WSPolyp-SAM: Weakly Supervised and Self-Guided Fine-Tuning of SAM for Colonoscopy Polyp Segmentation
Abstract
:1. Introduction
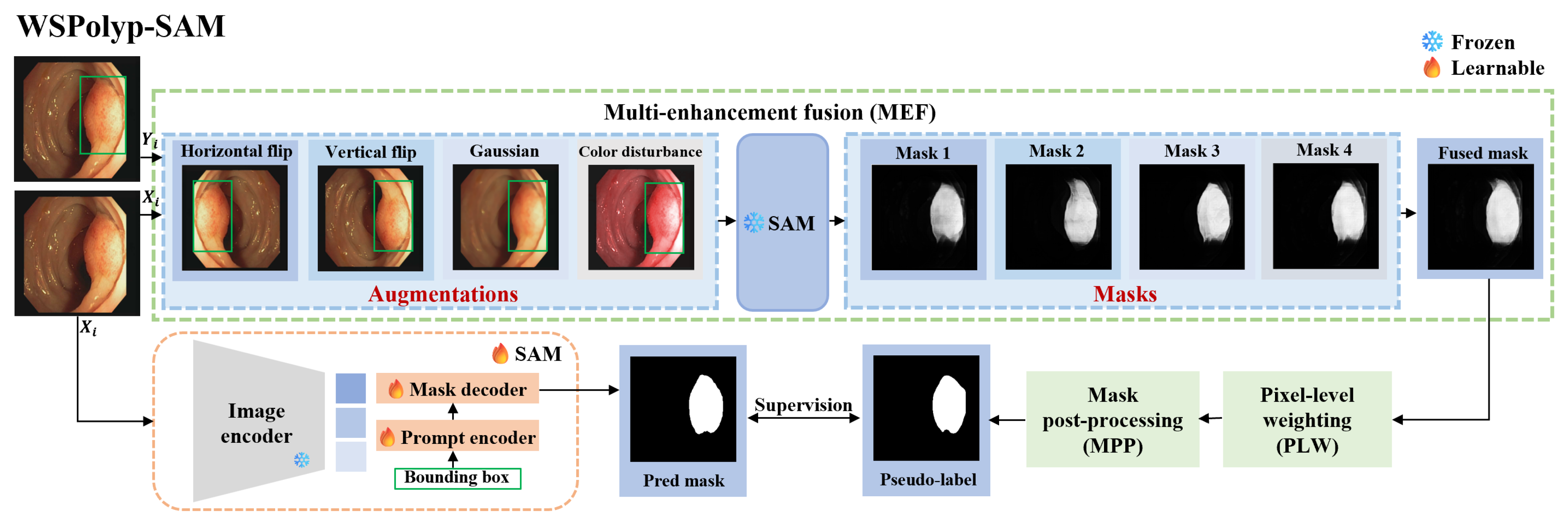
- The proposal of a novel weakly supervised and self-guided fine-tuning method of SAM for colonoscopy polyp segmentation (abbreviated as WSPolyp-SAM). This method reduces the dependence on precise annotations by fully utilizing SAM’s zero-shot ability to use weak annotations for guiding the generation of segmentation masks, which are then utilized as pseudo-labels for self-guided fine-tuning, avoiding the need to introduce additional segmentation models.
- The introduction of a series of pseudo-label enhancement strategies to generate high-quality pseudo-labels. These enhancement strategies, including multi-enhancement fusion, pixel-level weighting, and mask post-processing techniques, enable the acquisition of more accurate pseudo-labels.
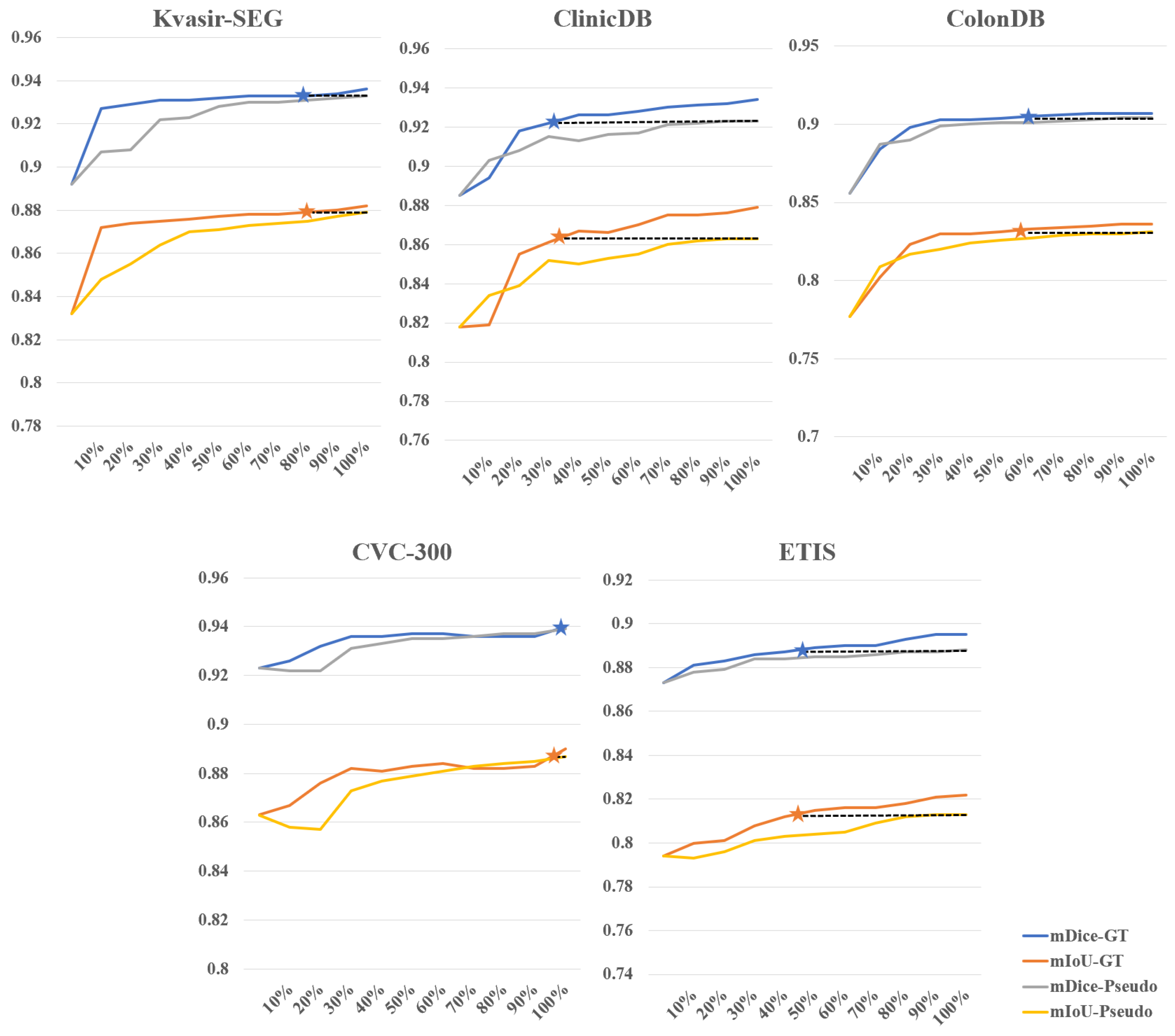
- Experimental results on five medical image datasets demonstrate that WSPolyp-SAM outperforms current fully supervised mainstream polyp segmentation networks on the Kvasir-SEG, ColonDB, CVC-300, and ETIS datasets. Particularly, on the ColonDB dataset, our method demonstrated an improvement of 9.4% in mDice score and 9.6% in mIoU score compared to state-of-the-art networks, representing a significant breakthrough in the field of colonoscopy polyp segmentation. Furthermore, by using different amounts of training data in weakly supervised and fully supervised experiments, it is found that weakly supervised fine-tuning can save 70% to 73% of the cost of annotation time compared to fully supervised fine-tuning.
2. Related Work
2.1. Polyp Segmentation
2.2. Segment Anything Model (SAM) Related Work
3. Methods
3.1. Background
3.2. Prompt Selection
3.3. Self-Guided Pseudo-Label Generation
3.4. Weakly Supervised Fine-Tuning
4. Experiments
4.1. Datasets
- 1.
- Kvasir-SEG [46]: This dataset, gathered by the Vestre Viken Health Trust in Norway, comprises 1000 colonoscopy video sequences showcasing polyp images, each accompanied by its respective annotations. The image resolution ranges from 332 × 487 to 1920 × 1072 pixels. Annotations are meticulously labeled by medical professionals and validated by seasoned gastroenterologists.
- 2.
- ClinicDB [47]: The collection of images within this dataset, totaling 612 and sourced from 29 distinct colonoscopy procedures, features a pixel dimension of 288 × 384. This initiative was developed collaboratively with the renowned Hospital Clinic of Barcelona, located in Spain.
- 3.
- ColonDB [48]: The dataset encompasses 380 images of polyps, complete with their respective annotations, all captured at a resolution of 500 × 570 pixels. These images are extracted from 15 distinct videos, with frames meticulously chosen by experts to showcase diverse perspectives. The annotations are manually crafted with precision.
- 4.
- CVC-300 [49]: The dataset includes 60 images of polyps, each with a resolution of 500 × 574 pixels.
- 5.
- ETIS [50]: This dataset was released by the MIC-CAI Polyp Detection Subchallenge in 2017, encompassing 196 polyp images extracted from colonoscopy videos, each paired with its respective label. The images boast a resolution of 966 × 1225 pixels.
4.2. Implementation Details
4.3. Results
4.3.1. Quantitative Results
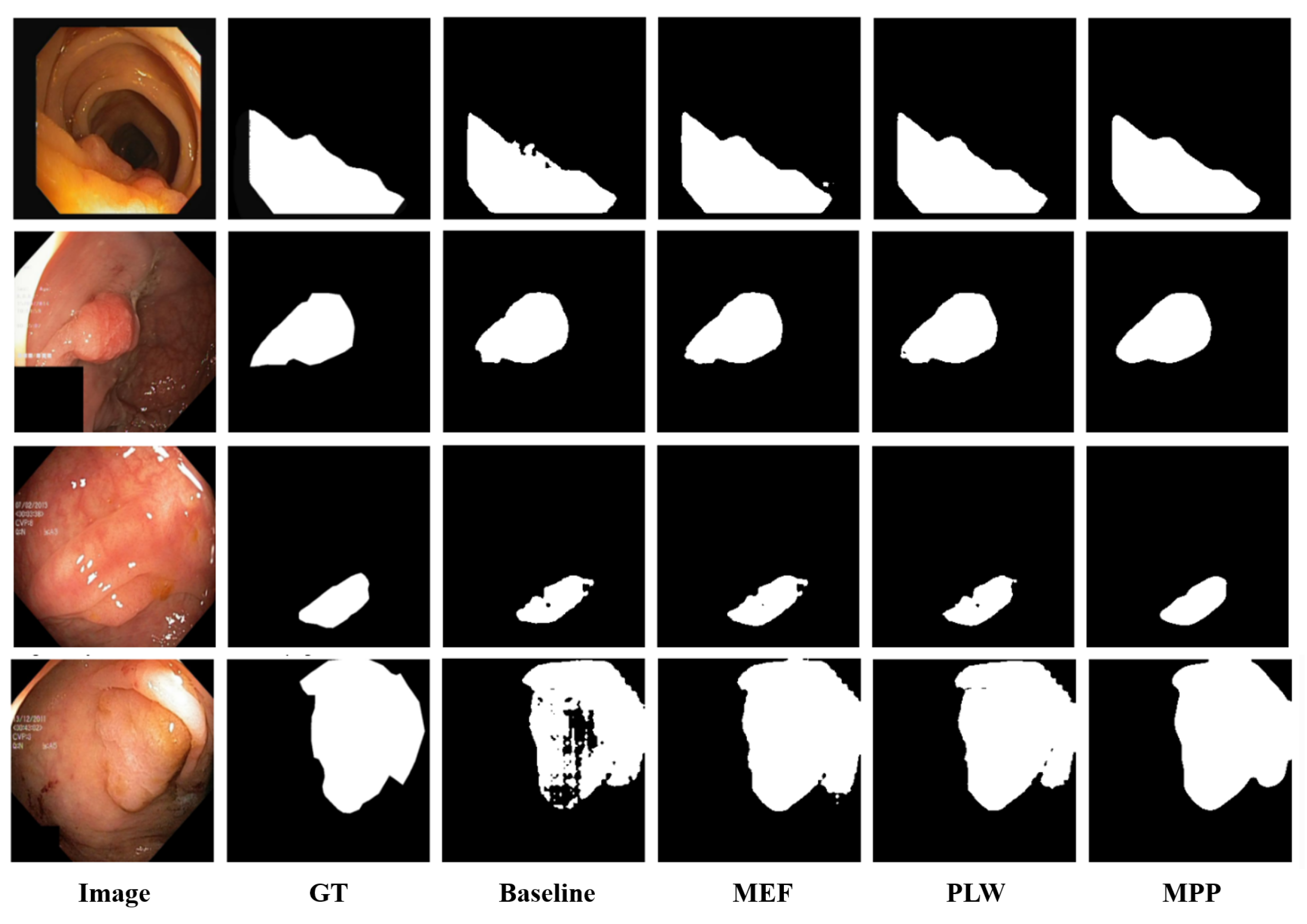
4.3.2. Qualitative Results
4.3.3. Ablation Study
4.3.4. Further Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gschwantler, M.; Kriwanek, S.; Langner, E.; Göritzer, B.; Schrutka-Kölbl, C.; Brownstone, E.; Feichtinger, H.; Weiss, W. High-grade dysplasia and invasive carcinoma in colorectal adenomas: A multivariate analysis of the impact of adenoma and patient characteristics. Eur. J. Gastroenterol. Hepatol. 2002, 14, 183. [Google Scholar] [CrossRef] [PubMed]
- Arnold, M.; Sierra, M.S.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global patterns and trends in colorectal cancer incidence and mortality. Gut J. Br. Soc. Gastroenterol. 2017, 66, 683–691. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Shi, J.; Qi, X.; Wang, X.; Jia, J. Pyramid scene parsing network. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 2881–2890. [Google Scholar]
- Chen, L.C.; Papandreou, G.; Kokkinos, I.; Murphy, K.; Yuille, A.L. Deeplab: Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected crfs. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 40, 834–848. [Google Scholar] [CrossRef] [PubMed]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; Van Der Laak, J.A.; Van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef]
- Asgari Taghanaki, S.; Abhishek, K.; Cohen, J.P.; Cohen-Adad, J.; Hamarneh, G. Deep semantic segmentation of natural and medical images: A review. Artif. Intell. Rev. 2021, 54, 137–178. [Google Scholar] [CrossRef]
- Zhang, R.; Lai, P.; Wan, X.; Fan, D.J.; Gao, F.; Wu, X.J.; Li, G. Lesion-aware dynamic kernel for polyp segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Singapore, 18–22 September 2022; Springer: Cham, Switzerland, 2022; pp. 99–109. [Google Scholar]
- Zhou, T.; Zhou, Y.; Gong, C.; Yang, J.; Zhang, Y. Feature aggregation and propagation network for camouflaged object detection. IEEE Trans. Image Process. 2022, 31, 7036–7047. [Google Scholar] [CrossRef]
- Ma, J.; He, Y.; Li, F.; Han, L.; You, C.; Wang, B. Segment anything in medical images. Nat. Commun. 2024, 15, 654. [Google Scholar] [CrossRef]
- Ding, N.; Qin, Y.; Yang, G.; Wei, F.; Yang, Z.; Su, Y.; Hu, S.; Chen, Y.; Chan, C.M.; Chen, W.; et al. Parameter-efficient fine-tuning of large-scale pre-trained language models. Nat. Mach. Intell. 2023, 5, 220–235. [Google Scholar] [CrossRef]
- Kirillov, A.; Mintun, E.; Ravi, N.; Mao, H.; Rolland, C.; Gustafson, L.; Xiao, T.; Whitehead, S.; Berg, A.C.; Lo, W.Y.; et al. Segment anything. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Paris, France, 2–3 October 2023; pp. 4015–4026. [Google Scholar]
- Hu, M.; Li, Y.; Yang, X. Skinsam: Empowering skin cancer segmentation with segment anything model. arXiv 2023, arXiv:2304.13973. [Google Scholar]
- Wu, J.; Fu, R.; Fang, H.; Liu, Y.; Wang, Z.; Xu, Y.; Jin, Y.; Arbel, T. Medical sam adapter: Adapting segment anything model for medical image segmentation. arXiv 2023, arXiv:2304.12620. [Google Scholar]
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 3431–3440. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the 18th International Conference of Medical Image Computing and Computer-Assisted Intervention (MICCAI 2015), Munich, Germany, 5–9 October 2015; Proceedings—Part III 18; Springer: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar]
- Zhou, Z.; Siddiquee, M.M.R.; Tajbakhsh, N.; Liang, J. Unet++: Redesigning skip connections to exploit multiscale features in image segmentation. IEEE Trans. Med. Imaging 2019, 39, 1856–1867. [Google Scholar] [CrossRef]
- Yang, X.; Li, X.; Ye, Y.; Lau, R.Y.; Zhang, X.; Huang, X. Road detection and centerline extraction via deep recurrent convolutional neural network U-Net. IEEE Trans. Geosci. Remote. Sens. 2019, 57, 7209–7220. [Google Scholar] [CrossRef]
- Qin, X.; Zhang, Z.; Huang, C.; Dehghan, M.; Zaiane, O.R.; Jagersand, M. U2-Net: Going deeper with nested U-structure for salient object detection. Pattern Recognit. 2020, 106, 107404. [Google Scholar] [CrossRef]
- Sun, X.; Zhang, P.; Wang, D.; Cao, Y.; Liu, B. Colorectal polyp segmentation by U-Net with dilation convolution. In Proceedings of the 2019 18th IEEE International Conference on Machine Learning and Applications (ICMLA), Boca Raton, FL, USA, 16–19 December 2019; pp. 851–858. [Google Scholar]
- Chen, L.C.; Zhu, Y.; Papandreou, G.; Schroff, F.; Adam, H. Encoder-decoder with atrous separable convolution for semantic image segmentation. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 801–818. [Google Scholar]
- Liu, S.; Huang, D. Receptive field block net for accurate and fast object detection. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 385–400. [Google Scholar]
- Fang, Y.; Chen, C.; Yuan, Y.; Tong, K.y. Selective feature aggregation network with area-boundary constraints for polyp segmentation. In Proceedings of the 22nd International Conference of Medical Image Computing and Computer Assisted Intervention (MICCAI 2019), Shenzhen, China, 13–17 October 2019; Proceedings—Part I 22; Springer: Cham, Switzerland, 2019; pp. 302–310. [Google Scholar]
- Zhang, R.; Li, G.; Li, Z.; Cui, S.; Qian, D.; Yu, Y. Adaptive context selection for polyp segmentation. In Proceedings of the 23rd International Conference of Medical Image Computing and Computer Assisted Intervention (MICCAI 2020), Lima, Peru, 4–8 October 2020; Proceedings—Part VI 23; Springer: Cham, Switzerland, 2020; pp. 253–262. [Google Scholar]
- Nguyen, T.C.; Nguyen, T.P.; Diep, G.H.; Tran-Dinh, A.H.; Nguyen, T.V.; Tran, M.T. CCBANet: Cascading context and balancing attention for polyp segmentation. In Proceedings of the 24th International Conference of Medical Image Computing and Computer Assisted Intervention (MICCAI 2021), Strasbourg, France, 27 September–1 October 2021; Proceedings—Part I 24; Springer: Cham, Switzerland, 2021; pp. 633–643. [Google Scholar]
- Deng, R.; Cui, C.; Liu, Q.; Yao, T.; Remedios, L.W.; Bao, S.; Landman, B.A.; Wheless, L.E.; Coburn, L.A.; Wilson, K.T.; et al. Segment anything model (sam) for digital pathology: Assess zero-shot segmentation on whole slide imaging. arXiv 2023, arXiv:2304.04155. [Google Scholar]
- Hu, C.; Li, X. When sam meets medical images: An investigation of segment anything model (sam) on multi-phase liver tumor segmentation. arXiv 2023, arXiv:2304.08506. [Google Scholar]
- He, S.; Bao, R.; Li, J.; Grant, P.E.; Ou, Y. Accuracy of segment-anything model (sam) in medical image segmentation tasks. arXiv 2023, arXiv:2304.09324. [Google Scholar]
- Roy, S.; Wald, T.; Koehler, G.; Rokuss, M.R.; Disch, N.; Holzschuh, J.; Zimmerer, D.; Maier-Hein, K.H. Sam. md: Zero-shot medical image segmentation capabilities of the segment anything model. arXiv 2023, arXiv:2304.05396. [Google Scholar]
- Zhou, T.; Zhang, Y.; Zhou, Y.; Wu, Y.; Gong, C. Can sam segment polyps? arXiv 2023, arXiv:2304.07583. [Google Scholar]
- Mohapatra, S.; Gosai, A.; Schlaug, G. Brain extraction comparing segment anything model (sam) and fsl brain extraction tool. arXiv 2023, arXiv:2304.04738. [Google Scholar]
- Mazurowski, M.A.; Dong, H.; Gu, H.; Yang, J.; Konz, N.; Zhang, Y. Segment anything model for medical image analysis: An experimental study. Med. Image Anal. 2023, 89, 102918. [Google Scholar] [CrossRef]
- Chen, J.; Bai, X. Learning to “segment anything” in thermal infrared images through knowledge distillation with a large scale dataset satir. arXiv 2023, arXiv:2304.07969. [Google Scholar]
- Tang, L.; Xiao, H.; Li, B. Can sam segment anything? When sam meets camouflaged object detection. arXiv 2023, arXiv:2304.04709. [Google Scholar]
- Ji, G.P.; Fan, D.P.; Xu, P.; Cheng, M.M.; Zhou, B.; Van Gool, L. SAM Struggles in Concealed Scenes–Empirical Study on “Segment Anything”. arXiv 2023, arXiv:2304.06022. [Google Scholar] [CrossRef]
- Ji, W.; Li, J.; Bi, Q.; Li, W.; Cheng, L. Segment anything is not always perfect: An investigation of sam on different real-world applications. arXiv 2023, arXiv:2304.05750. [Google Scholar] [CrossRef]
- Cheng, J.; Ye, J.; Deng, Z.; Chen, J.; Li, T.; Wang, H.; Su, Y.; Huang, Z.; Chen, J.; Jiang, L.; et al. Sam-med2d. arXiv 2023, arXiv:2308.16184. [Google Scholar]
- Jiang, P.T.; Yang, Y. Segment anything is a good pseudo-label generator for weakly supervised semantic segmentation. arXiv 2023, arXiv:2305.01275. [Google Scholar]
- He, C.; Li, K.; Zhang, Y.; Xu, G.; Tang, L.; Zhang, Y.; Guo, Z.; Li, X. Weakly-supervised concealed object segmentation with sam-based pseudo labeling and multi-scale feature grouping. In Proceedings of the Advances in Neural Information Processing Systems, Vancouver, BC, Canada, 13 February 2024; Volume 36. [Google Scholar]
- Reedha, R.; Dericquebourg, E.; Canals, R.; Hafiane, A. Transformer neural network for weed and crop classification of high resolution UAV images. Remote. Sens. 2022, 14, 592. [Google Scholar] [CrossRef]
- Brown, T.; Mann, B.; Ryder, N.; Subbiah, M.; Kaplan, J.D.; Dhariwal, P.; Neelakantan, A.; Shyam, P.; Sastry, G.; Askell, A.; et al. Language models are few-shot learners. Adv. Neural Inf. Process. Syst. 2020, 33, 1877–1901. [Google Scholar]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An image is worth 16x16 words: Transformers for image recognition at scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- He, K.; Chen, X.; Xie, S.; Li, Y.; Dollár, P.; Girshick, R. Masked autoencoders are scalable vision learners. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 18–24 June 2022; pp. 16000–16009. [Google Scholar]
- Zhang, W.; Fu, C.; Zheng, Y.; Zhang, F.; Zhao, Y.; Sham, C.W. HSNet: A hybrid semantic network for polyp segmentation. Comput. Biol. Med. 2022, 150, 106173. [Google Scholar] [CrossRef]
- Isensee, F.; Jaeger, P.F.; Kohl, S.A.; Petersen, J.; Maier-Hein, K.H. nnU-Net: A self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods 2021, 18, 203–211. [Google Scholar] [CrossRef]
- Ma, J.; Chen, J.; Ng, M.; Huang, R.; Li, Y.; Li, C.; Yang, X.; Martel, A.L. Loss odyssey in medical image segmentation. Med. Image Anal. 2021, 71, 102035. [Google Scholar] [CrossRef] [PubMed]
- Jha, D.; Smedsrud, P.H.; Riegler, M.A.; Halvorsen, P.; De Lange, T.; Johansen, D.; Johansen, H.D. Kvasir-seg: A segmented polyp dataset. In Proceedings of the 26th International Conference of MultiMedia Modeling (MMM 2020), Daejeon, Republic of Korea, 5–8 January 2020; Proceedings—Part II 26; Springer: Cham, Switzerland, 2020; pp. 451–462. [Google Scholar]
- Bernal, J.; Sánchez, F.J.; Fernández-Esparrach, G.; Gil, D.; Rodríguez, C.; Vilariño, F. WM-DOVA maps for accurate polyp highlighting in colonoscopy: Validation vs. saliency maps from physicians. Comput. Med. Imaging Graph. 2015, 43, 99–111. [Google Scholar] [CrossRef] [PubMed]
- Tajbakhsh, N.; Gurudu, S.R.; Liang, J. Automated polyp detection in colonoscopy videos using shape and context information. IEEE Trans. Med. Imaging 2015, 35, 630–644. [Google Scholar] [CrossRef] [PubMed]
- Vázquez, D.; Bernal, J.; Sánchez, F.J.; Fernández-Esparrach, G.; López, A.M.; Romero, A.; Drozdzal, M.; Courville, A. A benchmark for endoluminal scene segmentation of colonoscopy images. J. Healthc. Eng. 2017, 2017, 4037190. [Google Scholar] [CrossRef] [PubMed]
- Silva, J.; Histace, A.; Romain, O.; Dray, X.; Granado, B. Toward embedded detection of polyps in wce images for early diagnosis of colorectal cancer. Int. J. Comput. Assist. Radiol. Surg. 2014, 9, 283–293. [Google Scholar] [CrossRef] [PubMed]
- Fan, D.P.; Ji, G.P.; Zhou, T.; Chen, G.; Fu, H.; Shen, J.; Shao, L. Pranet: Parallel reverse attention network for polyp segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Lima, Peru, 4–8 October 2020; Springer: Cham, Switzerland, 2020; pp. 263–273. [Google Scholar]
- Dong, B.; Wang, W.; Fan, D.P.; Li, J.; Fu, H.; Shao, L. Polyp-pvt: Polyp segmentation with pyramid vision transformers. arXiv 2021, arXiv:2108.06932. [Google Scholar] [CrossRef]
- Sun, Y.; Chen, G.; Zhou, T.; Zhang, Y.; Liu, N. Context-aware cross-level fusion network for camouflaged object detection. arXiv 2021, arXiv:2105.12555. [Google Scholar]


| Models | Kvasir-SEG | ClinicDB | ColonDB | CVC-300 | ETIS | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| mDice | mIoU | mDice | mIoU | mDice | mIoU | mDice | mIoU | mDice | mIoU | |
| U-Net | 0.818 | 0.746 | 0.823 | 0.755 | 0.504 | 0.436 | 0.710 | 0.627 | 0.398 | 0.335 |
| U-Net++ | 0.821 | 0.744 | 0.794 | 0.729 | 0.482 | 0.408 | 0.707 | 0.624 | 0.401 | 0.344 |
| SFA | 0.723 | 0.611 | 0.700 | 0.607 | 0.456 | 0.337 | 0.467 | 0.329 | 0.297 | 0.271 |
| PraNet | 0.898 | 0.840 | 0.899 | 0.849 | 0.712 | 0.640 | 0.871 | 0.797 | 0.628 | 0.567 |
| C2FNet | 0.886 | 0.831 | 0.919 | 0.872 | 0.724 | 0.650 | 0.874 | 0.801 | 0.699 | 0.624 |
| Polyp-PVT | 0.917 | 0.864 | 0.948 | 0.905 | 0.808 | 0.727 | 0.900 | 0.833 | 0.787 | 0.706 |
| HSNet | 0.926 | 0.877 | 0.937 | 0.887 | 0.810 | 0.735 | 0.903 | 0.839 | 0.808 | 0.734 |
| SAM-B | 0.892 | 0.832 | 0.884 | 0.812 | 0.856 | 0.777 | 0.922 | 0.862 | 0.873 | 0.794 |
| WSPolyp-SAM-B | 0.933 | 0.879 | 0.920 | 0.858 | 0.904 | 0.831 | 0.933 | 0.878 | 0.888 | 0.813 |
| 0.041 | 0.047 | 0.036 | 0.046 | 0.048 | 0.054 | 0.011 | 0.016 | 0.015 | 0.019 | |
| Baseline | MEF | PLW | MPP | Kvasir-SEG | ClinicDB | ColonDB | CVC-300 | ETIS | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| mDice | mIoU | mDice | mIoU | mDice | mIoU | mDice | mIoU | mDice | mIoU | ||||
| - | - | - | - | 0.892 | 0.832 | 0.884 | 0.812 | 0.856 | 0.777 | 0.922 | 0.862 | 0.873 | 0.794 |
| √ | 0.928 | 0.873 | 0.914 | 0.850 | 0.895 | 0.820 | 0.931 | 0.875 | 0.876 | 0.796 | |||
| √ | √ | 0.928 | 0.875 | 0.914 | 0.851 | 0.897 | 0.822 | 0.931 | 0.875 | 0.880 | 0.797 | ||
| √ | √ | √ | 0.932 | 0.878 | 0.916 | 0.851 | 0.903 | 0.830 | 0.933 | 0.878 | 0.885 | 0.805 | |
| √ | √ | √ | √ | 0.933 | 0.879 | 0.920 | 0.858 | 0.904 | 0.831 | 0.933 | 0.878 | 0.888 | 0.813 |
| Model-ViT | Mask | Kvasir-SEG | ClinicDB | ColonDB | CVC-300 | ETIS | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| mDice | mIoU | mDice | mIoU | mDice | mIoU | mDice | mIoU | mDice | mIoU | ||
| SAM-B | - | 0.892 | 0.832 | 0.884 | 0.812 | 0.856 | 0.777 | 0.922 | 0.862 | 0.873 | 0.794 |
| SAM-L | - | 0.921 | 0.865 | 0.884 | 0.817 | 0.897 | 0.823 | 0.926 | 0.868 | 0.855 | 0.782 |
| SAM-H | - | 0.923 | 0.868 | 0.890 | 0.826 | 0.895 | 0.822 | 0.914 | 0.852 | 0.866 | 0.732 |
| SAM-B | Pseudo | 0.933 | 0.879 | 0.920 | 0.858 | 0.904 | 0.831 | 0.933 | 0.878 | 0.888 | 0.813 |
| SAM-L | Pseudo | 0.932 | 0.884 | 0.925 | 0.867 | 0.906 | 0.837 | 0.934 | 0.879 | 0.881 | 0.812 |
| SAM-B | GT | 0.936 | 0.882 | 0.935 | 0.882 | 0.907 | 0.836 | 0.934 | 0.879 | 0.895 | 0.822 |
| SAM-L | GT | 0.934 | 0.886 | 0.926 | 0.876 | 0.908 | 0.842 | 0.934 | 0.880 | 0.890 | 0.815 |
| Methods | Kvasir-SEG | ClinicDB | ColonDB | CVC-300 | ETIS | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| mDice | mIoU | mDice | mIoU | mDice | mIoU | mDice | mIoU | mDice | mIoU | |
| SAM | 0.892 | 0.832 | 0.884 | 0.812 | 0.856 | 0.777 | 0.922 | 0.862 | 0.873 | 0.794 |
| Pseudo-label Generation | 0.899 | 0.844 | 0.912 | 0.844 | 0.890 | 0.814 | 0.932 | 0.876 | 0.884 | 0.810 |
| WSPolyp-SAM | 0.933 | 0.879 | 0.920 | 0.858 | 0.904 | 0.831 | 0.933 | 0.878 | 0.888 | 0.813 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cai, T.; Yan, H.; Ding, K.; Zhang, Y.; Zhou, Y. WSPolyp-SAM: Weakly Supervised and Self-Guided Fine-Tuning of SAM for Colonoscopy Polyp Segmentation. Appl. Sci. 2024, 14, 5007. https://doi.org/10.3390/app14125007
Cai T, Yan H, Ding K, Zhang Y, Zhou Y. WSPolyp-SAM: Weakly Supervised and Self-Guided Fine-Tuning of SAM for Colonoscopy Polyp Segmentation. Applied Sciences. 2024; 14(12):5007. https://doi.org/10.3390/app14125007
Chicago/Turabian StyleCai, Tingting, Hongping Yan, Kun Ding, Yan Zhang, and Yueyue Zhou. 2024. "WSPolyp-SAM: Weakly Supervised and Self-Guided Fine-Tuning of SAM for Colonoscopy Polyp Segmentation" Applied Sciences 14, no. 12: 5007. https://doi.org/10.3390/app14125007
APA StyleCai, T., Yan, H., Ding, K., Zhang, Y., & Zhou, Y. (2024). WSPolyp-SAM: Weakly Supervised and Self-Guided Fine-Tuning of SAM for Colonoscopy Polyp Segmentation. Applied Sciences, 14(12), 5007. https://doi.org/10.3390/app14125007






