Exploring the Potential of Four Novel Halotolerant Bacterial Strains as Plant-Growth-Promoting Rhizobacteria (PGPR) under Saline Conditions
Abstract
:1. Introduction
2. Materials and Methods
2.1. Characterization of Strains and Culture Conditions
2.2. Amplification and Sequencing of 16S rDNA
2.3. Identification of Bacteria by Analysis of 16S rDNA Sequences
2.4. Salt and Temperature Tolerance
2.5. Plant-Growth-Promoting Features
2.5.1. IAA Production
2.5.2. ACC Deaminase Quantitative Analysis
2.5.3. Protein Extract Quantization
2.5.4. Production of Siderophores
2.5.5. Phosphate Solubilization Capacity
2.5.6. Ammonia Production
3. Results
3.1. Genetic Characterization
3.2. Morphological and Biochemical Characterization
3.3. PGP Features
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Egamberdieva, D.; Wirth, S.; Bellingrath-Kimura, S.D.; Mishra, J.; Arora, N.K. Salt-Tolerant Plant Growth Promoting Rhizobacteria for Enhancing Crop Productivity of Saline Soils. Front. Microbiol. 2019, 10, 2791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nosova, M.V.; Seredina, V.P.; Rybin, A.S. Ecological State of Technogeneous Saline Soil of Oil-Contaminated Alluvial Ecosystems and Their Remediation Techniques. IOP Conf. Ser. Mater. Sci. Eng. 2020, 921, 012018. [Google Scholar] [CrossRef]
- Moharana, P.C.; Singh, R.S.; Singh, S.K.; Tailor, B.L.; Jena, R.K.; Meena, M.D. Development of secondary salinity and salt migration in the irrigated landscape of hot arid India. Environ. Earth Sci. 2019, 78, 454. [Google Scholar] [CrossRef]
- Stavi, I.; Thevs, N.; Priori, S. Soil Salinity and Sodicity in Drylands: A Review of Causes, Effects, Monitoring, and Restoration Measures. Front. Environ. Sci. 2021, 9, 330. [Google Scholar] [CrossRef]
- Ondrasek, G.; Rathod, S.; Manohara, K.K.; Gireesh, C.; Anantha, M.S.; Sakhare, A.S.; Parmar, B.; Yadav, B.K.; Bandumula, N.; Raihan, F.; et al. Salt Stress in Plants and Mitigation Approaches. Plants 2022, 11, 717. [Google Scholar] [CrossRef]
- Ondrasek, G.; Rengel, Z. Environmental salinization processes: Detection, implications & solutions. Sci. Total Environ. 2021, 754, 142432. [Google Scholar] [CrossRef]
- Safdar, H.; Amin, A.; Shafiq, Y.; Ali, A.; Yasin, R.; Sarwar, M.I. Abbas Shoukat, Maqsood Ul Hussan, Muhammad Ishtiaq Sarwar. A review: Impact of salinity on plant growth. Nat. Sci. 2019, 1, 34–40. [Google Scholar] [CrossRef]
- dos Santos, T.B.; Ribas, A.F.; de Souza, S.G.H.; Budzinski, I.G.F.; Domingues, D.S. Physiological Responses to Drought, Salinity, and Heat Stress in Plants: A Review. Stresses 2022, 2, 113–135. [Google Scholar] [CrossRef]
- Rossi, M.; Borromeo, I.; Capo, C.; Glick, B.R.; Del Gallo, M.; Pietrini, F.; Forni, C. PGPB Improve Photosynthetic Activity and Tolerance to Oxidative Stress in Brassica napus Grown on Salinized Soils. Appl. Sci. 2021, 11, 11442. [Google Scholar] [CrossRef]
- Bharti, N.; Barnawal, D. Chapter Five—Amelioration of Salinity Stress by PGPR: ACC Deaminase and ROS Scavenging Enzymes Activity. In PGPR Amelioration in Sustainable Agriculture; Singh, A.K., Kumar, A., Singh, P.K., Eds.; Woodhead Publishing: Sawston, UK, 2019; pp. 85–106. [Google Scholar] [CrossRef]
- de la Porte, A.; Schmidt, R.; Yergeau, É.; Constant, P. A Gaseous Milieu: Extending the Boundaries of the Rhizosphere. Trends Microbiol. 2020, 28, 536–542. [Google Scholar] [CrossRef]
- Dastogeer, K.M.G.; Tumpa, F.H.; Sultana, A.; Akter, M.A.; Chakraborty, A. Plant microbiome–an account of the factors that shape community composition and diversity. Curr. Plant Biol. 2020, 23, 100161. [Google Scholar] [CrossRef]
- Vocciante, M.; Grifoni, M.; Fusini, D.; Petruzzelli, G.; Franchi, E. The Role of Plant Growth-Promoting Rhizobacteria (PGPR) in Mitigating Plant’s Environmental Stresses. Appl. Sci. 2022, 12, 1231. [Google Scholar] [CrossRef]
- Ur Rehman, F.; Kalsoom, M.; Adnan, M.; Toor, M. Plant Growth Promoting Rhizobacteria and their Mechanisms Involved in Agricultural Crop Production: A Review. SunText Rev. Biotechnol. 2020, 1, 110. [Google Scholar] [CrossRef]
- Myo, E.M.; Ge, B.; Ma, J.; Cui, H.; Liu, B.; Shi, L.; Jiang, M.; Zhang, K. Indole-3-acetic acid production by Streptomyces fradiae NKZ-259 and its formulation to enhance plant growth. BMC Microbiol. 2019, 19, 155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Misra, S.; Chauhan, P.S. ACC deaminase-producing rhizosphere competent Bacillus spp. mitigate salt stress and promote Zea mays growth by modulating ethylene metabolism. 3 Biotech 2020, 10, 119. [Google Scholar] [CrossRef]
- Kramer, J.; Özkaya, Ö.; Kümmerli, R. Bacterial siderophores in community and host interactions. Nat. Rev. Microbiol. 2020, 18, 152–163. [Google Scholar] [CrossRef]
- Meena, M.; Swapnil, P.; Divyanshu, K.; Kumar, S.; Harish; Tripathi, Y.N.; Zehra, A.; Marwal, A.; Upadhyay, R.S. PGPR-mediated induction of systemic resistance and physiochemical alterations in plants against the pathogens: Current perspectives. J. Basic Microbiol. 2020, 60, 828–861. [Google Scholar] [CrossRef]
- Neshat, M.; Abbasi, A.; Hosseinzadeh, A.; Sarikhani, M.R.; Dadashi Chavan, D.; Rasoulnia, A. Plant growth promoting bacteria (PGPR) induce antioxidant tolerance against salinity stress through biochemical and physiological mechanisms. Physiol. Mol. Biol. Plants 2022, 28, 347–361. [Google Scholar] [CrossRef]
- Habib, S.H.; Kausar, H.; Saud, H.M. Plant Growth-Promoting Rhizobacteria Enhance Salinity Stress Tolerance in Okra through ROS-Scavenging Enzymes. BioMed Res. Int. 2016, 2016, e6284547. [Google Scholar] [CrossRef] [Green Version]
- Ilangumaran, G.; Smith, D.L. Plant Growth Promoting Rhizobacteria in Amelioration of Salinity Stress: A Systems Biology Perspective. Front. Plant Sci. 2017, 8, 1768. [Google Scholar] [CrossRef] [Green Version]
- Yousef, N.M.H. Capability Of Plant Growth-Promoting Rhizobacteria (Pgpr) For Producing Indole Acetic Acid (Iaa) Under Extreme Conditions. Eur. J. Biol. 2018, 8, 174–182. [Google Scholar] [CrossRef]
- Deshwal, V.K.; Kumar, P. Effect of Salinity on Growth and PGPR Activity of Pseudomonads. J. Acad. Ind. Res. 2013, 2, 353. [Google Scholar]
- Castiglione, S.; Oliva, G.; Vigliotta, G.; Novello, G.; Gamalero, E.; Lingua, G.; Cicatelli, A.; Guarino, F. Effects of Compost Amendment on Glycophyte and Halophyte Crops Grown on Saline Soils: Isolation and Characterization of Rhizobacteria with Plant Growth Promoting Features and High Salt Resistance. Appl. Sci. 2021, 11, 2125. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Silakari, O.; Singh, P. Statistical methods and parameters: Tools to generate and evaluate theoretical in silico models. In Concepts and Experimental Protocols of Modelling and Informatics in Drug Design; Academic Press: Cambridge, MA, USA, 2021; pp. 333–350. [Google Scholar] [CrossRef]
- Penrose, D.M.; Glick, B.R. Methods for isolating and characterizing ACC deaminase-containing plant growth-promoting rhizobacteria. Physiol. Plant. 2003, 118, 10–15. [Google Scholar] [CrossRef] [Green Version]
- Alexander, D.B.; Zuberer, D.A. Use of chrome azurol S reagents to evaluate siderophore production by rhizosphere bacteria. Biol. Fertil. Soils 1991, 12, 39–45. [Google Scholar] [CrossRef]
- Nautiyal, C.S. An efficient microbiological growth medium for screening phosphate solubilizing microorganisms. FEMS Microbiol. Lett. 1999, 170, 265–270. [Google Scholar] [CrossRef]
- Gang, S.; Sharma, S.; Saraf, M.; Buck, M.; Schumacher, J. Analysis of Indole-3-acetic Acid (IAA) Production in Klebsiellaby LC-MS/MS and the Salkowski Method. Bio-Protocol 2019, 9, e3230. [Google Scholar] [CrossRef]
- Li, H.; La, S.; Zhang, X.; Gao, L.; Tian, Y. Salt-induced recruitment of specific root-associated bacterial consortium capable of enhancing plant adaptability to salt stress. ISME J. 2021, 15, 2865–2882. [Google Scholar] [CrossRef]
- Lagier, J.-C.; Dubourg, G.; Million, M.; Cadoret, F.; Bilen, M.; Fenollar, F.; Levasseur, A.; Rolain, J.-M.; Fournier, P.-E.; Raoult, D. Culturing the human microbiota and culturomics. Nat. Rev. Microbiol. 2018, 16, 540–550. [Google Scholar] [CrossRef] [Green Version]
- Parks, D.H.; Chuvochina, M.; Waite, D.W.; Rinke, C.; Skarshewski, A.; Chaumeil, P.-A.; Hugenholtz, P. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat. Biotechnol. 2018, 36, 996–1004. [Google Scholar] [CrossRef] [PubMed]
- Bejaoui, Z. Optimization of the effect of PGPR–based biofertlizer on wheat growth and yield. Biom. Biostat. Int. J. 2018, 3, 226–232. [Google Scholar] [CrossRef] [Green Version]
- Bharti, N.; Barnawal, D.; Awasthi, A.; Yadav, A.; Kalra, A. Plant growth promoting rhizobacteria alleviate salinity induced negative effects on growth, oil content and physiological status in Mentha arvensis. Acta Physiol. Plant. 2014, 36, 45–60. [Google Scholar] [CrossRef]
- Zeng, X.; Alain, K.; Shao, Z. Microorganisms from deep-sea hydrothermal vents. Mar. Life Sci. Technol. 2021, 3, 204–230. [Google Scholar] [CrossRef]
- Orhan, F.; Demirci, A. Salt Stress Mitigating Potential of Halotolerant/Halophilic Plant Growth Promoting. Geomicrobiol. J. 2020, 37, 663–669. [Google Scholar] [CrossRef]
- Khan, A.L.; Halo, B.A.; Elyassi, A.; Ali, S.; Al-Hosni, K.; Hussain, J.; Al-Harrasi, A.; Lee, I.-J. Indole acetic acid and ACC deaminase from endophytic bacteria improves the growth of Solanum lycopersicum. Electron. J. Biotechnol. 2016, 21, 58–64. [Google Scholar] [CrossRef] [Green Version]
- Fu, S.-F.; Wei, J.-Y.; Chen, H.-W.; Liu, Y.-Y.; Lu, H.-Y.; Chou, J.-Y. Indole-3-acetic acid: A widespread physiological code in interactions of fungi with other organisms. Plant Signal. Behav. 2015, 10, e1048052. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, A.; Hasnain, S. Extraction and evaluation of indole acetic acid from indigenous auxin-producing rhizosphere bacteria. J. Anim. Plant Sci. 2020, 30, 1024–1036. [Google Scholar] [CrossRef] [Green Version]
- Karamat, M.; Ahmed, A. Impact of Arthrobacter mysorens, Kushneria avicenniae, Halomonas spp. and Bacillus sp. on Helianthus annuus L. for growth enhancement. J. Anim. Plant Sci. 2018, 28, 1629–1634. [Google Scholar]
- Thilagar, G.; Bagyaraj, D.J.; Podile, A.R.; Vaikuntapu, P.R. Bacillus sonorensis, a Novel Plant Growth Promoting Rhizobacterium in Improving Growth, Nutrition and Yield of Chilly (Capsicum annuum L.). Proc. Natl. Acad. Sci. India Sect. B Biol. Sci. 2018, 88, 813–818. [Google Scholar] [CrossRef]
- Nabti, E.H.; Mokrane, N.; Ghoul, M.; Manyani, H.; Dary, M.; Megias, M.G. Isolation and Characterization of Two Halophilic Bacillus (B. licheniformis and Bacillus sp.) with Antifungal Activity. J. Ecol. Health Environ. 2013, 1, 13–17. [Google Scholar] [CrossRef]
- Kwon, J.-H.; Won, S.-J.; Moon, J.-H.; Lee, U.; Park, Y.-S.; Maung, C.E.H.; Ajuna, H.B.; Ahn, Y.S. Bacillus licheniformis PR2 Controls Fungal Diseases and Increases Production of Jujube Fruit under Field Conditions. Horticulturae 2021, 7, 49. [Google Scholar] [CrossRef]

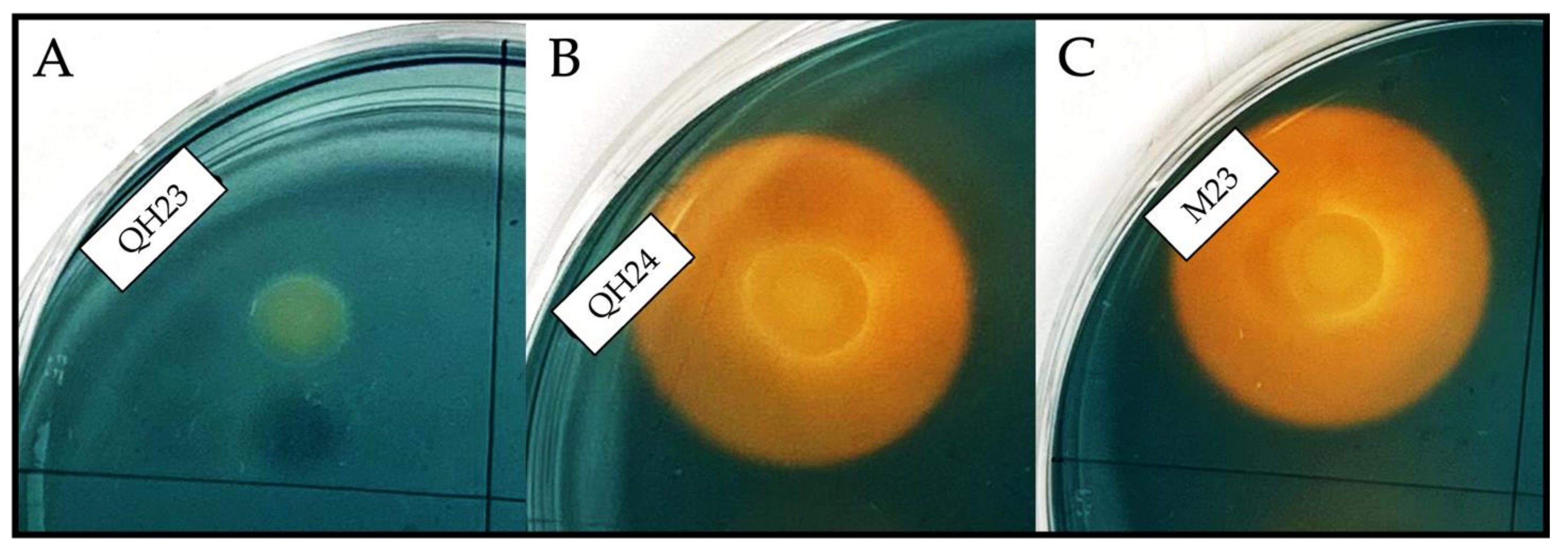
| Colony Characteristics | Microscopy Characteristics | Temperature | NaCl Concentration | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Strain | Shape | Margin | Elevation | Texture | Pigment | Shape | Arrangements | Gram | 28 | 37 | 45 | 50 | 55 | 1.5 | 2.0 | 2.5 | 3.0 | 3.5 |
| QH23 | Circular | Entire | Flat | Mucoid | White-Cream | Rod | Single or in pairs | - | + | + | - | - | - | + | + | + | - | - |
| QH24 | Circular | Entire | Flat | Mucoid | White-Cream | Rod | Single or in pairs | - | + | + | - | - | - | + | + | + | - | - |
| M21 | Circular | Irregular | Raised | Leathery | White-Cream | Rod | Single or in chain | + | + | + | + | + | - | + | + | - | - | - |
| M23 | Circular | Irregular | Raised | Leathery | White-Cream | Rod | Single or in chain | + | + | + | + | + | - | + | + | - | - | - |
| Strain | NO3− | TRP | GLU | ADH | URE | ESC | GEL | PNPG | |GLU| | |ARA| | |MNE| | |MAN| | |NAG| | |MAL| | |GNT| | |CAP| | |ADI| | |MLT| | |CIT| | |PAC| | OX | CAT |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| QH23 | + | + | - | - | - | - | - | - | + | - | - | - | + | - | + | - | - | + | - | - | + | + |
| QH24 | + | - | - | - | - | + | - | + | + | + | + | - | - | + | - | - | - | + | - | - | + | + |
| M21 | + | - | - | - | - | + | + | + | + | + | + | + | + | + | + | - | - | + | + | - | + | + |
| M23 | + | - | - | + | - | + | - | + | + | + | + | + | + | + | + | - | - | + | + | - | + | + |
| Strain | NaCl 0 mM | NaCl 75 mM | NaCl 600 mM |
|---|---|---|---|
| QH23 | 2.23 ± 0.16 | 2.19 ± 0.11 | 1.57 ± 0.04 |
| QH24 | - | 1.46 ± 0.33 | 2.01 ± 0.16 |
| M21 | 1.89 ± 0.10 | 1.75 ± 0.05 | 0.96 ± 0.06 |
| M23 | 1.58 ± 0.05 | 1.95 ± 0.08 | 0.96 ± 0.04 |
| Strain | NaCl 0 mM | NaCl 75 mM | NaCl 600 mM |
|---|---|---|---|
| QH23 | 8.20 ± 0.12 | 7.22 ± 0.20 | 5.16 ± 0.06 |
| QH24 | - | 3.67 ± 0.04 | 5.48 ± 0.25 |
| M21 | 3.08 ± 0.11 | 2.80 ± 0.15 | 1.17 ± 0.23 |
| M23 | 2.56 ± 0.18 | 2.94 ± 0.25 | 0.97 ± 0.07 |
| Siderophore Production Index | Phosphate Solubilization Index | Ammonium Production | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Strain | ACC | 0 mM | 75 mM | 600 mM | 0 mM | 75 mM | 600 mM | 0 mM | 75 mM | 600 mM |
| QH23 | - | n.d. | n.d. | - | - | - | - | - | 7.4 ± 0.4 | 138.9 ± 3.2 |
| QH24 | 79.6 ± 18.7 | n.d. | n.d. | 3.1 ± 0.2 | - | - | - | - | 51.5 ± 0.3 | 363.0 ± 21.1 |
| M21 | - | n.d. | n.d. | n.d. | - | - | - | 533.2 ± 11.4 | 153.9 ± 51.4 | 62.3 ± 2.3 |
| M23 | - | n.d. | n.d. | 3.3 ± 0.5 | - | - | - | 536.8 ± 6.3 | 290.5 ± 48.4 | 48.4 ± 3.9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oliva, G.; Di Stasio, L.; Vigliotta, G.; Guarino, F.; Cicatelli, A.; Castiglione, S. Exploring the Potential of Four Novel Halotolerant Bacterial Strains as Plant-Growth-Promoting Rhizobacteria (PGPR) under Saline Conditions. Appl. Sci. 2023, 13, 4320. https://doi.org/10.3390/app13074320
Oliva G, Di Stasio L, Vigliotta G, Guarino F, Cicatelli A, Castiglione S. Exploring the Potential of Four Novel Halotolerant Bacterial Strains as Plant-Growth-Promoting Rhizobacteria (PGPR) under Saline Conditions. Applied Sciences. 2023; 13(7):4320. https://doi.org/10.3390/app13074320
Chicago/Turabian StyleOliva, Gianmaria, Luca Di Stasio, Giovanni Vigliotta, Francesco Guarino, Angela Cicatelli, and Stefano Castiglione. 2023. "Exploring the Potential of Four Novel Halotolerant Bacterial Strains as Plant-Growth-Promoting Rhizobacteria (PGPR) under Saline Conditions" Applied Sciences 13, no. 7: 4320. https://doi.org/10.3390/app13074320
APA StyleOliva, G., Di Stasio, L., Vigliotta, G., Guarino, F., Cicatelli, A., & Castiglione, S. (2023). Exploring the Potential of Four Novel Halotolerant Bacterial Strains as Plant-Growth-Promoting Rhizobacteria (PGPR) under Saline Conditions. Applied Sciences, 13(7), 4320. https://doi.org/10.3390/app13074320









