multiclass gene selection on DNA microarray data. Bioinformatics 2007, 23, 1106–1114. [Google Scholar] [CrossRef] [Green Version]
- Kohonen, T. Self-Organizing Maps; Springer Science & Business Media: Heidelberg, Germany, 2012; Volume 30. [Google Scholar]
- Anke, Z.; Xinjian, Q.; Guojian, C. Clustering analysis of gene data based on PCA and SOM neural networks. In Proceedings of the 2014 Fifth International Conference on Intelligent Systems Design and Engineering Applications, Hunan, China, 15–16 June 2014; pp. 284–287. [Google Scholar]
- Wehrens, R.; Buydens, L.M. Self-and super-organizing maps in R: The Kohonen package. J. Stat. Softw. 2007, 21, 1–19. [Google Scholar] [CrossRef] [Green Version]
- Bro, R.; Smilde, A.K. Principal component analysis. Anal. Methods 2014, 6, 2812–2831. [Google Scholar] [CrossRef] [Green Version]
- Jolliffe, I.T.; Cadima, J. Principal component analysis: A review and recent developments. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 2016, 374, 20150202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paraskevopoulou, M.D.; Georgakilas, G.; Kostoulas, N.; Vlachos, I.S.; Vergoulis, T.; Reczko, M.; Filippidis, C.; Dalamagas, T.; Hatzigeorgiou, A.G. DIANA-microT web server v5. 0: Service integration into miRNA functional analysis workflows. Nucleic Acids Res. 2013, 41, W169–W173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vlachos, I.S.; Zagganas, K.; Paraskevopoulou, M.D.; Georgakilas, G.; Karagkouni, D.; Vergoulis, T.; Dalamagas, T.; Hatzigeorgiou, A.G. DIANA-miRPath v3. 0: Deciphering microRNA function with experimental support. Nucleic Acids Res. 2015, 43, W460–W466. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Ge, X.; Yao, T.; Zhang, C.; Wang, Q.; Wang, X.; Xu, L.-C. Human microRNA-4433 (hsa-miR-4443) Targets 18 Genes to be a Risk Factor of Neurodegenerative Diseases. Curr. Alzheimer Res. 2022, 19, 511–522. [Google Scholar]
- Sproviero, D.; Gagliardi, S.; Zucca, S.; Arigoni, M.; Giannini, M.; Garofalo, M.; Olivero, M.; Dell’Orco, M.; Pansarasa, O.; Bernuzzi, S. Different miRNA profiles in plasma derived small and large extracellular vesicles from patients with neurodegenerative diseases. Int. J. Mol. Sci. 2021, 22, 2737. [Google Scholar] [CrossRef]
- Peña-Bautista, C.; Álvarez-Sánchez, L.; Cañada-Martínez, A.J.; Baquero, M.; Cháfer-Pericás, C. Epigenomics and Lipidomics Integration in Alzheimer Disease: Pathways Involved in Early Stages. Biomedicines 2021, 9, 1812. [Google Scholar] [CrossRef]
- Cheng, á.; Doecke, J.D.; Sharples, R.; Villemagne, V.L.; Fowler, C.J.; Rembach, A.; Martins, R.N.; Rowe, C.C.; Macaulay, S.L.; Masters, C.L. Prognostic serum miRNA biomarkers associated with Alzheimer’s disease shows concordance with neuropsychological and neuroimaging assessment. Mol. Psychiatry 2015, 20, 1188–1196. [Google Scholar] [CrossRef] [Green Version]
- Hara, N.; Kikuchi, M.; Miyashita, A.; Hatsuta, H.; Saito, Y.; Kasuga, K.; Murayama, S.; Ikeuchi, T.; Kuwano, R. Serum microRNA miR-501-3p as a potential biomarker related to the progression of Alzheimer’s disease. Acta Neuropathol. Commun. 2017, 5, 10. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Shui, X.; Mei, Y.; Xia, Y.; Lan, G.; Hu, L.; Zhang, M.; Gan, C.-L.; Li, R.; Tian, Y. miR-143-3p Inhibits Aberrant Tau Phosphorylation and Amyloidogenic Processing of APP by Directly Targeting DAPK1 in Alzheimer’s Disease. Int. J. Mol. Sci. 2022, 23, 7992. [Google Scholar] [CrossRef] [PubMed]
- Mayr, J.A.; Havlíčková, V.; Zimmermann, F.; Magler, I.; Kaplanova, V.; Ješina, P.; Pecinová, A.; Nůsková, H.; Koch, J.; Sperl, W. Mitochondrial ATP synthase deficiency due to a mutation in the ATP5E gene for the F1 ε subunit. Hum. Mol. Genet. 2010, 19, 3430–3439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lunnon, K.; Keohane, A.; Pidsley, R.; Newhouse, S.; Riddoch-Contreras, J.; Thubron, E.B.; Devall, M.; Soininen, H.; Kłoszewska, I.; Mecocci, P. Mitochondrial genes are altered in blood early in Alzheimer’s disease. Neurobiol. Aging 2017, 53, 36–47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chowdhury, U.N.; Islam, M.B.; Ahmad, S.; Moni, M.A. Network-based identification of genetic factors in ageing, lifestyle and type 2 diabetes that influence to the progression of Alzheimer’s disease. Inform. Med. Unlocked 2020, 19, 100309. [Google Scholar] [CrossRef]
- Chiocco, M.J.; Zhu, X.; Walther, D.; Pletnikova, O.; Troncoso, J.C.; Uhl, G.R.; Liu, Q.-R. Fine mapping of calcineurin (PPP3CA) gene reveals novel alternative splicing patterns, association of 5′ UTR trinucleotide repeat with addiction vulnerability, and differential isoform expression in Alzheimer’s disease. Subst. Use Misuse 2010, 45, 1809–1826. [Google Scholar] [CrossRef] [Green Version]
- Sadick, J.S.; Liddelow, S.A. Don’t forget astrocytes when targeting Alzheimer’s disease. Br. J. Pharmacol. 2019, 176, 3585–3598. [Google Scholar] [CrossRef]
- Griswold, A.J.; Celis, K.; Bussies, P.L.; Rajabli, F.; Whitehead, P.L.; Hamilton-Nelson, K.L.; Beecham, G.W.; Dykxhoorn, D.M.; Nuytemans, K.; Wang, L. Increased APOE ε4 expression is associated with the difference in Alzheimer’s disease risk from diverse ancestral backgrounds. Alzheimer’s Dement. 2021, 17, 1179–1188. [Google Scholar] [CrossRef]
- Morabito, S.; Miyoshi, E.; Michael, N.; Swarup, V. Integrative genomics approach identifies conserved transcriptomic networks in Alzheimer’s disease. Hum. Mol. Genet. 2020, 29, 2899–2919. [Google Scholar] [CrossRef]
- Sesele, K.; Thanopoulou, K.; Paouri, E.; Tsefou, E.; Klinakis, A.; Georgopoulos, S. Conditional inactivation of nicastrin restricts amyloid deposition in an Alzheimer’s disease mouse model. Aging Cell 2013, 12, 1032–1040. [Google Scholar] [CrossRef]
- Herrera-Rivero, M.; Hernández-Aguilar, M.E.; Aranda-Abreu, G.E. A strategy focused on MAPT, APP, NCSTN and BACE1 to build blood classifiers for Alzheimer׳ s disease. J. Theor. Biol. 2015, 376, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Andreoli, V.; De Marco, E.V.; Trecroci, F.; Cittadella, R.; Di Palma, G.; Gambardella, A. Potential involvement of GRIN2B encoding the NMDA receptor subunit NR2B in the spectrum of Alzheimer’s disease. J. Neural Transm. 2014, 121, 533–542. [Google Scholar] [CrossRef]
- Wong, T.H.; van der Lee, S.J.; van Rooij, J.G.; Meeter, L.H.; Frick, P.; Melhem, S.; Seelaar, H.; Ikram, M.A.; Rozemuller, A.J.; Holstege, H. EIF2AK3 variants in Dutch patients with Alzheimer’s disease. Neurobiol. Aging 2019, 73, 229.e11–229.e18. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.-Y.; Yu, J.-T.; Miao, D.; Ma, X.-Y.; Wang, H.-F.; Wang, W.; Tan, L. An exploratory study on STX6, MOBP, MAPT, and EIF2AK3 and late-onset Alzheimer’s disease. Neurobiol. Aging 2013, 34, 1519.e13–1519.e17. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Liu, H.-P.; Lin, W.-Y.; Tsai, F.-J. Machine learning compensates fold-change method and highlights oxidative phosphorylation in the brain transcriptome of Alzheimer’s disease. Sci. Rep. 2021, 11, 13704. [Google Scholar] [CrossRef]
- Pinto, M.; Pickrell, A.M.; Fukui, H.; Moraes, C.T. Mitochondrial DNA damage in a mouse model of Alzheimer’s disease decreases amyloid beta plaque formation. Neurobiol. Aging 2013, 34, 2399–2407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Randa, N.; Bora, E.; Ataman, E.; Öz, O.; Yener, G. Identification of PSEN1 and PSEN2 Gene Variants and Clinical Findings with the Literature. Int. J. Neurodegener. Dis. 2019, 2, 7. [Google Scholar]
- Raut, S.; Patel, R.; Al-Ahmad, A.J. Presence of a mutation in PSEN1 or PSEN2 gene is associated with an impaired brain endothelial cell phenotype in vitro. Fluids Barriers CNS 2021, 18, 3. [Google Scholar] [CrossRef]
- Du, Y.; Du, Y.; Zhang, Y.; Huang, Z.; Fu, M.; Li, J.; Pang, Y.; Lei, P.; Wang, Y.T.; Song, W. MKP-1 reduces Aβ generation and alleviates cognitive impairments in Alzheimer’s disease models. Signal Transduct. Target. Ther. 2019, 4, 58. [Google Scholar] [CrossRef] [Green Version]
- Gee, M.S.; Son, S.H.; Jeon, S.H.; Do, J.; Kim, N.; Ju, Y.-J.; Lee, S.J.; Chung, E.K.; Inn, K.-S.; Kim, N.-J. A selective p38α/β MAPK inhibitor alleviates neuropathology and cognitive impairment, and modulates microglia function in 5XFAD mouse. Alzheimer’s Res. Ther. 2020, 12, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Liu, P.; Wang, J.; Yu, H.; Zhang, Z.; Liu, J.; Chen, X.; Zhu, F.; Yang, X. Dauricine attenuates spatial memory impairment and alzheimer-like pathologies by enhancing mitochondrial function in a mouse model of Alzheimer’s disease. Front. Cell Dev. Biol. 2021, 8, 624339. [Google Scholar] [CrossRef] [PubMed]
- Adav, S.S.; Park, J.E.; Sze, S.K. Quantitative profiling brain proteomes revealed mitochondrial dysfunction in Alzheimer’s disease. Mol. Brain 2019, 12, 8. [Google Scholar] [CrossRef] [Green Version]
- Gong, S.; Su, B.B.; Tovar, H.; Mao, C.; Gonzalez, V.; Liu, Y.; Lu, Y.; Wang, K.-S.; Xu, C. Polymorphisms within RYR3 gene are associated with risk and age at onset of hypertension, diabetes, and Alzheimer’s disease. Am. J. Hypertens. 2018, 31, 818–826. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Del Prete, D.; Checler, F.; Chami, M. Ryanodine receptors: Physiological function and deregulation in Alzheimer disease. Mol. Neurodegener. 2014, 9, 21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Supnet, C.; Noonan, C.; Richard, K.; Bradley, J.; Mayne, M. Up-regulation of the type 3 ryanodine receptor is neuroprotective in the TgCRND8 mouse model of Alzheimer’s disease. J. Neurochem. 2010, 112, 356–365. [Google Scholar] [CrossRef]
- Das, B.; Yan, R. Role of BACE1 in Alzheimer’s synaptic function. Transl. Neurodegener. 2017, 6, 1–8. [Google Scholar] [CrossRef]
- Cervellati, C.; Valacchi, G.; Zuliani, G. BACE1 role in Alzheimer’s disease and other dementias: From the theory to the practice. Neural Regen. Res. 2021, 16, 2407. [Google Scholar] [CrossRef]
- McDade, E.; Voytyuk, I.; Aisen, P.; Bateman, R.J.; Carrillo, M.C.; De Strooper, B.; Haass, C.; Reiman, E.M.; Sperling, R.; Tariot, P.N. The case for low-level BACE1 inhibition for the prevention of Alzheimer disease. Nat. Rev. Neurol. 2021, 17, 703–714. [Google Scholar] [CrossRef]
- Bi, R.; Zhang, W.; Zhang, D.-F.; Xu, M.; Fan, Y.; Hu, Q.-X.; Jiang, H.-Y.; Tan, L.; Li, T.; Fang, Y. Genetic association of the cytochrome c oxidase-related genes with Alzheimer’s disease in Han Chinese. Neuropsychopharmacology 2018, 43, 2264–2276. [Google Scholar] [CrossRef] [Green Version]
- Wilkins, H.M.; Koppel, S.J.; Bothwell, R.; Mahnken, J.; Burns, J.M.; Swerdlow, R.H. Platelet cytochrome oxidase and citrate synthase activities in APOE ε4 carrier and non-carrier Alzheimer’s disease patients. Redox Biol. 2017, 12, 828–832. [Google Scholar] [CrossRef]
- Alldred, M.J.; Lee, S.H.; Stutzmann, G.E.; Ginsberg, S.D. Oxidative phosphorylation is dysregulated within the basocortical circuit in a 6-month old mouse model of down syndrome and alzheimer’s disease. Front. Aging Neurosci. 2021, 13, 484. [Google Scholar] [CrossRef]
- Misrani, A.; Tabassum, S.; Yang, L. Mitochondrial dysfunction and oxidative stress in Alzheimer’s disease. Front. Aging Neurosci. 2021, 13, 617588. [Google Scholar] [CrossRef] [PubMed]
- Bell, S.M.; Barnes, K.; De Marco, M.; Shaw, P.J.; Ferraiuolo, L.; Blackburn, D.J.; Venneri, A.; Mortiboys, H. Mitochondrial dysfunction in Alzheimer’s disease: A biomarker of the future? Biomedicines 2021, 9, 63. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Zhao, F.; Ma, X.; Perry, G.; Zhu, X. Mitochondria dysfunction in the pathogenesis of Alzheimer’s disease: Recent advances. Mol. Neurodegener. 2020, 15, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Gao, H.; Zaikin, A.; Chen, S. Unraveling Aβ-mediated multi-pathway calcium dynamics in astrocytes: Implications for Alzheimer’s disease treatment from simulations. Front. Physiol. 2021, 12, 1918. [Google Scholar] [CrossRef]
- Popugaeva, E.; Pchitskaya, E.; Bezprozvanny, I. Dysregulation of intracellular calcium signaling in Alzheimer’s disease. Antioxid. Redox Signal. 2018, 29, 1176–1188. [Google Scholar] [CrossRef]
- Workgroup, A.s.A.C.H.; Khachaturian, Z.S. Calcium hypothesis of Alzheimer’s disease and brain aging: A framework for integrating new evidence into a comprehensive theory of pathogenesis. Alzheimer’s Dement. 2017, 13, 178–182.e17. [Google Scholar]


| DEMi | Log2FC | p-Value | Regulation |
|---|---|---|---|
| hsa-miR-941 | 0.752891 | 0.020286 | Up |
| hsa-miR-1273c | 0.72872 | 0.016058 | Up |
| hsa-miR-150-5p | 0.685625 | 0.049536 | Up |
| hsa-miR-10a-5p | 0.597925 | 0.023684 | Up |
| hsa-miR-9-5p | 0.563954 | 0.011416 | Up |
| hsa-miR-1307-3p | −0.59803 | 0.024833 | Down |
| hsa-miR-877-5p | −0.61671 | 0.048433 | Down |
| hsa-miR-4433b-5p | −0.67039 | 0.036891 | Down |
| hsa-miR-143-3p | −0.69404 | 0.048802 | Down |
| hsa-miR-1296-5p | −0.85825 | 0.018019 | Down |
| hsa-miR-5100 | −1.21725 | 0.046714 | Down |
| hsa-miR-6501-5p | −1.34159 | 0.003957 | Down |
| DEMi | Gene ID | Gene Name | miTG Score |
|---|---|---|---|
| hsa-miR-6501-5p | ENSG00000124172 | ATP5E | 0.824031785 |
| ENSG00000138814 | PPP3CA | 0.793189081 | |
| hsa-miR-4433b-5p | ENSG00000161509 | GRIN2C | 0.754847813 |
| ENSG00000160014 | CALM3 | 0.746931533 | |
| ENSG00000162736 | NCSTN | 0.720055687 | |
| hsa-miR-143-3p | ENSG00000273079 | GRIN2B | 0.932644441 |
| ENSG00000172071 | EIF2AK3 | 0.898086248 | |
| ENSG00000147684 | NDUFB9 | 0.852775513 | |
| ENSG00000139180 | NDUFA9 | 0.84781014 | |
| ENSG00000080815 | PSEN1 | 0.825585469 | |
| ENSG00000100030 | MAPK1 | 0.756048436 | |
| ENSG00000023228 | NDUFS1 | 0.751173742 | |
| ENSG00000198838 | RYR3 | 0.744614559 | |
| ENSG00000186318 | BACE1 | 0.739327029 | |
| ENSG00000131143 | COX4I | 0.736374347 |
| KEGG Pathway | p-Value | DEMi Involved | Gene Involved |
|---|---|---|---|
| Alzheimer’s disease | 1.58 × 10−10 | hsa-miR-6501-5p | ATP5E |
| PPP3CA | |||
| hsa-miR-4433b-5p | GRIN2C | ||
| CALM3 | |||
| NCSTN | |||
| hsa-miR-143-3p | EIF2AK3 | ||
| NDUFB9 | |||
| NDUFA9 | |||
| PSEN1 | |||
| MAPK1 | |||
| NDUFS1 | |||
| RYR3 | |||
| BACE1 | |||
| COX4I1 | |||
| Oxidative phosphorylation | 9.67 × 10−6 | hsa-miR-6501-5p | ATP5E |
| NDUFB9 | |||
| hsa-miR-143-3p | NDUFA9 | ||
| NDUFS1 | |||
| COX4I1 | |||
| Circadian entrainment | 0.023972099 | hsa-miR-4433b-5p | GRIN2C |
| CALM3 | |||
| hsa-miR-143-3p | MAPK1 | ||
| RYR3 | |||
| Amphetamine addiction | 0.023972099 | hsa-miR-6501-5p | PPP3CA |
| hsa-miR-4433b-5p | GRIN2C | ||
| CALM3 | |||
| Long-term potentiation | 0.023972099 | hsa-miR-6501-5p | PPP3CA |
| hsa-miR-4433b-5p | GRIN2C | ||
| CALM3 | |||
| hsa-miR-143-3p | MAPK1 | ||
| Oxytocin signalling pathway | 0.023972099 | hsa-miR-6501-5p | PPP3CA |
| hsa-miR-4433b-5p | CALM3 | ||
| hsa-miR-143-3p | MAPK1 | ||
| RYR3 |
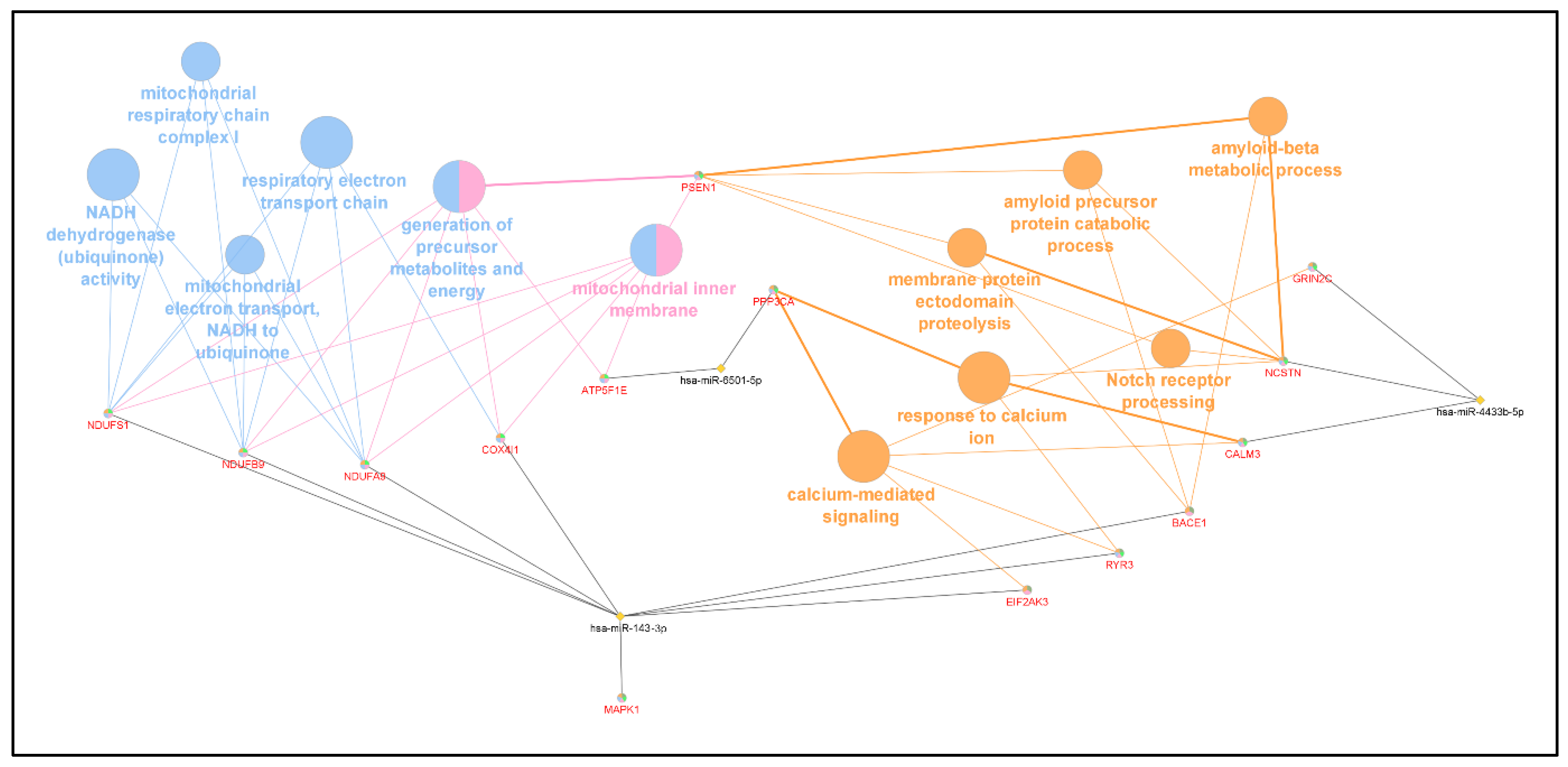
| GO Category | p-Value | DEMi Involved | Genes Involved |
|---|---|---|---|
| respiratory electron transport chain | 1.19 × 10−5 | hsa-miR-6501-5p | ATP5E |
| NDUFB9 | |||
| NDUFA9 | |||
| NDUFS1 | |||
| hsa-miR-143-3p | COX4I1 | ||
| generation of precursor metabolites and energy | 3.03 × 10−5 | hsa-miR-6501-5p | ATP5E |
| hsa-miR-4433b-5p | CALM3 | ||
| hsa-miR-143-3p | NDUFB9 | ||
| NDUFA9 | |||
| NDUFS1 | |||
| COX4I1 | |||
| beta-amyloid metabolic process | 0.001017955 | hsa-miR-4433b-5p | NCSTN |
| hsa-miR-143-3p | PSEN1 | ||
| BACE1 | |||
| calcium-mediated signalling | 0.001521237 | hsa-miR-6501-5p | PPP3CA |
| hsa-miR-4433b-5p | CALM3 | ||
| hsa-miR-143-3p | EIF2AK3 | ||
| membrane protein ectodomain proteolysis | 0.002220614 | hsa-miR-143-3p | PSEN1 |
| BACE1 | |||
| hsa-miR-4433b-5p | NCSTN | ||
| mitochondrial inner membrane | 0.005473536 | hsa-miR-143-3p | NDUFB9 |
| NDUFA9 | |||
| PSEN1 | |||
| NDUFS1 | |||
| COX4I1 | |||
| mitochondrial electron transport, NADH to ubiquinone | 0.005473536 | hsa-miR-143-3p | NDUFB9 |
| NDUFA9 | |||
| NDUFS1 | |||
| NADH dehydrogenase (ubiquinone) activity | 0.01130008 | hsa-miR-143-3p | NDUFB9 |
| NDUFA9 | |||
| NDUFS1 | |||
| mitochondrial respiratory chain complex I | 0.01133565 | hsa-miR-143-3p | NDUFB9 |
| NDUFA9 | |||
| NDUFS1 | |||
| amyloid precursor protein catabolic process | 0.0124875 | hsa-miR-4433b-5p | NCSTN |
| hsa-miR-143-3p | PSEN1 | ||
| Notch-receptor processing | 0.01577116 | hsa-miR-4433b-5p | NCSTN |
| hsa-miR-143-3p | PSEN1 | ||
| response to calcium ion | 0.01746326 | hsa-miR-6501-5p | PPP3CA |
| hsa-miR-4433b-5p | CALM3 |
| Gene | Roles and Functions as Related to AD |
|---|---|
| ATP synthase F1 subunit epsilon (ATP5E) | |
| Protein phosphatase 3 catalytic subunit alpha (PPP3CA) |
|
| Glutamate Ionotropic Receptor NMDA Type Subunit 2C (GRIN2C) |
|
| Calmodulin 3 (CALM3) | |
| Nicastrin (NCSTN) |
|
| Glutamate Ionotropic Receptor NMDA Type Subunit 2B (GRIN2B) | |
| Eukaryotic translation initiation factor 2 alpha kinase 3 (EIF2AK3) | |
| NADH:ubiquinone oxidoreductase subunit B9 (NDUFB9) | |
| NADH:ubiquinone oxidoreductase subunit A9 (NDUFA9) | |
| Presenilin 1 (PSEN1) |
|
| Mitogen-activated protein kinase 1 (MAPK1) |
|
| NADH:ubiquinone oxidoreductase subunit S1 (NDUFS1) | |
| Ryanodine receptor 3 (RYR3) |
|
| β-secretase cleaving enzyme 1 (BACE1) | |
| Cytochrome c oxidase subunit 4I1 (COX4I1) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tan, M.S.; Cheah, P.-L.; Chin, A.-V.; Looi, L.-M.; Chang, S.-W. Differential Expression Analysis of Blood MicroRNA in Identifying Potential Genes Relevant to Alzheimer’s Disease Pathogenesis, Using an Integrated Bioinformatics and Machine Learning Approach. Appl. Sci. 2023, 13, 3071. https://doi.org/10.3390/app13053071
Tan MS, Cheah P-L, Chin A-V, Looi L-M, Chang S-W. Differential Expression Analysis of Blood MicroRNA in Identifying Potential Genes Relevant to Alzheimer’s Disease Pathogenesis, Using an Integrated Bioinformatics and Machine Learning Approach. Applied Sciences. 2023; 13(5):3071. https://doi.org/10.3390/app13053071
Chicago/Turabian StyleTan, Mei Sze, Phaik-Leng Cheah, Ai-Vyrn Chin, Lai-Meng Looi, and Siow-Wee Chang. 2023. "Differential Expression Analysis of Blood MicroRNA in Identifying Potential Genes Relevant to Alzheimer’s Disease Pathogenesis, Using an Integrated Bioinformatics and Machine Learning Approach" Applied Sciences 13, no. 5: 3071. https://doi.org/10.3390/app13053071
APA StyleTan, M. S., Cheah, P.-L., Chin, A.-V., Looi, L.-M., & Chang, S.-W. (2023). Differential Expression Analysis of Blood MicroRNA in Identifying Potential Genes Relevant to Alzheimer’s Disease Pathogenesis, Using an Integrated Bioinformatics and Machine Learning Approach. Applied Sciences, 13(5), 3071. https://doi.org/10.3390/app13053071






