Ultrasound Intima-Media Complex (IMC) Segmentation Using Deep Learning Models
Abstract
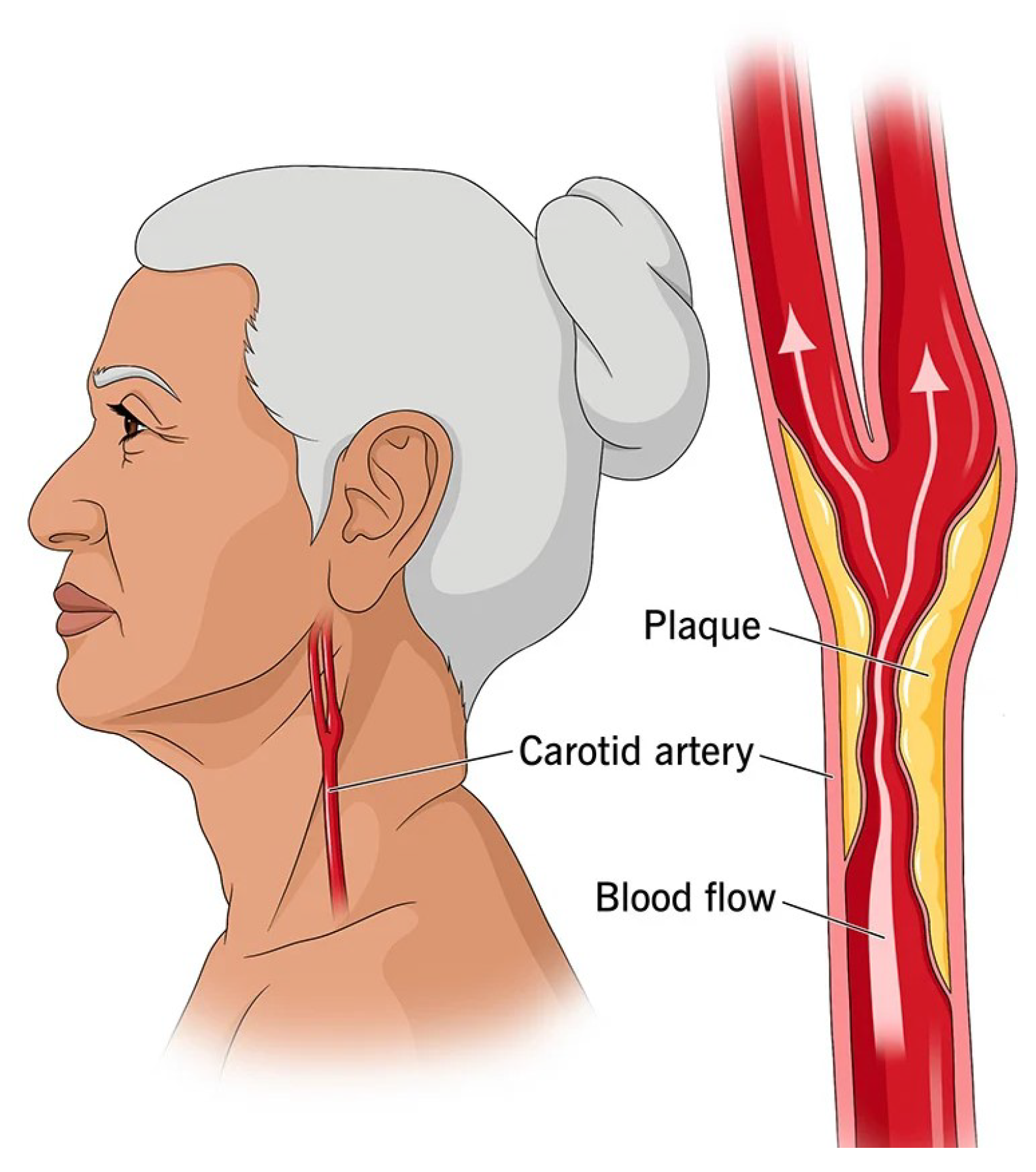
1. Introduction
- We develop and investigate various recent deep learning models for the segmentation of IMC in B-mode ultrasound images of the carotid artery.
- We propose a pioneer application for self-organized operational neural networks (self-ONNs) for IMC segmentation.
- We investigate the level of non-linearity for operational layers required to achieve a better segmentation performance.
2. Related Works
3. Methods
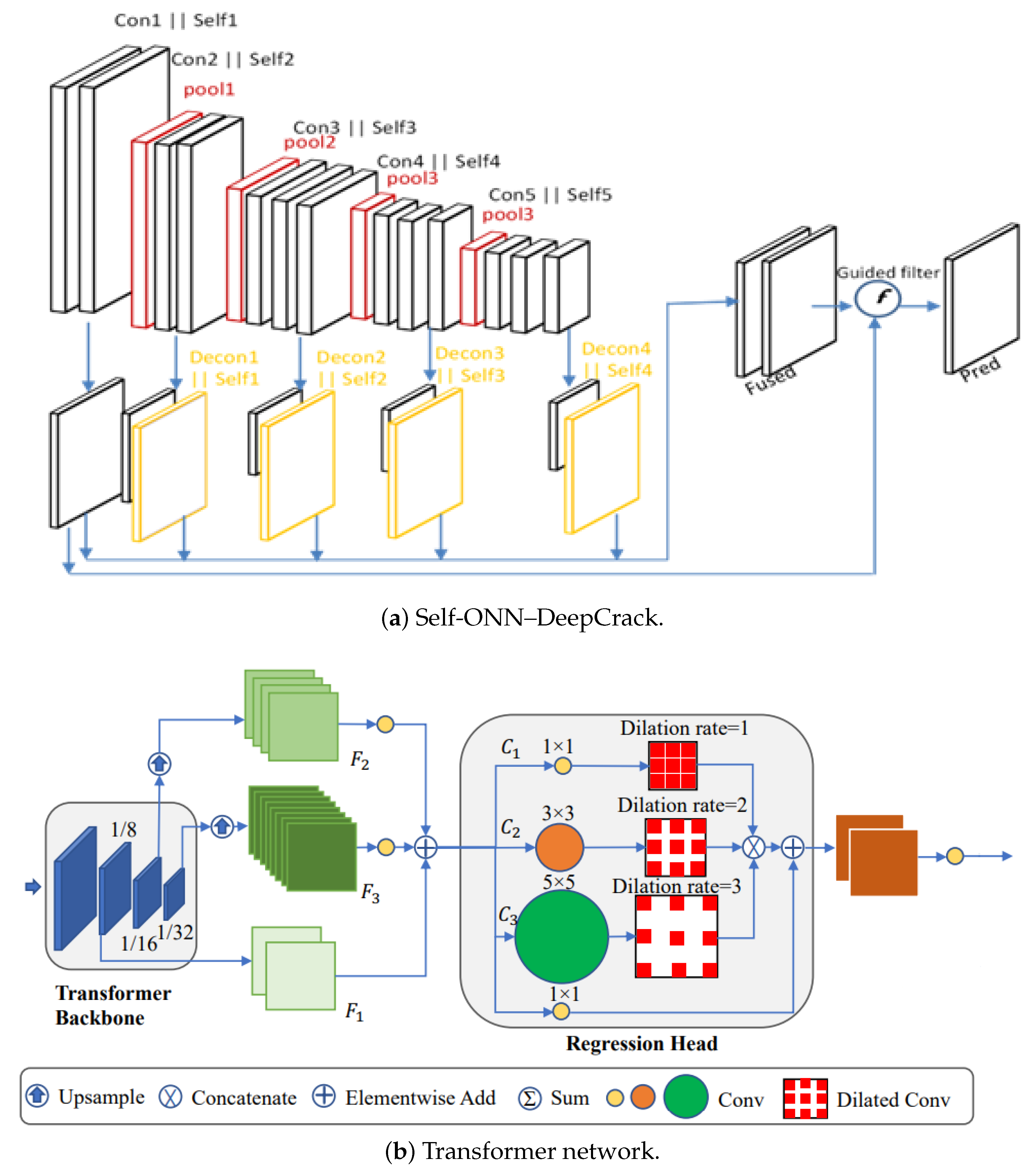
3.1. Self-Operational Neural Network-Based Model
3.2. Pixel Difference-Based Model
3.3. Transformer-Based Model
3.4. Post-Processing
4. Experimental Results
4.1. Implementation Details
4.2. Dataset and Evaluation Metrics
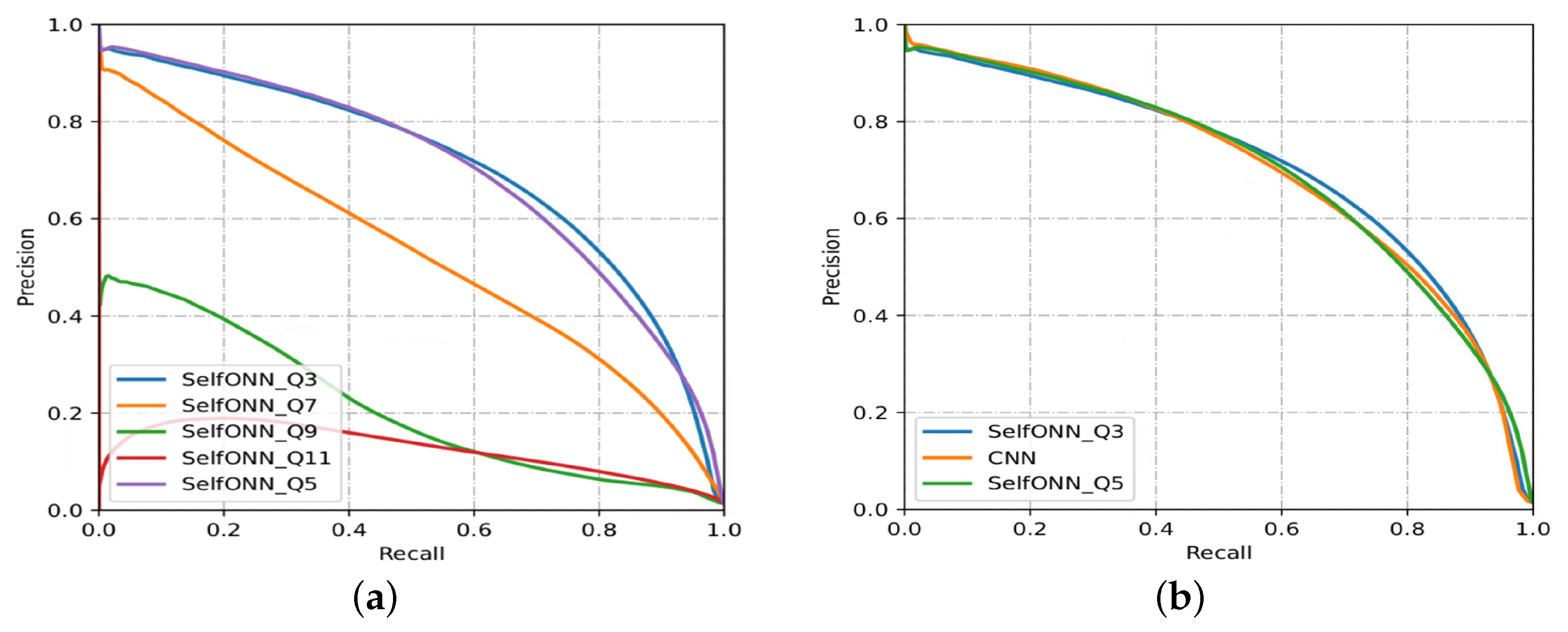
4.3. Evaluation
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| DR | Diabetic retinopathy |
| DL | Deep learning |
| AI | Artificial intelligence |
| CNN | Convolutional neural network |
References
- Latha, S.; Samiappan, D.; Kumar, R. Carotid artery ultrasound image analysis: A review of the literature. Proc. Inst. Mech. Eng. Part H J. Eng. Med. 2020, 234, 417–443. [Google Scholar] [CrossRef]
- Vila, M.D.M.; Remeseiro, B.; Grau, M.; Elosua, R.; Igual, L. Last Advances on Automatic Carotid Artery Analysis in Ultrasound Images: Towards Deep Learning. In Handbook of Artificial Intelligence in Healthcare; Springer: Berlin/Heidelberg, Germany, 2022; pp. 215–247. [Google Scholar]
- Riahi, A.; Elharrouss, O.; Al-Maadeed, S. BEMD-3DCNN-based method for COVID-19 detection. Comput. Biol. Med. 2022, 142, 105188. [Google Scholar] [CrossRef]
- Loizou, C.P.; Kasparis, T.; Spyrou, C.; Pantziaris, M. Integrated system for the complete segmentation of the common carotid artery bifurcation in ultrasound images. In IFIP International Conference on Artificial Intelligence Applications and Innovations; Springer: Berlin/Heidelberg, Germany, 2013; Volume 9, pp. 292–301. [Google Scholar]
- Christodoulou, L.; Loizou, C.P.; Spyrou, C.; Kasparis, T.; Pantziaris, M. Full-automated system for the segmentation of the common carotid artery in ultrasound images. In Proceedings of the 2012 IEEE 5th International Symposium on Communications, Control and Signal Processing, Rome, Italy, 2–4 May 2012; pp. 1–6. [Google Scholar]
- Ikeda, N.; Dey, N.; Sharma, A.; Gupta, A.; Bose, S.; Acharjee, S.; Shafique, S.; Cuadrado-Godia, E.; Araki, T.; Saba, L.; et al. Automated segmental-IMT measurement in thin/thick plaque with bulb presence in carotid ultrasound from multiple scanners: Stroke risk assessment. Comput. Methods Programs Biomed. 2017, 141, 73–81. [Google Scholar] [CrossRef]
- Madipalli, P.; Kotta, S.; Dadi, H.; Nagaraj, Y.; Asha, C.S.; Narasimhadhan, A.V. Automatic Segmentation of Intima Media Complex in Common Carotid Artery using Adaptive Wind Driven Optimization. In Proceedings of the 2018 Twenty Fourth National Conference on Communications (NCC), Hyderbad, India, 25–28 February 2018; pp. 1–6. [Google Scholar]
- Nagaraj, Y.; Teja, A.; Narasimha, D. Automatic Segmentation of Intima Media Complex in Carotid Ultrasound Images Using Support Vector Machine. Arab. J. Sci. Eng. 2019, 44, 3489–3496. [Google Scholar] [CrossRef]
- Biswas, M.; Saba, L.; Chakrabartty, S.; Khanna, N.N.; Song, H.; Suri, H.S.; Sfikakis, P.P.; Mavrogeni, S.; Viskovic, K.; Laird, J.R.; et al. Two-stage artificial intelligence model for jointly measurement of atherosclerotic wall thickness and plaque burden in carotid ultrasound: A screening tool for cardiovascular/stroke risk assessment. Comput. Biol. Med. 2020, 123, 103847. [Google Scholar] [CrossRef]
- Su, Z.; Liu, W.; Yu, Z.; Hu, D.; Liao, Q.; Tian, Q.; Pietikainen, M.; Liu, L. Pixel difference networks for efficient edge detection. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, BC, Canada, 11–17 October 2021; pp. 5117–5127. [Google Scholar]
- Kiranyaz, S.; Malik, J.; Abdallah, H.B.; Ince, T.; Iosifidis, A.; Gabbouj, M. Self-organized operational neural networks with generative neurons. Neural Netw. 2021, 140, 294–308. [Google Scholar] [CrossRef]
- Gabbouj, M.; Kiranyaz, S.; Malik, J.; Zahid, M.U.; Ince, T.; Chowdhury, M.E.; Khandakar, A.; Tahir, A. Robust peak detection for holter ECGs by self-organized operational neural networks. IEEE Trans. Neural Netw. Learn. Syst. 2022. Early Access. [Google Scholar] [CrossRef]
- Malik, J.; Kiranyaz, S.; Gabbouj, M. Operational vs. convolutional neural networks for image denoising. arXiv 2020, arXiv:2009.00612. [Google Scholar]
- Malik, J.; Devecioglu, O.C.; Kiranyaz, S.; Ince, T.; Gabbouj, M. Real-time patient-specific ECG classification by 1D self-operational neural networks. IEEE Trans. Biomed. Eng. 2021, 69, 1788–1801. [Google Scholar] [CrossRef]
- Rahman, A.; Chowdhury, M.E.; Khandakar, A.; Tahir, A.M.; Ibtehaz, N.; Hossain, M.S.; Kiranyaz, S.; Malik, J.; Monawwar, H.; Kadir, M.A. Robust biometric system using session invariant multimodal EEG and keystroke dynamics by the ensemble of self-ONNs. Comput. Biol. Med. 2022, 142, 105238. [Google Scholar] [CrossRef]
- Soltanian, M.; Malik, J.; Raitoharju, J.; Iosifidis, A.; Kiranyaz, S.; Gabbouj, M. Speech command recognition in computationally constrained environments with a quadratic self-organized operational layer. In Proceedings of the 2021 IEEE International Joint Conference on Neural Networks (IJCNN), Shenzhen, China, 18–22 July 2021; pp. 1–6. [Google Scholar]
- Kiranyaz, S.; Ince, T.; Iosifidis, A.; Gabbouj, M. Operational neural networks. Neural Comput. Appl. 2020, 32, 6645–6668. [Google Scholar] [CrossRef]
- Khan, S.; Naseer, M.; Hayat, M.; Zamir, S.W.; Khan, F.S.; Shah, M. Transformers in vision: A survey. ACM Comput. Surv. (CSUR) 2022, 54, 1–41. [Google Scholar] [CrossRef]
- Liu, Y.; Yao, J.; Lu, X.; Xie, R.; Li, L. DeepCrack: A deep hierarchical feature learning architecture for crack segmentation. Neurocomputing 2019, 338, 139–153. [Google Scholar] [CrossRef]
- Elharrouss, O.; Hmamouche, Y.; Idrissi, A.K.; El Khamlichi, B.; El Fallah-Seghrouchni, A. Refined edge detection with cascaded and high-resolution convolutional network. Pattern Recognit. 2023, 138, 109361. [Google Scholar] [CrossRef]
- Petroudi, S.; Loizou, C.; Pantziaris, M.; Pattichis, C. Segmentation of the common carotid intima-media complex in ultrasound images using active contours. IEEE Trans. Biomed. Eng. 2012, 59, 3060–3069. [Google Scholar] [CrossRef]
- Molinari, F.; Meiburger, K.M.; Saba, L.; Acharya, U.R.; Ledda, M.; Nicolaides, A.; Suri, J.S. Constrained snake vs. conventional snake for carotid ultrasound automated IMT measurements on multi-center data sets. Ultrasonics 2012, 52, 949–961. [Google Scholar] [CrossRef]
- Ceccarelli, M.; Luca, N.D.; Morganella, A. An active contour approach to automatic detection of the intima-media thickness. In Proceedings of the 2006 IEEE International Conference on Acoustics Speech and Signal Processing, Toulouse, France, 14–19 May 2006; Volume 2, p. II. [Google Scholar]
- Loizou, C.P.; Pattichis, C.S.; Pantziaris, M.; Tyllis, T.; Nicolaides, A. Snakes based segmentation of the common carotid artery intima media. Med. Biol. Eng. Comput. 2007, 45, 35–49. [Google Scholar] [CrossRef]
- Gutierrez, M.A.; Pilon, P.E.; Lage, S.G.; Kopel, L.; Carvalho, R.T.; Furuie, S.S. Automatic measurement of carotid diameter and wall thickness in ultrasound images. In Proceedings of the Computers in Cardiology, Memphis, TN, USA, 22–25 September 2002; pp. 359–362. [Google Scholar]
- Chan, R.C.; Kaufhold, J.; Hemphill, L.C.; Lees, R.S.; Karl, W.C. Anisotropic edge-preserving smoothing in carotid B-mode ultrasound for improved segmentation and intima-media thickness (IMT) measurement. In Proceedings of the Computers in Cardiology 2000. Vol.27 (Cat. 00CH37163), Cambridge, MA, USA, 24–27 September 2000; pp. 37–40. [Google Scholar]
- Delsanto, S.; Molinari, F.; Giustetto, P.; Liboni, W.; Badalamenti, S.; Suri, J.S. Characterization of a completely user-independent algorithm for carotid artery segmentation in 2-D ultrasound images. IEEE Trans. Instrum. Meas. 2007, 56, 1265–1274. [Google Scholar] [CrossRef]
- Gagan, J.H.; Shirsat, H.S.; Mathias, G.P.; Mallya, B.V.; Andrade, J.; Rajagopal, K.V.; Kumar, J.H. Automated Segmentation of Common Carotid Artery in Ultrasound Images. IEEE Access 2022, 10, 58419–58430. [Google Scholar] [CrossRef]
- Liang, Q.; Wendelhag, I.; Wikstrand, J.; Gustavsson, T. A multiscale dynamic programming procedure for boundary detection in ultrasonic artery images. IEEE Trans. Med. Imaging 2000, 19, 127–142. [Google Scholar] [CrossRef]
- Wendelhag, I.; Liang, Q.; Gustavsson, T.; Wikstrand, J. A new automated computerized analyzing system simplifies readings and reduces the variability in ultrasound measurement of intima-media thickness. Stroke 1997, 28, 2195–2200. [Google Scholar] [CrossRef]
- Cheng, D.-C.; Jiang, X. Detections of arterial wall in sonographic artery images using dual dynamic programming. IEEE Trans. Inf. Technol. Biomed. 2008, 12, 792–799. [Google Scholar] [CrossRef]
- Gustavsson, T.; Wendelhag, Q.L.I.; Wikstrand, J. A dynamic programming procedure for automated ultrasonic measurement of the carotid artery. In Proceedings of the Computers in Cardiology, Bethesda, MD, USA, 25–28 September 1994; pp. 297–300. [Google Scholar]
- Santhiyakumari, N.; Madheswaran, M. Non-invasive evaluation of carotid artery wall thickness using improved dynamic programming technique. Signal Image Video Process. 2008, 2, 183–193. [Google Scholar] [CrossRef]
- Lee, Y.-B.; Choi, Y.-J.; Kim, M.-H. Boundary detection in carotid ultrasound images using dynamic programming and a directional Haar-like filter. Comput. Biol. Med. 2010, 40, 687–697. [Google Scholar] [CrossRef]
- Liguori, C.; Paolillo, A.; Pietrosanto, A. An automatic measurement system for the evaluation of carotid intima-media thickness. IEEE Trans. Instrum. Meas. 2001, 50, 1684–1691. [Google Scholar] [CrossRef]
- Selzer, R.H.; Mack, W.J.; Lee, P.L.; Kwong-Fu, H.; Hodis, H.N. Improved common carotid elasticity and intima-media thickness measurements from computer analysis of sequential ultrasound frames. Atherosclerosis 2002, 154, 185–193. [Google Scholar] [CrossRef]
- Pramulen, A.S.; Yuniarno, E.M.; Nugroho, J.; Sunarya, I.M.G.; Purnama, I.K.E. Carotid Artery Segmentation on Ultrasound Image using Deep Learning based on Non-Local Means-based Speckle Filtering. In Proceedings of the 2020 International Conference on Computer Engineering, Network and Intelligent Multimedia (CENIM), Surabaya, Indonesia, 17–18 November 2020; pp. 360–365. [Google Scholar]
- Jain, P.K.; Sharma, N.; Saba, L.; Paraskevas, K.I.; Kalra, M.K.; Johri, A.; Nicolaides, A.N.; Suri, J.S. Automated deep learning-based paradigm for high-risk plaque detection in B-mode common carotid ultrasound scans: An asymptomatic Japanese cohort study. Int. Angiol. 2021, 41, 9–23. [Google Scholar] [CrossRef]
- Lainé, N.; Liebgott, H.; Zahnd, G.; Orkisz, M. Carotid artery wall segmentation in ultrasound image sequences using a deep convolutional neural network. arXiv 2022, arXiv:2201.12152. [Google Scholar]
- Radovanovic, N.; Dašić, L.; Blagojevic, A.; Sustersic, T.; Filipovic, N. Carotid Artery Segmentation Using Convolutional Neural Network in Ultrasound Images. 2022. Available online: https://scidar.kg.ac.rs/bitstream/123456789/16643/4/p8.pdf (accessed on 1 January 2023).
- Park, J.H.; Seo, E.; Choi, W.; Lee, S.J. Ultrasound deep learning for monitoring of flow–vessel dynamics in murine carotid artery. Ultrasonics 2022, 120, 106636. [Google Scholar] [CrossRef]
- Jain, P.K.; Sharma, N.; Kalra, M.K.; Johri, A.; Saba, L.; Suri, J.S. Far wall plaque segmentation and area measurement in common and internal carotid artery ultrasound using U-series architectures: An unseen Artificial Intelligence paradigm for stroke risk assessment. Comput. Biol. Med. 2022, 149, 106017. [Google Scholar] [CrossRef]
- Fu, H.; Xu, J.C.Y.; Wong, D.W.K.; Liu, J.; Cao, X. Joint optic disc and cup segmentation based on multi-label deep network and polar transformation. IEEE Trans. Med. Imaging 2018, 37, 1597–1605. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Li, C.; Xu, L.; Zhu, S.; Hua, Y.; Zhang, J. CSM-Net: Automatic joint segmentation of intima-media complex and lumen in carotid artery ultrasound images. Comput. Biol. Med. 2022, 150, 106119. [Google Scholar] [CrossRef]
- Chu, X.; Tian, Z.; Wang, Y.; Zhang, B.; Ren, H.; Wei, X.; Xia, H.; Shen, C. Twins: Revisiting the design of spatial attention in vision transformers. Adv. Neural Inf. Process. Syst. 2021, 34, 9355–9366. [Google Scholar]
- Elharrouss, O.; Akbari, Y.; Almaadeed, N.; Al-Maadeed, S. Backbones-review: Feature extraction networks for deep learning and deep reinforcement learning approaches. arXiv 2022, arXiv:2206.08016. [Google Scholar]
- Meiburger, K.M.; Zahnd, G.; Faita, F.; Loizou, C.P.; Carvalho, C.; Steinman, D.A.; Gibello, L.; Bruno, R.M.; Marzola, F.; Clarenbach, R.; et al. Carotid ultrasound boundary study (CUBS): An open multicenter analysis of computerized intima-media thickness measurement systems and their clinical impact. Ultrasound Med. Biol. 2021, 47, 2442–2455. [Google Scholar] [CrossRef]


| Method | Learning Rate | Optimizer | Epochs | Training Parameters |
|---|---|---|---|---|
| DeepCrack | 0.0001 | Adam | 100 | 14.720 M |
| DeepCrack_Self_ONN | 0.0001 | Adam | 100 | 44.144 M |
| PidiNet | 0.005 | Adam | 70 | 1.150 MB |
| Transformer | 0.00001 | Adam | 70 | 104.609 M |
| Model | Precision | Recall | F-Measure | Dice | Jaccard | FPS |
|---|---|---|---|---|---|---|
| DeepCrack_CNN | 0.631 | 0.675 | 0.652 | 0.652 | 0.484 | 17.074 |
| DeepCrack_CNN + Post-processing | 0.834 | 0.618 | 0.697 | 0.697 | 0.544 | 17.074 |
| DeepCrack_Self (q = 3) | 0.652 | 0.688 | 0.669 | 0.669 | 0.503 | 13.45 |
| DeepCrack_Self + Post-processing | 0.792 | 0.691 | 0.721 | 0.721 | 0.571 | 13.45 |
| PiDiNet | 0.687 | 0.825 | 0.750 | 0.750 | 0.60 | 20.62 |
| PiDiNet + Post-processing | 0.876 | 0.740 | 0.791 | 0.791 | 0.661 | 20.62 |
| Transformer | 0.68 | 0.826 | 0.746 | 0.746 | 0.595 | 11.427 |
| Transformer + Post-processing | 0.882 | 0.849 | 0.801 | 0.801 | 0.656 | 11.427 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hassen Mohammed, H.; Elharrouss, O.; Ottakath, N.; Al-Maadeed, S.; Chowdhury, M.E.H.; Bouridane, A.; Zughaier, S.M. Ultrasound Intima-Media Complex (IMC) Segmentation Using Deep Learning Models. Appl. Sci. 2023, 13, 4821. https://doi.org/10.3390/app13084821
Hassen Mohammed H, Elharrouss O, Ottakath N, Al-Maadeed S, Chowdhury MEH, Bouridane A, Zughaier SM. Ultrasound Intima-Media Complex (IMC) Segmentation Using Deep Learning Models. Applied Sciences. 2023; 13(8):4821. https://doi.org/10.3390/app13084821
Chicago/Turabian StyleHassen Mohammed, Hanadi, Omar Elharrouss, Najmath Ottakath, Somaya Al-Maadeed, Muhammad E. H. Chowdhury, Ahmed Bouridane, and Susu M. Zughaier. 2023. "Ultrasound Intima-Media Complex (IMC) Segmentation Using Deep Learning Models" Applied Sciences 13, no. 8: 4821. https://doi.org/10.3390/app13084821
APA StyleHassen Mohammed, H., Elharrouss, O., Ottakath, N., Al-Maadeed, S., Chowdhury, M. E. H., Bouridane, A., & Zughaier, S. M. (2023). Ultrasound Intima-Media Complex (IMC) Segmentation Using Deep Learning Models. Applied Sciences, 13(8), 4821. https://doi.org/10.3390/app13084821








