Vertebral Center Points Locating and Cobb Angle Measurement Based on Deep Learning
Abstract
1. Introduction
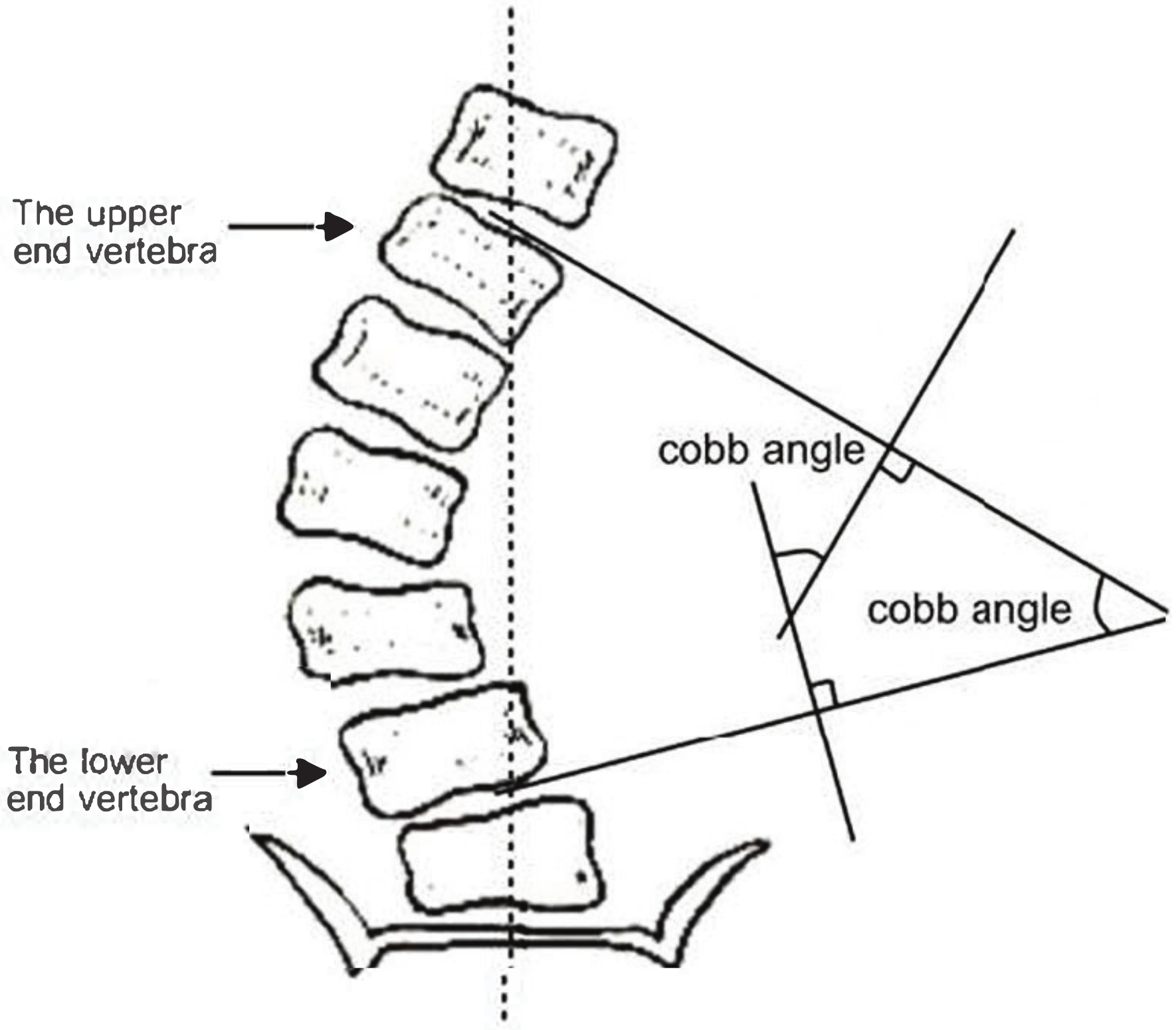
2. Manual Measurement of Scoliosis
3. Materials and Methods
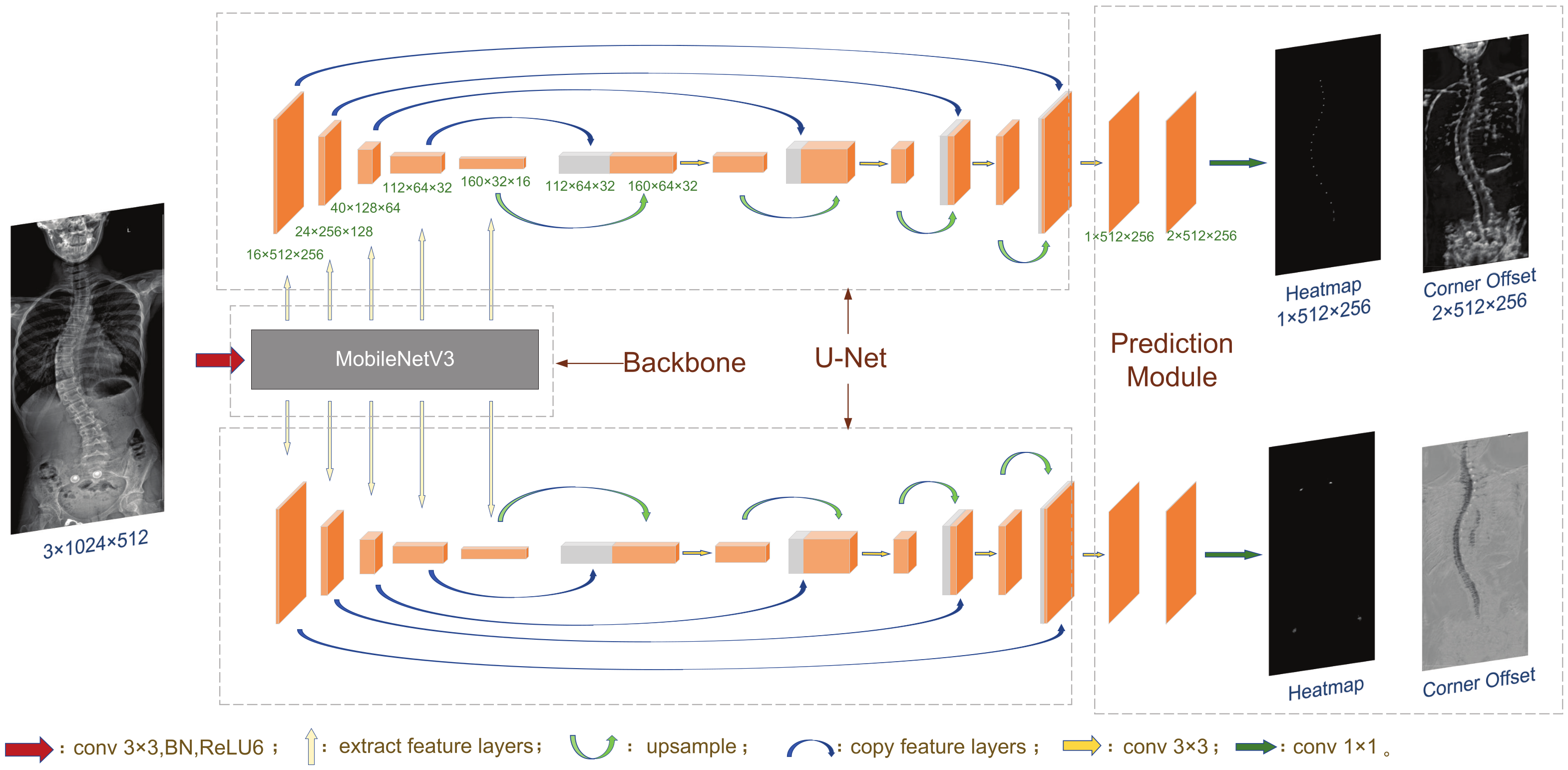
3.1. Network Structure
3.2. Loss Calculation
3.2.1. Heat Map Loss
3.2.2. Center Offset
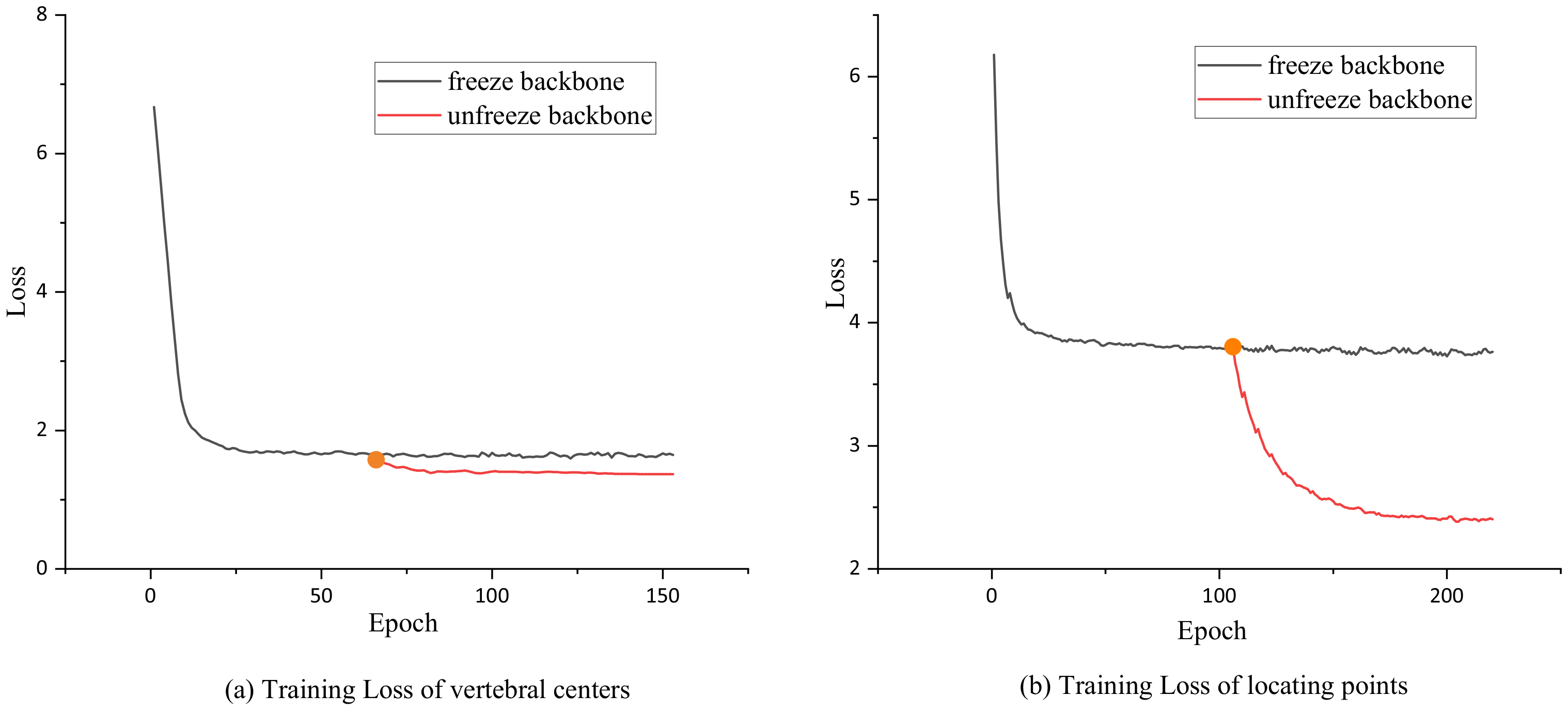
3.3. Network Training
3.3.1. Experimental Environment
3.3.2. Training Process
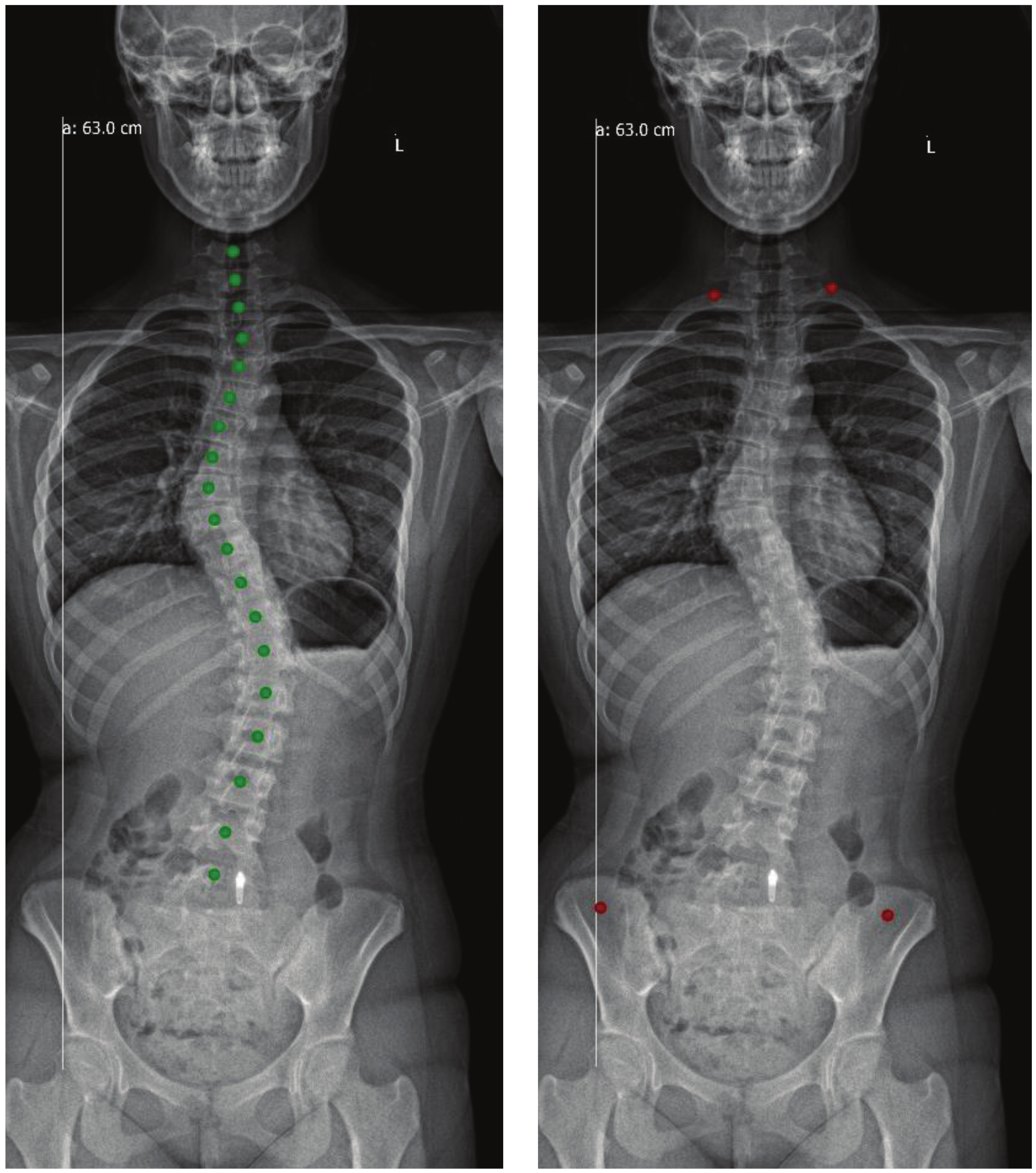
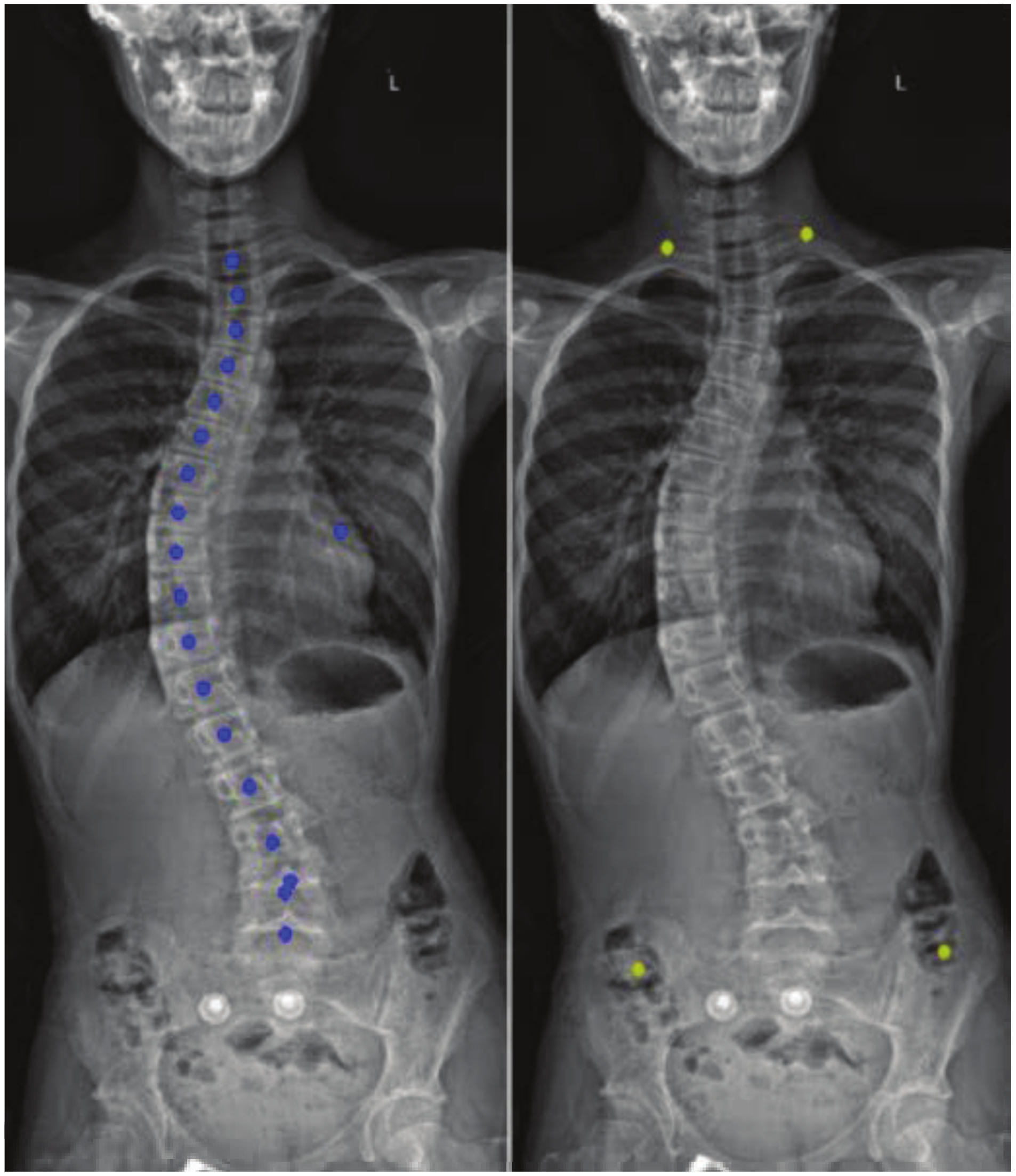
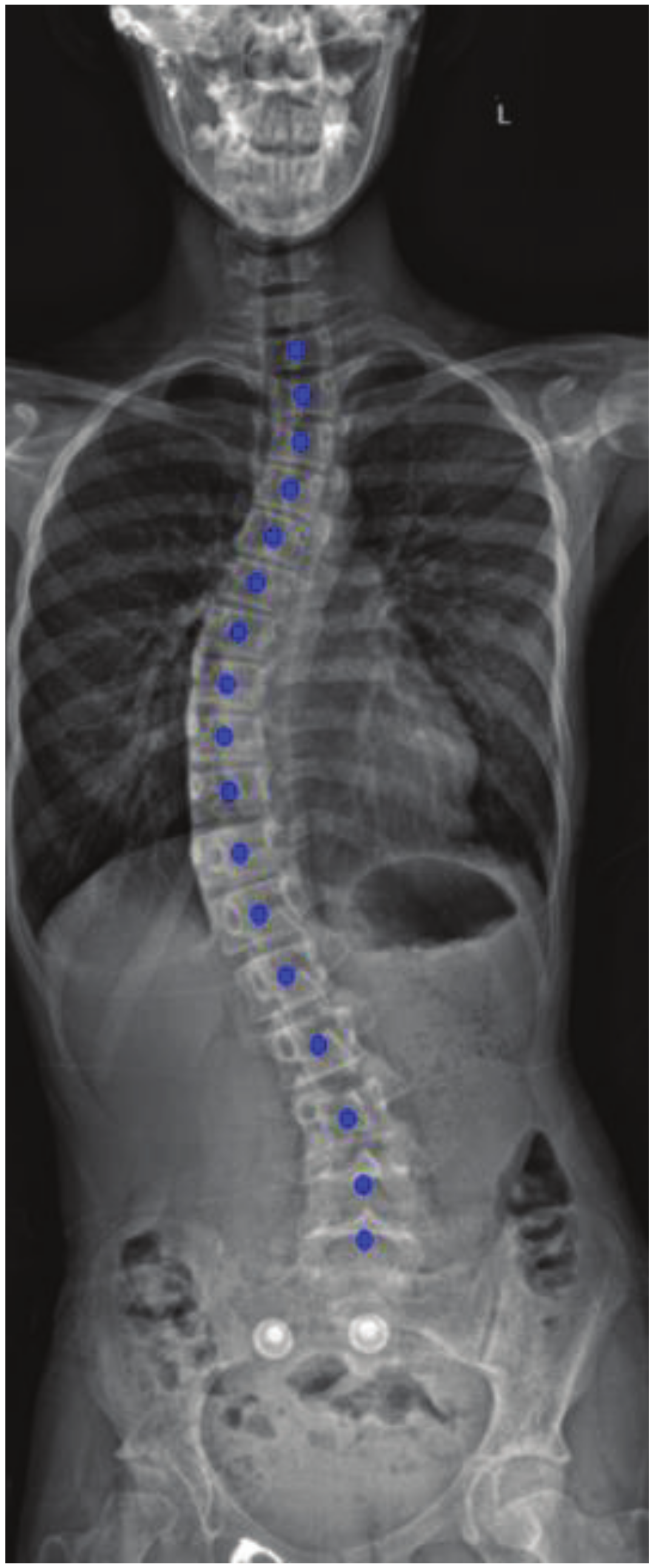
3.4. Post-Processing
3.4.1. Noise Removal
3.4.2. Spine Curve Fitting and Cobb Angle Calculation
4. Results
5. Discussion
6. Conclusions
- (1)
- It can predict the entire X-ray input without the need for cropping while only keeping the thoracic and lumbar vertebrae that need to be predicted, which meets the practical application requirements.
- (2)
- It is more stable, rigorous, and rapid than traditional manual measurement.
- (3)
- The number of network parameters is small, saving memory resources and improving the detection speed while ensuring the accuracy.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alman, B. Overview and Comparison of Idiopathic, Neuromuscular, and Congenital Forms of Scoliosis. In The Genetics and Development of Scoliosis; Kusumi, K., Dunwoodie, S.L., Eds.; Springer: Cham, Switzerland, 2018; pp. 73–79. [Google Scholar]
- Haleem, S.; Nnadi, C. Scoliosis: A review. Paediatr. Child Health 2018, 28, 209–217. [Google Scholar] [CrossRef]
- Du, C.P.; Yu, J.D.; Zhang, J.Q.; Jiang, J.; Lai, H.; Liu, W.; Liu, Y.; Li, H.; Wang, P. Relevant areas of functioning in patients with adolescent idiopathic scoliosis on the International Classification of Functioning, Disability and Health: The patients’ perspective. J. Rehabil. Med. 2016, 48, 806–814. [Google Scholar] [CrossRef] [PubMed]
- Lang, C.D.; Wang, R.J.; Chen, Z.M.; He, S.; Zou, Q.; Wu, J.; Zhu, X. Incidence and Risk Factors of Cardiac Abnormalities in Patients with Idiopathic Scoliosis. World Neurosurg. 2019, 125, 824–828. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Guo, H.; Chen, C.; Tan, H.; Lin, Y.; Li, Z.; Shen, J. Does Scoliosis Affect Sleep Breathing? World Neurosurg. 2018, 118, 946–950. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Lou, E.; Le, L.H.; Hill, D.L.; Raso, J.V.; Wang, Y. Automatic Cobb measurement of scoliosis based on fuzzy Hough Transform with vertebral shape prior. J. Digit. Imaging 2009, 22, 463–472. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, J.; Kundu, R.; Chakrabarti, A. Variability of Cobb angle measurement from digital X-ray image based on different de-noising techniques. Int. J. Biomed. Eng. Technol. 2014, 16, 113–134. [Google Scholar] [CrossRef]
- Mehmood, A.; Akram, M.U.; Akhtar, M.; Usman, A. Separation of Vertebrae Regions from Cervical Radiographs Using Inter-Vertebra Distance and Orientation. In Proceedings of the 16th International Conference on Hybrid Intelligent Systems (HIS 2016), Marrakech, Morocco, 21–23 November 2016; Springer: Berlin/Heidelberg, Germany, 2017; pp. 29–37. [Google Scholar]
- Al-Bashir, A.K.; Al-Abed, M.A.; Amari, H.K.; Al-Rousan, F.; Bashmaf, O.; Al-Basheer, A. Computer-based Cobb angle measurement using deflection points in adolescence idiopathic scoliosis from radiographic images. Neural Comput. Appl. 2019, 31, 1547–1561. [Google Scholar] [CrossRef]
- Zhang, J.H.; Li, H.J.; Lv, L.; Zhang, Y. Computer-aided cobb measurement based on automatic detection of vertebral slopes using deep neural network. Int. J. Biomed. Imaging 2017, 2017, 9083916. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.L.; Zhen, X.T.; Bailey, C.; Rasoulinejad, P.; Yin, Y.; Li, S. Direct estimation of spinal cobb angles by structured multi-output regression. In Proceedings of the Medical Imaging: 25th International Conference, Boone, NC, USA, 25–30 June 2017; Volume 10265, pp. 529–540. [Google Scholar]
- Wang, J.; Zhang, J.; Xu, R.; Chen, T.G.; Zhou, K.S.; Zhang, H.H. Measurement of scoliosis Cobb angle by end vertebra tilt angle method. J. Orthop. Surg. Res. 2018, 13, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Xu, Q.H.; Wang, L.S.; Leung, S.; Chung, J.; Li, S. An automated and accurate spine curve analysis system. IEEE Access 2019, 7, 124596–124609. [Google Scholar] [CrossRef]
- Sun, Y.; Xing, Y.Z.; Zhao, Z.; Meng, X.; Xu, G.; Hai, Y. Comparison of manual versus automated measurement of Cobb angle in idiopathic scoliosis based on a deep learning keypoint detection technology. Eur. Spine J. 2022, 31, 1969–1978. [Google Scholar] [CrossRef] [PubMed]
- McLeod, C.B. Anesthesia for Pediatric Spinal Deformity. In Multidisciplinary Spine Care; Noe, C.E., Ed.; Publishing House: Zurich, Switzerland, 2022; pp. 667–710. [Google Scholar]
- Nisser, J.; Smolenski, U.C.; Sliwinski, G.E.; Krueger, P.; Heinke, A.; Malberg, H.; Werner, M.; Drossel, W.G.; Sliwinski, Z.; Derlien, S. Scoliosis Specific Physiotherapy Approach to Adolescent Idiopathic Scoliosis (AIS)—A Narrative Review. Phys. Med. Rehab. Kuror. 2018, 28, 88–102. [Google Scholar]
- Glotzbecker, M.P.; Emans, J.B.; Hresko, M.T. Orthotic Management for Early Onset Scoliosis. In The Growing Spine; Akbarnia, B.A., Thompson, G.H., Yazici, M., El-Hawary, R., Eds.; Publishing House: Zurich, Switzerland, 2022; pp. 509–527. [Google Scholar]
- Dubousset, J. Idiopathic Scoliosis. In Essentials of Spine Surgery; Şenköylü, A., Canavese, F., Eds.; Publishing House: Zurich, Switzerland, 2022; pp. 101–105. [Google Scholar]
- Howard, A.; Sandler, M.; Chu, G.; Chen, L.C.; Chen, B.; Tan, M.; Wang, W.; Zhu, Y.; Pang, R.; Vasudevan, V.; et al. Searching for mobilenetv3. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Seoul, Republic of Korea, 20–26 October 2019; pp. 1314–1324. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015: 18th International Conference, Munich, Germany, 5–9 October 2015; pp. 234–241. [Google Scholar]
- Duan, K.; Bai, S.; Xie, L.X.; Qi, H.; Huang, Q.; Tian, Q. CenterNet: Keypoint Triplets for Object Detection. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Seoul, Republic of Korea, 20–26 October 2019; pp. 6569–6578. [Google Scholar]
- Hu, X.M.; Tian, J.H.; Qiao, H.X.; Wang, M. Multi-Person Pose Estimation via Learning Feature Integration. J. Phys. Conf. Ser. 2019, 1302, 32015. [Google Scholar] [CrossRef]
- Xu, J.J.; Song, B.; Yang, X.; Nan, X. An Improved Deep Keypoint Detection Network for Space Targets Pose Estimation. Remote Sens. 2020, 12, 3857. [Google Scholar] [CrossRef]
- Lin, T.Y.; Goyal, P.; Girshick, R.; He, K.; Dollár, P. Focal Loss for Dense Object Detection. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 24–27 October 2017; pp. 2980–2988. [Google Scholar]
- Girshick, R. Fast r-cnn. In Proceedings of the IEEE International Conference on Computer Vision, Santiago, Chile, 13–16 December 2015; pp. 1440–1448. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A Method for Stochastic Optimization. In Proceedings of the International Conference on Learning Representations, Banff, AB, Canada, 14–16 April 2014; pp. 1440–1448. [Google Scholar]

| Parameters (M) | MAPE (%) | FPS (Hz) | |
|---|---|---|---|
| Hourglass | 193.62 | 9.78 | 8.43 |
| HrNet | 242.60 | 10.03 | 6.65 |
| Proposed method | 13.46 | 9.24 | 14.44 |
| Locating Points | Modifier Formula | MAPE (%) | MAPE (%) | MAPE (%) | Accuracy of End Vertebrae Selection (%) | |
|---|---|---|---|---|---|---|
| 1 | 42.96 | 35.64 | 38.03 | 20.6 | ||
| 2 | √ | 26.73 | 18.58 | 21.06 | 34.4 | |
| 3 | √ | 25.45 | 33.19 | 28.27 | 78.1 | |
| 4 | √ | √ | 15.32 | 10.87 | 11.36 | 83.8 |
| ICC (95% CI) | MAD () | ||
|---|---|---|---|
| Intra-observer | Observer1 | 0.951 | 2.24 |
| Observer2 | 0.958 | 2.16 | |
| Inter-observer | 1st | 0.940 | 2.63 |
| 2nd | 0.942 | 2.55 |
| ICC (95% CI) | MAD () | |
|---|---|---|
| Observer1 & Auto | 0.897 | 3.13 |
| Observer2 & Auto | 0.901 | 3.04 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, Z.; Zhu, J.; Yao, C. Vertebral Center Points Locating and Cobb Angle Measurement Based on Deep Learning. Appl. Sci. 2023, 13, 3817. https://doi.org/10.3390/app13063817
Zhou Z, Zhu J, Yao C. Vertebral Center Points Locating and Cobb Angle Measurement Based on Deep Learning. Applied Sciences. 2023; 13(6):3817. https://doi.org/10.3390/app13063817
Chicago/Turabian StyleZhou, Zhifeng, Jia Zhu, and Chengxian Yao. 2023. "Vertebral Center Points Locating and Cobb Angle Measurement Based on Deep Learning" Applied Sciences 13, no. 6: 3817. https://doi.org/10.3390/app13063817
APA StyleZhou, Z., Zhu, J., & Yao, C. (2023). Vertebral Center Points Locating and Cobb Angle Measurement Based on Deep Learning. Applied Sciences, 13(6), 3817. https://doi.org/10.3390/app13063817





