Long Eared Owls (Asio otus Linnaeus, 1758) as Field-Assistants in an Integrative Taxonomy Survey of a Peculiar Microtus savii (Rodentia, Cricetidae) Population
Abstract
:1. Introduction
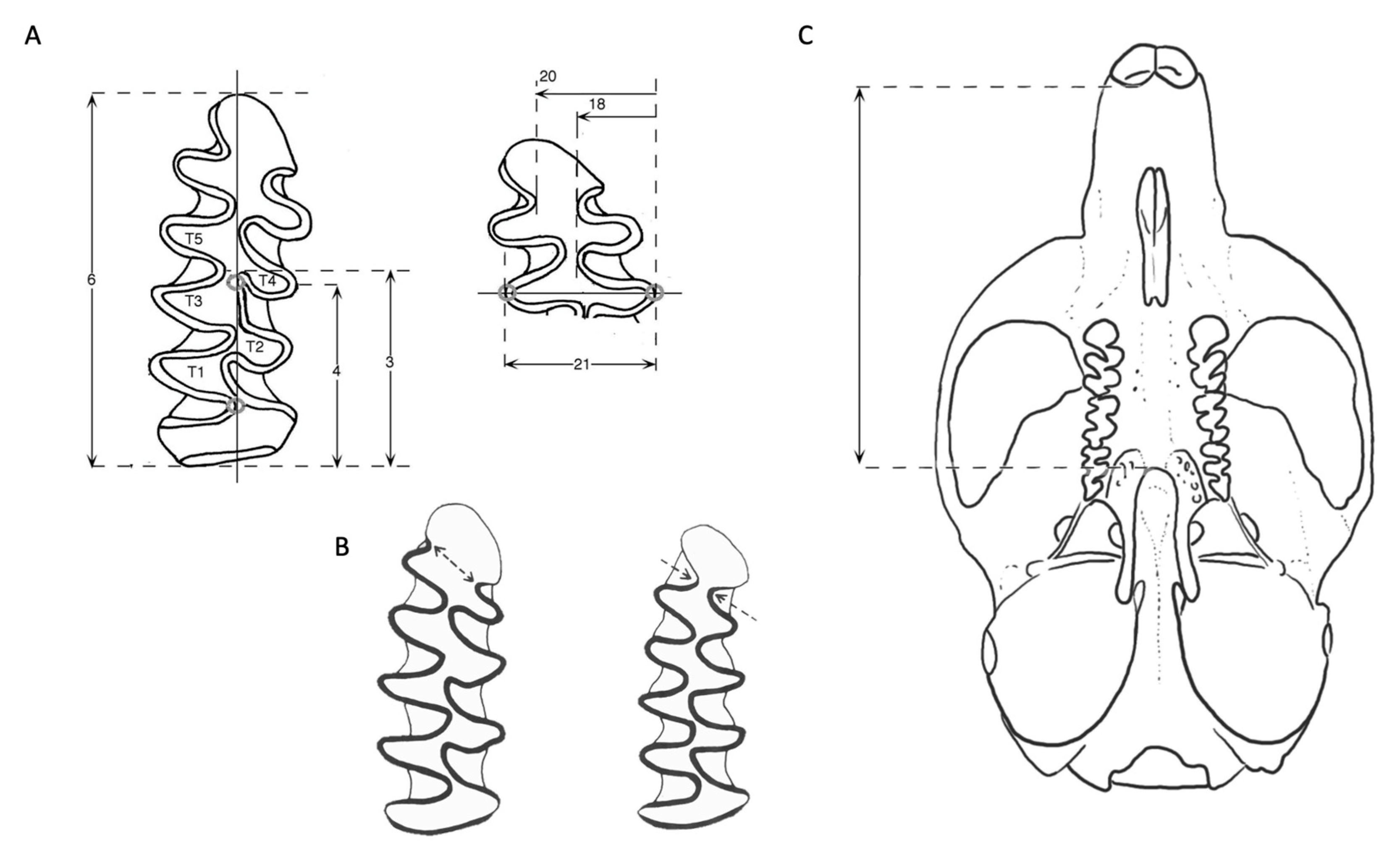
2. Materials and Methods
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Abramson, N.I.; Bodrov, S.Y.; Bondareva, O.V.; Genelt-Yanovskiy, E.A.; Petrova, T.V. A mitochondrial genome phylogeny of voles and lemmings (Rodentia: Arvicolinae): Evolutionary and taxonomic implications. PLoS ONE 2021, 16, e0248198. [Google Scholar] [CrossRef] [PubMed]
- Amori, G.; Castiglia, R. Mammal endemism in Italy: A review. Biogeographia 2017, 33, 19–31. [Google Scholar] [CrossRef] [Green Version]
- Castiglia, R.; Annesi, F.; Aloise, G.; Amori, G. Systematics of the Microtus savii complex (Rodentia, Cricetidae) via mitochondrial DNA analyses: Paraphyly and pattern of sex chromosome evolution. Mol. Phylogenet. Evol. 2008, 46, 1157–1164. [Google Scholar] [CrossRef] [PubMed]
- Galleni, L.; Stanyon, R.; Tellini, A.; Giordano, G.; Santini, L. Taxonomy of Microtus savii (Rodentia, Arvicolidae) in Italy: Cytogenetic and hybridization data. J. Mammal. 1994, 75, 1040–1044. [Google Scholar] [CrossRef]
- Galleni, L.; Stanyon, R.; Contadini, L.; Tellini, A. Biogeographical and kariological data of the Microtus savii group (Rodentia, Arvicolidae) in Italy. Bonn. Zool. Beitr. 1998, 47, 277–282. [Google Scholar]
- Bezerra, A.M.R.; Annesi, F.; Aloise, G.; Amori, G.; Giustini, L.; Castiglia, R. Integrative taxonomy of the Italian pine voles, Microtus savii group (Cricetidae, Arvicolinae). Zool. Scr. 2015, 45, 225–236. [Google Scholar] [CrossRef]
- Ellerman, R.; Morrison-Scott, T.C.S. Checklist of Palearctic and Indian mammals 1758–1946, 2nd ed.; British Museum of Natural History: London, UK, 1966; 810p. [Google Scholar]
- Corbet, G.B. The Mammals of the Palearctic Region: A Taxonomic Review; British Museum of Natural History Press: London, UK; Cornell University: Ithaca, NY, USA, 1978. [Google Scholar]
- Saint Girons, M.C. Les Mammifères de France et du Benelux (Faune Marine Exceptée); Doin: Paris, France, 1973. [Google Scholar]
- Petrov, B.M.; Zivkovic, S.; Rimsa, D. Uber die Arteigenstandingkeit der Kleinwühlmaus, Pitymys felteni. Senckenberg. Biol. 1976, 57, 1–10. [Google Scholar]
- Amori, G.; Contoli, L.; Nappi, A. Mammalia II. Erinaceomorpha, Soricomorpha, Lagomorpha, Rodentia. In “Fauna d’Italia” Series; Edizioni Calderini: Milano, Italy, 2008; Volume XLIV. [Google Scholar]
- Pardiñas, M.F.J.; Myers, P.; León-Paniagua, L.; Garza, N.O.; Cook, J.A.; Kryštufek, B.; Haslauer, R.; Bradley, R.D.; Shenbrot, G.I.; Patton, J.L. Family Cricetidae (true Hamsters, Voles, Lemmings and New World Rats and Mice). In Handbook of the Mammals of the World Vol. 7. Rodents II; Wilson, D.E., Lacher, T.E., Jr., Mittermeier, R.A., Eds.; Lynx Edicions: Barcelona, Spain, 2017; pp. 204–535. [Google Scholar]
- Loy, A.; Aloise, G.; Ancillotto, L.; Angelici, F.M.; Bertolino, S.; Capizzi, D.; Castiglia, R.; Colangelo, P.; Contoli, L.; Cozzi, B.; et al. Mammals of Italy: An annotated checklist. Hystrix 2019, 30, 87–106. [Google Scholar]
- Petty, S. Diet of tawny owls (Strix aluco) in relation to field vole (Microtus agrestis) abundance in a conifer forest in northern England. J. Zool. 1999, 248, 451–465. [Google Scholar] [CrossRef]
- Cecere, F.; Vicini, G. Micromammals in the diet of the Long-eared Owl (Asio otus) at the W.W.F.’s Oasi San Giuliano (Matera, South Italy). Hystrix 2000, 11, 2. [Google Scholar]
- Sergio, F.; Marchesi, L.; Pedrini, P. Density, diet and productivity of Long-eared Owls Asio otus in the Italian Alps: The importance of Microtus voles. Bird Study 2008, 55, 321–328. [Google Scholar] [CrossRef] [Green Version]
- Taberlet, P.; Fumagalli, L. Owl pellets as a source of DNA for genetic studies of small mammals. Mol. Ecol. 1996, 5, 301–305. [Google Scholar] [CrossRef]
- Turpen, E.R.; Vrla, S.C.; Matlack, R.S.; Bradley, R.D. Evidence from Owl Pellets Indicate that Prairie Voles (Microtus ochrogaster) are Broadly Distributed in the Texas Panhandle. In Occasional Papers; Museum of Texas Tech University: Lubbock, TX, USA, 2022; n. 382. [Google Scholar]
- Nappi, A.; Brunet-Lecomte, P.; Montuire, S. The systematics of Microtus (Terricola) savii group: An odonthometrical perspective (Mammalia, Rodentia, Cricetidae). J. Nat. Hist. 2019, 53, 2855–2867. [Google Scholar] [CrossRef]
- Angermann, R. Die Zahnvariabilität bei Microtinen im Liechte von Vavilov’s “Gesetz der homologen Serien“. In Proceedings of the Symposium Theriologicum II—International Symposium on Species and Zoogeography of European Mammals, Brno, Czech Republic, 22–26 November 1971; Accademia, Publishing house of the Czechoslovak Academy of Sciences: Praha, Czech Republic, 1974; pp. 61–73. [Google Scholar]
- Contoli, L.; Amori, G.; Nazzaro, C. Tooth diversity in Arvicolidae (Mammalia, Rodentia): Ecochorological factors and speciation time. Hystrix 1993, 4, 2. [Google Scholar]
- Navarro, N.; Montuire, S.; Laffont, R.; Steimetz, E.; Onofrei, C.; Royer, A. Identifying past remains of morphologically similar vole species using molar shapes. Quaternary 2018, 1, 20. [Google Scholar] [CrossRef] [Green Version]
- Brunet-Lecomte, P. Les Campagnols Souterrains (Terricola, Arvicolidae, Rodentia) Actuels et Fossiles d’Europe Occidentale. Ph.D. Dissertation, Université de Bourgogne, Dijon, France, 1988. [Google Scholar]
- Laplana, C.; Montuire, S.; Brunet-Lecomte, P.; Chaline, J. Révision des Allophaiomys (Arvicolidae, Rodentia, Mammalia) des Valerots (Côte-d’Or, France). Geodiversitas 2000, 22, 255–267. [Google Scholar]
- Nappi, A.; Brunet-Lecomte, P.; Montuire, S. Intraspecific morphological tooth variability and geographical distribution: Application to the Savi’s vole, Microtus (Terricola) savii (Rodentia, Arvicolinae). J. Nat. Hist. 2006, 40, 345–358. [Google Scholar] [CrossRef]
- Iudica, C.A.; Whitten, W.M.; Williams, N.H. Small bones from dried mammal museum specimens as a reliable source of DNA. BioTechniques 2001, 30, 732–736. [Google Scholar] [CrossRef] [PubMed]
- Brustenga, L.; Chiesa, S.; Maltese, S.; Consales, G.; Marsili, L.; Lauriano, G.; Scarano, V.; Scarpato, A.; Lucentini, L. A Reliable and Cost-Efficient PCR-RFLP Tool for the Rapid Identification of Cetaceans in the Mediterranean Sea. Sustainability 2022, 14, 16763. [Google Scholar] [CrossRef]
- Fontaneto, D.; Viola, P.; Pizzirani, C.; Chiesa, S.; Rossetti, A.; Amici, A.; Lucentini, L. Mismatches between Morphology and DNA in Italian Partridges May Not Be Explained Only by Recent Artificial Release of Farm-Reared Birds. Animals 2022, 12, 541. [Google Scholar] [CrossRef] [PubMed]
- Irwin, D.M.; Kocher, T.D.; Wilson, A.C. Evolution of the cytochrome b gene of mammals. J. Mol. Evol. 1991, 32, 128–144. [Google Scholar] [CrossRef]
- Jaarola, M.; Searle, J.B. Phylogeography of field voles (Microtus agrestis) in Eurasia inferred from mitochondrial DNA sequences. Mol. Ecol. 2002, 11, 2613–2621. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA 11: Molecular Evolutionary Genetics Analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinf. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Templeton, A.R.; Crandall, K.A.; Sing, C.F. A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA sequence data. III. Cladogram estimation. Genetics 1992, 132, 619–633. [Google Scholar] [CrossRef] [PubMed]
- Clement, M.; Posada, D.; Crandall, K.A. TCS: A computer program to estimate gene genealogies. Mol. Ecol. 2000, 9, 1657–1659. [Google Scholar] [CrossRef] [Green Version]
- Chiesa, S.; Lucentini, L.; Chainho, P.; Plazzi, F.; Angélico, M.M.; Ruano, F.; Freitas, R.; Costa, J.L. One in a Million: Genetic Diversity and Conservation of the Reference Crassostrea angulata Population in Europe from the Sado Estuary (Portugal). Life 2021, 11, 1173. [Google Scholar] [CrossRef]
- dos Santos, A.M.; Cabezas, M.P.; Tavares, A.I.; Xavier, R.; Branco, M. tcsBU: A tool to extend TCS network layout and visualization. Bioinformatics 2016, 32, 627–628. [Google Scholar] [CrossRef] [Green Version]
- Guindon, S.; Gascuel, O. A simple, fast and accurate method to estimate large phylogenies by maximum-likelihood. Syst. Biol. 2003, 52, 696–704. [Google Scholar] [CrossRef] [Green Version]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [Green Version]
- Tajima, F. Statistical methods to test for nucleotide mutation hypothesis by DNA polymorphism. Genetics 1989, 123, 585–595. [Google Scholar] [CrossRef]
- Sorenson, M.D.; Quinn, T.W. Numts: A Challenge for Avian Systematics and Population Biology. Ornithology 1998, 115, 214–221. [Google Scholar] [CrossRef]
- Triant, D.A.; DeWoody, J.A. Extensive mitochondrial DNA transfer in a rapidly evolving rodent has been mediated by independent insertion events and by duplications. Gene 2007, 401, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Feng, Z.; Liu, Y. Nucleotide Substitution Pattern in Mitochondrial Cytochrome b Pseudogenes of Ten Species in Galliformes. In Information Technology and Agricultural Engineering. Advances in Intelligent and Soft Computing; Zhu, E., Sambath, S., Eds.; Springer: Berlin/Heidelberg, Germany, 2012; Volume 134, pp. 269–270. [Google Scholar]
- Jukes, T.H.; Cantor, C.R. Evolution of protein molecules. In Mammalian Protein Metabolism; Munro, H.N., Ed.; Academic Press: New York, NY, USA, 1969; pp. 21–132. [Google Scholar]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Dąbrowski, M.J.; Pomorski, J.J.; Gliwicz, J. Cytochrome b gene (cytb) sequence diversity in a Microtus oeconomus population from Bialowieza Primeval Forest. Acta Theriol. 2013, 58, 119–126. [Google Scholar] [CrossRef] [Green Version]
- Gandolfi, A.; Crestanello, B.; Fagotti, A.; Simoncelli, F.; Chiesa, S.; Girardi, M.; Giovagnoli, E.; Marangoni, C.; Di Rosa, I.; Lucentini, L. New Evidences of Mitochondrial DNA Heteroplasmy by Putative Paternal Leakage between the Rock Partridge (Alectoris graeca) and the Chukar Partridge (Alectoris chukar). PLoS ONE 2017, 12, e0170507. [Google Scholar] [CrossRef] [PubMed]
- Kovalskaya, Y.M.; Savinetskaya, L.E.; Aksenova, T.G. Experimental hybridization of voles of the genus Microtus s. l. M. socialis with species of the group arvalis (Mammalia, Rodentia). Biol. Bull. Russ. Acad. Sci. 2014, 41, 559–563. [Google Scholar] [CrossRef]
- Galleni, L.; Tellini, A.; Cicalò, A.; Fantini, C.; Fabiani, O. Histological examination of the male gonad of hybrid specimens: Microtus savii x Microtus brachycercus (Rodentia-Arvicolinae). Bonn. Zool. Beitr. 1998, 48, 1–8. [Google Scholar]
- Tougard, C.; Renvoisé, E.; Petitjean, A.; Quéré, J.P. New insight into the colonization processes of common voles: Inferences from molecular and fossil evidence. PLoS ONE 2008, 3, e3532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stojak, J.; McDevitt, A.D.; Herman, J.S.; Kryštufek, B.; Uhlíková, J.; Purger, J.J.; Lavrenchenko, L.A.; Searle, J.B.; Wójcik, J.M. Between the Balkans and the Baltic:Phylogeography of a Common Vole Mitochondrial DNA Lineage Limited to Central Europe. PLoS ONE 2016, 11, e0168621. [Google Scholar] [CrossRef] [Green Version]
- Francl, K.E.; Glenn, T.C.; Castleberry, S.B.; Ford, W.M. Genetic relationships of meadow vole (Microtus pennsylvanicus) populations in central Appalachian wetlands. Can. J. Zool. 2008, 86, 344–355. [Google Scholar] [CrossRef]
- Emin, D.; Toxopeus, A.G.; Groen, I.T.A.; Kontogeorgos, I.; Georgopoulou, E.; Xirouchakis, S. Home Range and Habitat Selection of Long-Eared Owls (Asio otus) in Mediterranean Agricultural Landscapes (Crete, Greece). Avian. Biol. Res. 2018, 11, 204–218. [Google Scholar] [CrossRef]
- Nappi, A. L’analisi delle borre degli uccelli: Metodiche, applicazioni e informazioni. Un lavoro monografico. Picus 2011, 37, 106–120. [Google Scholar]
- Buś, M.M.; Żmihorski, M.; Romanowski, J.; Balčiauskienė, L.; Cichocki, J.; Balčiauskas, L. High efficiency protocol of DNA extraction from Micromys minutus mandibles from owl pellets: A tool for molecular research of cryptic mammal species. Acta Theriol. 2013, 59, 99–109. [Google Scholar] [CrossRef] [Green Version]
- Gomes-Rocha, R.; Justino, J.; Reis-Leite, Y.L.; Pires-Costa, L. DNA from owl pellet bones uncovers hidden biodiversity. Syst. Biodiver. 2015, 13, 403–412. [Google Scholar] [CrossRef]
- Gaggi, A.; Paci, A.M. Atlante degli Erinaceomorfi, dei Soricomorfi e dei piccoli Roditori dell’Umbria; Regione Umbria: Perugia, Italy, 2014; 211p. [Google Scholar]
- Nappi, A.; Brunet-Lecomte, P.; Montuire, S. Dental morphology of Microtus (Terricola) voles from Calabria (Southern Italy) and relationships with M. (T.) savii (Rodentia, Arvicolidae). Hystrix 2005, 16, 75–85. [Google Scholar]
- Yletyinen, S.; Norrdahl, K. Habitat use of field voles (Microtus agrestis) in wide and narrow buffer zones. Agric. Ecosyst. Environ. 2008, 123, 194–200. [Google Scholar] [CrossRef]
- Barili, A.; Gentili, S.; Lucentini, L. DNA, archivi della biodiversità e musei scientifici: Alcune esperienze presso la Galleria di Storia Naturale dell’Università degli Studi di Perugia. Museol. Sci. Mem. 2017, 17, 97–100. [Google Scholar]







Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lucentini, L.; Brunet-Lecomte, P.; Brustenga, L.; La Porta, G.; Barili, A.; Gaggi, A.; Gentili, S.; Nappi, A.; Paci, A.M. Long Eared Owls (Asio otus Linnaeus, 1758) as Field-Assistants in an Integrative Taxonomy Survey of a Peculiar Microtus savii (Rodentia, Cricetidae) Population. Appl. Sci. 2023, 13, 4703. https://doi.org/10.3390/app13084703
Lucentini L, Brunet-Lecomte P, Brustenga L, La Porta G, Barili A, Gaggi A, Gentili S, Nappi A, Paci AM. Long Eared Owls (Asio otus Linnaeus, 1758) as Field-Assistants in an Integrative Taxonomy Survey of a Peculiar Microtus savii (Rodentia, Cricetidae) Population. Applied Sciences. 2023; 13(8):4703. https://doi.org/10.3390/app13084703
Chicago/Turabian StyleLucentini, Livia, Patrick Brunet-Lecomte, Leonardo Brustenga, Gianandrea La Porta, Angelo Barili, Angela Gaggi, Sergio Gentili, Armando Nappi, and Andrea Maria Paci. 2023. "Long Eared Owls (Asio otus Linnaeus, 1758) as Field-Assistants in an Integrative Taxonomy Survey of a Peculiar Microtus savii (Rodentia, Cricetidae) Population" Applied Sciences 13, no. 8: 4703. https://doi.org/10.3390/app13084703
APA StyleLucentini, L., Brunet-Lecomte, P., Brustenga, L., La Porta, G., Barili, A., Gaggi, A., Gentili, S., Nappi, A., & Paci, A. M. (2023). Long Eared Owls (Asio otus Linnaeus, 1758) as Field-Assistants in an Integrative Taxonomy Survey of a Peculiar Microtus savii (Rodentia, Cricetidae) Population. Applied Sciences, 13(8), 4703. https://doi.org/10.3390/app13084703








