1. Introduction
Scoliosis is a spinal deformity where one or more spinal segments deviate from the center line of the body and curve laterally; it can also be accompanied by spinal rotation [
1]. Scoliosis can occur in any age group, especially during adolescence, which is known as adolescent idiopathic scoliosis (AIS). The worldwide prevalence of AIS is approximately 0.5–5.2%, causing it to be the most common spinal deformity in adolescents [
2]. Scoliosis not only changes in the spine’s shape and function, but can, in severe cases, lead to a range of diseases, such as nerve damage, arrhythmia, cardiopulmonary dysfunction, pulmonary failure, and even paralysis.
Whole spine X-rays are the most common imaging examination for diagnosing, treating, and prognosis of scoliosis. Evaluating the Cobb angle (a type of measurement of the lateral curvature of the spine), vertebral rotation, and other parameters in X-rays can effectively reflect scoliosis severity and provide a basis for establishing the best treatment plan [
3]. Lenke et al. [
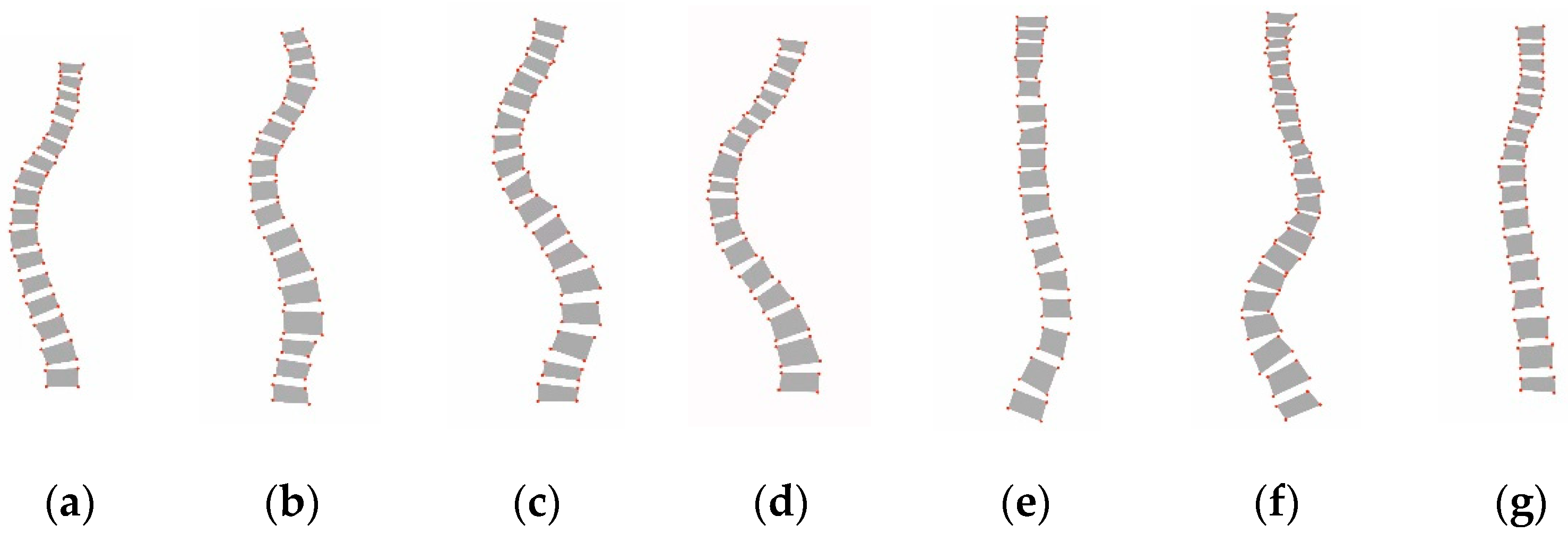
4] proposed a new classification system for assessing scoliosis severity, which is known as the Lenke classification criteria, and this has since been the standard guideline for evaluating scoliosis in clinical practice. As shown in
Figure 1, the Lenke classification criteria divide scoliosis into six types, from Lenke type 1 to type 6. Obtaining an accurate Lenke classification of scoliosis is integral for selecting the treatment modality, especially the surgical strategy.
The key steps for the Lenke classification of scoliosis include determining the location of each vertebra, computing the Cobb angle, and extracting the characteristic of the curve. In traditional methods, the key anatomical features of the spine are located and manually measured on the X-ray image using a ruler, protractor, and marker; then, using the Lenke classification criteria, the Lenke type is further determined by professional radiologists. However, manual measurement has many disadvantages, such as being time-consuming and having a large amount of errors, strong subjectivity, and high reliance on physician experience.
With the rapid development of artificial intelligence, computer vision application provides a more efficient way of diagnosing spinal diseases. Based on a comprehensive literature survey, three key visual techniques were identified as being involved in the Lenke classification of scoliosis.
The first is segmenting the spine image, that is, segmenting each vertebra from the spine image. Unet [
5] is a remarkable deep learning model that employs an “encoder–decoder” architecture based on fully convolutional networks (FCN) [
6]. Since then, numerous variants of Unet, such as UNet++ [
7], MAnet [
8], RMS-Unet [
9], X-Net [
10], and MQANet [
11], have been developed for use in medical image segmentation and other scenarios. Many researchers have also attempted to improve Unet for spinal image segmentation [
12]. For example, Sunetra et al. [
13] proposed an SIUNet with an improvised inception block and new dense skip pathways, which achieved promising performance in spine image segmentation. Christian et al. [
14] developed a coarse-to-fine approach for vertebrae localization and segmentation in CT images based on Unet, which achieved the best results in the MICCAI 2019 Vertebrae Segmentation Challenge [
15]. Recently, another deep learning method, transformers and their variants, has gained great attention and achieved significant success in image segmentation [
16]. Segmenter [
17] is a superior transformer model for segmentation that is built on top of the recent vision transformer [
18], which achieved state of the art in two scene image datasets. Recently, Rong et al. [
19] proposed a new transformer for labeling and segmenting of 3D vertebrae for arbitrary field-of-view CT images, thus demonstrating the great potential of transformer methods in spine image segmentation.
The second is Cobb angle measurement or spinal curvature estimation with geometric calculation, which is generally based on the segmentation results [
20]. Cheng et al. [
21] proposed a novel method to compute the Cobb angles monitoring the connection relationships among the segmented vertebrae in the X-ray images. Yi et al. [
22] proposed a spinal curvature estimation method built on top of the segmentation model. Kang et al. [
23] developed a spine curve assessment method by using a confidence map and vertebral tilt field, which also provided local and global spine structural information. Although these methods provide an excellent means to automatically compute the Cobb angle, there is still room for improvement as they heavily rely on the segmentation results. However, accurately segmenting vertebra is extremely difficult owing to the unclear vertebral boundary in the X-ray images. In addition, these methods are difficult to directly apply to the Lenke classification of scoliosis, as they have high requirements for the Cobb angle accuracy and need more additional features.
The third is feature representation for spinal images, which are beneficial for evaluating scoliosis severity. These features include the shape, texture, spatial relationship, and others. For example, Bayat et al. [
24] proposed a label method for the cervical, thoracic, and lumbar vertebrae using both texture and inter vertebral spatial information. Zhi et al. [
25] proposed to describe the spine using curve fitting, with the parameters of the fitted curve used for shape classification.
Recently, some work has focused on the classifying scoliosis using the above visual techniques. For example, Yang et al. [
26] proposed a classification scheme of mild AIS by using the bending asymmetry index (BAI) based on 3D ultrasound imaging. In [
27], another Lenke classification system based on BAI and cobb angle measurement was designed and achieved promising performance in X-ray images. However, in the above method, the BAI calculations were semi-automatic and relied on manual annotation. Gardner et al. [
28] proposed an effective method for the cluster analysis of Lenke types based on spinal and torso shape representation, but only Lenke type 1 was involved in this study. Rothstock et al. [
29] designed a semi-automatic classification framework for 3D surface trunk data by analyzing the asymmetry distance of the complete trunk as a predictor for scoliosis severity. Zhang et al. [
30] proposed a computerized Lenke classification approach based on Cobb angle measurement, which required the user to click the mouse to locate the vertebrae. To sum up, although various approaches have been proposed for classifying scoliosis with spine images, these methods still face many problems. First, most methods mainly provide some characteristics of the scoliosis as the output, such as the Cobb angle and most tilted vertebra [
31,
32]. Some recent work has focused on complete classification systems, but most are semi-automatic or rely on three-dimensional data; a fully automatic Lenke classification system with only X-ray images is rarely reported. In addition, the accuracy of Lenke classification greatly depends on the segmentation performance, and the design and selection of segmentation networks is challenging. Finally, the visual feature representation of scoliosis is another issue that must be carefully considered, as scoliosis has visually similar shapes and is difficult to distinguish in X-ray images. Therefore, it would be necessary to develop a complete automatic Lenke classification system that provides an accurate and efficient Lenke diagnosis in a visually interpretable manner.
To this end, we propose an automatic Lenke classification algorithm for scoliosis based on a segmentation network and an adaptive shape descriptor. Specifically, four important steps should be noted. First, a deep network based on Unet++ is employed to segment each vertebra in the spine X-ray images, and a post-processing approach is further used to enhance the segmentation effect. Thereafter, an automatic measurement system of the Cobb angle is designed, and then, an alternative Lenke classification solution for scoliosis is obtained. In addition, we propose an adaptive shape descriptor for segmented spine images to capture the discriminating shape features. Finally, a new Lenke classification algorithm for scoliosis is proposed by using the shape description and a KNN classifier. Performing rigorous experimental evaluations on a public dataset demonstrated that the proposed method achieved an accurate and robust performance for the Lenke classification of scoliosis. In summary, our contributions are fourfold:
- (1)
A novel clinician-friendly automatic Lenke classification framework for scoliosis based on a segmentation network and shape feature description is proposed.
- (2)
A new shape descriptor will be designed for segmented vertebrae, which can effectively describe the microscopic shape distribution of the vertebrae and adaptively adjust the matching weights.
- (3)
A simple and efficient post-processing approach for spinal image segmentation is proposed to avoid segmentation errors.
- (4)
A comprehensive evaluation will be performed for scoliosis, such as evaluating the impact of segmentation networks on Lenke classification and comparing of the Lenke classification frameworks based on the Cobb angle measurement and shape feature extraction, respectively.
The remainder of this paper is structured as follows:
Section 2 describes the proposed Lenke classification method of scoliosis and the implementation detail,
Section 3 presents the experimental evaluation and results, and
Section 4 presents the conclusion and future perspectives.
3. Experiments and Analysis
3.1. Experimental Datasets and Preprocessing
We employ a public dataset [
35] for the experimental evaluation, which was collected from the London Health Sciences Center in Canada that consists of 609 coronal spinal X-ray images with sizes ranging from 359 × 973 to 1386 × 2678. To ensure the deep learning framework’s effectiveness, we scale all images to a uniform size of 512 × 1536. Since the cervical vertebrae are rarely involved in spinal deformity, 12 thoracic and 5 lumbar vertebrae for each X-ray image are annotated by two professional radiologists. Each vertebra is labeled by four landmarks with reference to four corners, resulting in 68 points per spine image, which is also considered as the ground truth (GT) of vertebrae segmentation. With the landmarks, the Cobb angles can be further calculated. After the Cobb angle of each spine is determined, the Lenke type of scoliosis is annotated. For experimental data, we randomly selected 80% for training, 10% for validation, and 10% for testing. Some original image samples are shown in
Figure 13.
3.2. Experimental Setup and Evaluation
For the experimental environment, we used hardware platform with AMD R7-4800H CPU, NVIDIA GeForce GTX1650 GPU and SAMSUNG 16 GB DDR4 memory. The open-source PyTorch framework, the MMSegmentation toolkit [
36], and Segmentation Model PyTrorch toolkit [
37] were employed as the software environment. For the experimental parameter, the batch size and initial learning rate for the segmentation network were assigned as 4 and
, respectively. The Adam optimizer was used to change the learning rate. The number
n of the circular direction template of the shape descriptor was set as 24; the weight factors
,
, and
were set as 0.4, 0.4, and 0.2, respectively; and the parameter of the KNN classifier was set as 10.
Since our method involves two types of vision tasks, i.e., semantic segmentation and image classification, we used two groups of objective evaluation metrics for different vision tasks. For the semantic segmentation task, we employed five types of widely used metrics for evaluation, i.e., accuracy, sensitivity, specificity, dice, and MIoU [
38]. For the image classification task, the accuracy, precision, recall, and F1-score were selected for quantitative comparison. The equations of all the metrics are presented in
Table 2. In addition, TP, TN, FP, and FN were calculated at the pixel level for semantic segmentation, while they were calculated at the image level for the image classification.
3.3. Evaluation of Vertebrae Segmentation
In this section, we mainly evaluated the performance of the deep networks for segmenting the vertebrae and the proposed post-processing method. Six popular and recent methods were employed for this purpose, i.e., FPN [
39], Unet [
5], MAnet [
8], PSPNet [
40], Unet++ [
7], and Segmenter [
17]. For a fair comparison, the open-source MM segmentation toolkit was employed for all methods.
The quantitative experimental results of the compared methods are presented in
Table 3, where ⊗ denotes that post-processing was not used for the segmentation results; √ denotes that post-processing was used; and + indicates the increase with the post processing used. To intuitively present the visual comparison, the segmentation effects of the different methods are shown in
Figure 14, and some examples that illustrate the effects of post-processing are presented in
Figure 15.
From the above results, the following conclusions can be drawn.
First, as shown in
Table 3, the Segmenter achieved the best results with accuracy (0.946), sensitivity (0.915), specificity (0.997), dice (0.915), and MIoU (0.848), while Unet++ achieved the second-best results with accuracy (0.940), sensitivity (0.743), specificity (0.983), dice (0.805), and MIoU (0.679). In general, the accuracy and specificity are relatively superior among these methods, but the sensitivity, dice, and MIoU could be improved. This illustrates that the segmentation network approach is limited in terms of dealing with the details of the segmented vertebrae. More intuitive examples can be found in
Figure 14.
Second, by observing the visual comparison in
Figure 14, various errors such speckles, adhesions, and redundances exist in the segmentation results for different methods. For example, PSPNet has more speckle errors, and MANet has more adhesion errors. However, Unet++ achieved the best subjective results, which are reflected in the clearer segmentation contour and less adhesion errors. Segmenter has more speckles and adhesions in the subjective presentation than Unet++, even though Segmenter achieves high quantitative evaluation values. This greatly affects the Lenke classification performance of scoliosis, which partly why we use Unet++ as the recommended segmentation network.
Third, by observing the results in
Table 3 on whether post-processing was used, we note that the post-processing operation improved all methods under all indicators. This demonstrates that post-processing can effectively improve segmentation. Moreover, these improvements on quantitative segmentation indicators are not significant, as post-processing is primarily designed for subjective adhesions and speckles. From the visual comparison in
Figure 15, each method can successfully remove some subjective segmentation errors, such as adhesions, speckles, and holes, which significantly contributes to the Lenke classification of scoliosis. In addition, it is worth noting that the post-processing strategy improved Unet++ and Unet more significantly than the other approaches from both objective indicator and subjective observation. This also causes Unet++ to be more advantageous in the Lenke classification of scoliosis. In the next section, we conduct more experiments to verify the performance of the segmentation networks involved in the proposed Lenke classification framework.
In summary, the Unet++ and Segmenter models achieved relatively superior segmentation results with post-processing. In addition, in the next sections, we further test the performance of these segmentation models that are incorporated into the proposed Lenke classification framework of scoliosis.
3.4. Ablation Experiment for the Proposed Shape Descriptor
To examine the effectiveness of each module of the proposed adaptive shape descriptor, we conducted a series of ablation experiments to evaluate each strategy’s contribution. Specifically, we selected Unet++ as the segmentation network, and the segmentation results are described according to the proposed shape descriptor and further used for the Lenke classification (see
Section 2.5.2). Four shape descriptors should be marked for ablation evaluation: (1) S1 descriptor, which only used outline representation and matching (see
Section 2.4.1); (2) S2 descriptor, which employed adaptive similarity matching weight based on S1 (see
Section 2.4.2); (3) S3 descriptor, which improved the shape representation by the upper and lower boundaries based on S2 (see
Section 2.4.3); and (4) S4 descriptor, which combined S3 and symmetric matching, i.e., the finally proposed shape descriptor for the X-ray spinal images (see
Section 2.4.4). The experimental results for the four shape descriptors incorporated into the Lenke classification framework of scoliosis are listed in
Table 4. Since the segmentation results generated by the different deep networks may affect the Lenke classification’s performance, we also compared the Lenke classification results based on different segmentation networks combined with the S4 descriptor; the results are presented in
Table 5.
Based on a comprehensive analysis of the above results, the following conclusions can be drawn:
First, regardless of whether post-processing is used,
Table 4 shows a steady increase in all metrics, which indicates that our proposed shape modules have a positive effect on the shape representation and Lenke classification of scoliosis. Specifically, S2 improved S1 in accuracy (⊗5.8% and √6%), precision (⊗2.5% and √2.4%), recall (⊗1.8% and √3.9%), and F1-score (⊗7.3% and √8.3%). This is mainly because the proposed adaptive weigh assignment strategy for shape matching in S2 uses the most curved segments as the key factors, which effectively reflects the most discriminative features of the Lenke shape. In addition, S3 led to an obvious increment on S2 in accuracy (⊗7.9% and √7.2%), precision (⊗31.2% and √31.7%), recall (⊗8.4% and √6.8%), and F1-score (⊗10.9% and √10.3%), which demonstrates that the proposed shape description of each vertebra’s horizontal tilt angle is a great feature supplement for the Lenke classification of scoliosis. Finally, our S4 method achieved the best results and improved S3 in all metrics. This not only indicates that our method overcomes the errors caused by rotation, but also shows that the organic combination of these shape representation strategies can play a significant role in the Lenke classification.
Second, from the results of
Table 5, we notice that the proposed framework combining Unet++ and S4 achieved the best performance in the Lenke classification, irrespective of whether the post-processing operation was used. This is because Unet++ provided superior segmentation results in both the objective indicators and subjective effects, which cause the proposed shape descriptor to be more effective. However, a particular phenomenon should be explained, which is that Segmenter + S4 achieved an inferior classification performance even though Segmenter surpassed Unet++ in the quantitative evaluation of the segmentation. This is mainly because the actual segmentation effect of Segmenter has some non-negligible errors in the boundaries of the vertebrae, which greatly influences the Lenke classification of scoliosis. Additionally, for Unet++, by broadening the main structure of Unet, it can capture the features of different levels and obtain clearer boundaries for the vertebrae, thus providing a more reliable benchmark for shape description. Furthermore, the post-processing operation plays a greater role in Unet++ than Segmenter by viewing
Table 3, which also contributes to improving the classification effect.
Finally, by comparing the results in
Table 4 on whether post-processing was used, the proposed post-processing strategy improves the classification performance under all evaluation indicators. For example, the post-processing operation gains accuracy improvement with 2.6%, 2.8%, 2.1%, and 2.1% for S1, S2, S3, and S4, respectively, and gains F1-score improvements with 2.5%, 3.5%, 2.9%, and 2.7% for those four methods. Indeed, the results in
Table 4 support a similar conclusion, i.e., that a post-processing operation improves the Lenke classification performance for different segmentation networks combined with S4. These improvements are even more significant, such as the post-processing operation gaining precision improvement with 13.8%, 14.7%, 15.6%, 13.7%, 14.0%, and 2.7% for FPN, PSPNet, MANet, Unet, Segmenter, and Unet++ combined with S4, respectively. This suggests that the proposed post-processing algorithm can effectively overcome the segmentation error and help to improve the scoliosis classification performance.
3.5. Comparison of the Representative and Latest Methods
For a complete evaluation, we selected two types of methods for comparison. The first type is the four representative shape descriptors involved in our classification framework: (1) shape context [
41,
42], which is a classic shape descriptor and has important applications in many fields; (2) TAR [
43], which has superior performance in overall and local shape description; (3) CBoW [
44], which provides a new strategy to describe shapes with bag of visual words; and (4) Fourier descriptor [
45], which has recently been used for shape representation in cerebral microbleed detection. The second type is the recently popular deep learning classification method that we performed on spinal X-ray images for Lenke classification, such as remarkable Resnet101 [
46], the latest vision transformer [
18], and Swim transformer [
47]. All these compared methods achieved state-of-the-art for shape representation or image classification. All the experimental results are presented in
Table 6, where the values marked in bold indicate the best performance.
To further provide a visual comparison of the shape representation, we also conducted a content-based image retrieval experiment for the five compared shape descriptors. We input an original spine X-ray image as the query and retrieved the seven most similar results in the dataset using the proposed shape matching framework based segmentation network. The two groups of retrieval results are presented in
Figure 16 and
Figure 17, respectively.
From the above result, we can draw the following conclusions.
First, the proposed Lenke classification framework embedded with our adaptive shape descriptor achieves best results in all the evaluation indicators, which shows its outstanding performance compared with the other representative and latest methods. Specifically, compared with the shape context descriptor, we achieved an improvement of 35.8% in accuracy, 48.4% in precision, 37.5% in recall, and 43.1% in F1-score; compared with the Fourier descriptor, we achieved an improvement of 21.5% in accuracy, 20.9% in precision, 20.8% in recall, and 22.3% in F1-score. We also surpassed the other shape descriptors including CBoW and TAR for all metrics, and the improvement was markedly significant. This is mainly because our shape descriptor is especially designed for describing the microscopic shape distribution of the vertebrae, while the existing shape descriptors were mainly developed for objects with larger shape changes.
Second, we achieved superior results compared with deep learning classification methods. Specifically, compared with Resnet101, we achieved an improvement of 5.3% in accuracy, 20.8% in precision, 17.1% in recall, and 19.1% in F1-score. Compared to the recent transformer methods that demonstrated conspicuous performance in image classification, such as Swin transformer and vision transformer, we achieved more significant performance improvements. This demonstrates that the Lenke classification of scoliosis in X-rays is indeed challenging. It may be difficult to directly use an end-to-end deep learning method to the learn the small shape changes between different Lenke types, and our hand-designed method can better describe such shape differences. In addition, the small number of data samples affects the performance of transformers and other deep learning methods. We used deep leaning to segment and manually design the shape description and matching method to achieve a more efficient scoliosis classification.
Finally, by observing the content-based image retrieval results in
Figure 16 and
Figure 17, we found the proposed shape descriptor achieved the best results. Specifically, our method yielded the results that had the most Lenke types in common with the query. Using the query marked Lenke 4 in
Figure 16 as an example, five of the seven most similar results by our method were Lenke 4, while, for the shape context, CBoW, TAR, and Fourier descriptor, there are four, four, three and two samples marked Lenke 4 were retrieved, respectively. For the results in
Figure 17, our method also retrieved the most images, consistent with the Lenke type of the query. In addition, from the perspective of subjective observation, the results obtained using our method are more visually similar to the query, i.e., the shape of the spine obtained by our method is similar to that of the query. Even those results with different Lenke types from the query still have a certain similarity in the general outline shape. This implies that the proposed shape descriptor can describe the spine’s overall shape and local details well.
In summary, the proposed method achieved competitive performance in the Lenke classification of scoliosis.
3.6. Comparison of the Classification Framework Based on Cobb Angle Measurement and Shape Description
As shown in
Figure 2, we present two alternative schemes for the Lenke classification of scoliosis. The first was to use Cobb angle measurement and classifying criteria (denoted as Cobb angle + criteria for short) based on the segmentation results, and the second was to use the shape description and KNN classifier (denoted as Shape + KNN for short) for the segmentation results. In this section, we conducted an experiment to compare the two schemes; the results are listed in
Table 7.
From
Table 7, we note that the scheme using the shape description and KNN classifier achieved better experimental results than that using the Cobb angle and classifying criteria, where most of the indicators are significant. This is because the Lenke classification criteria built on the Cobb angle is very strict and is extremely dependent on the accuracy of the segmentation. However, the proposed automatic Lenke classification method based on the shape descriptor and KNN classifier achieved a considerable improvement and overcame the influence of the segmentation error caused by deep learning to a great extent.
3.7. Computational Complexity Analysis
In this section, we focus on computational complexity analysis for the proposed Lenke classification framework of scoliosis, which consists of two parts: segmentation using deep learning and classification using the shape descriptor and classifier.
First, we report the parameter memory as the evaluation indicator for the segmentation networks in
Table 8, in which the input and output image are resized to 256 * 256. To provide a more intuitive and comprehensive comparison, a schematic diagram of the computational complexity requirements vs. the classification performance is presented in
Figure 18. From the above results, the MANet evidently has the smallest params. However, if we regard the classification performance as a comprehensive consideration, the Unet++ has a moderate param size and operation count. Therefore, based on a balance of the classification and time complexity, we recommend Unet++ as the preferred segmentation network.
Next, we performed computational complexity analysis for different classification methods, which mainly included feature extraction, training, and testing. From the results shown in
Table 9, the proposed adaptive shape descriptor had minimum time consumption in the feature extraction among all shape descriptors and a moderate testing time. It is worth noting that shape context, TAR, and our method directly extracted features from a single image. Thus, there is no training process. For CBoW, the visual dictionary needs to be trained from the dataset, and the training time is relatively long. For deep learning methods, the feature extraction is included in the training time. Considering the actual performance and time complexity, we concluded that the proposed method is the best choice in the Lenke classification of scoliosis.
4. Conclusions and Future Perspectives
In this study, we mainly investigated the Lenke classification problem for scoliosis and proposed a novel automatic Lenke classification framework. First, we used deep networks such as Unet++ to segment the vertebrae in the spine X-ray images and employed an effective post-processing strategy to overcome the spots and adhesions caused by the segmentation errors. We then focused on the shape representation of the segmented spine, and a new shape descriptor was designed to describe the details of the spinal curvature. With shape feature extraction and matching, a new Lenke classification algorithm for scoliosis was constructed with a classifier. For comparison and application, we also built another alternative Lenke classification option based on the automatic measurement of the Cobb angle. Finally, multiple experiments were conducted on a public dataset, including evaluating the segmentation networks, evaluating the shape descriptors, evaluating the classification strategies, and performing ablation experiments. The experimental results indicated that the proposed method achieved the best Lenke classification performance for scoliosis among the compared methods. The ablation experiments demonstrated that the modules in our shape descriptor were organically connected and supported by each other.
To further highlight the technical improvements and contribution compared with the state-of-the-art methods, three aspects should be noted. First, we improved the overall framework and viewed the Lenke classification of scoliosis from the perspective of its microscopic shape on the basis of segmentation network. Second, we improved the shape representation through three novel strategies: the adaptive similarity matching based on key segment calculation, the tilt angle description, and the symmetric matching. These strategies are carefully designed for the needs of the Lenke classification of scoliosis, which can be used to obtain more shape details. Finally, a general post-processing method for vertebrae segmentation is proposed, which improved the segmentation results for all compared deep networks.
Although the proposed method achieved superior results in the Lenke classification of scoliosis, there is still much room for improvement in terms of the objective evaluation indicators. This is attributed to two reasons. First, different Lenke types have very similar shapes, which is very hard to distinguish with human judgment. The rules of the Lenke classification dictate that it requires a very high level of segmentation and fine-grained shape representation, which is a great challenge. In addition, for fairness of the comparison and reproducibility of the results, we evaluated our method on a publicly available dataset that only included coronal plane X-rays of scoliosis. In fact, sagittal plane X-rays are an important supplement for diagnosing of scoliosis. However, under the present case, our method still significantly improved the existing methods and yielded satisfactory results with smaller data requirements, faster computation, and full automation. Moreover, the proposed Lenke classification framework and adaptive shape descriptor are also applicable to evaluating the spinal curvature of the sagittal plane X-rays. Combining the coronal plane and sagittal plane X-rays can further improve the proposed method’s performance. One alternative solution is to assign the weights to the coronal and sagittal plane X-rays in the similarity matching of the two scoliosis cases. In future work, we will construct a clinical scoliosis dataset with both sagittal and coronal plane X-rays and fully utilize two kinds of plane information to further improve the Lenke classification of scoliosis.