Visual Analysis of Carbendazim Residues in Carrot Tubers via Postionization Mass Spectrometry Imaging
Abstract
:1. Introduction
2. Materials and Methods
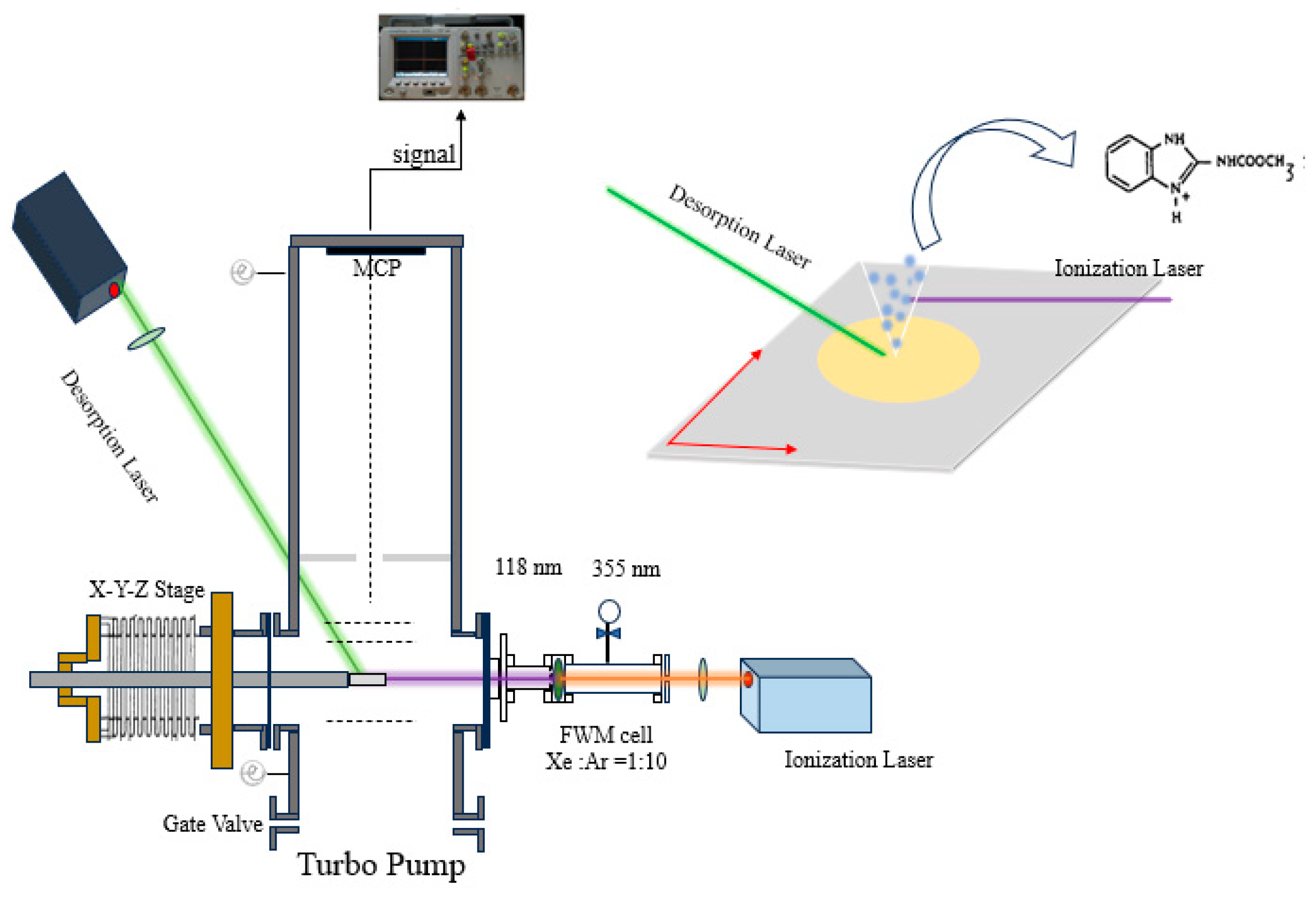
2.1. LDPI-TOF-MSI Apparatus
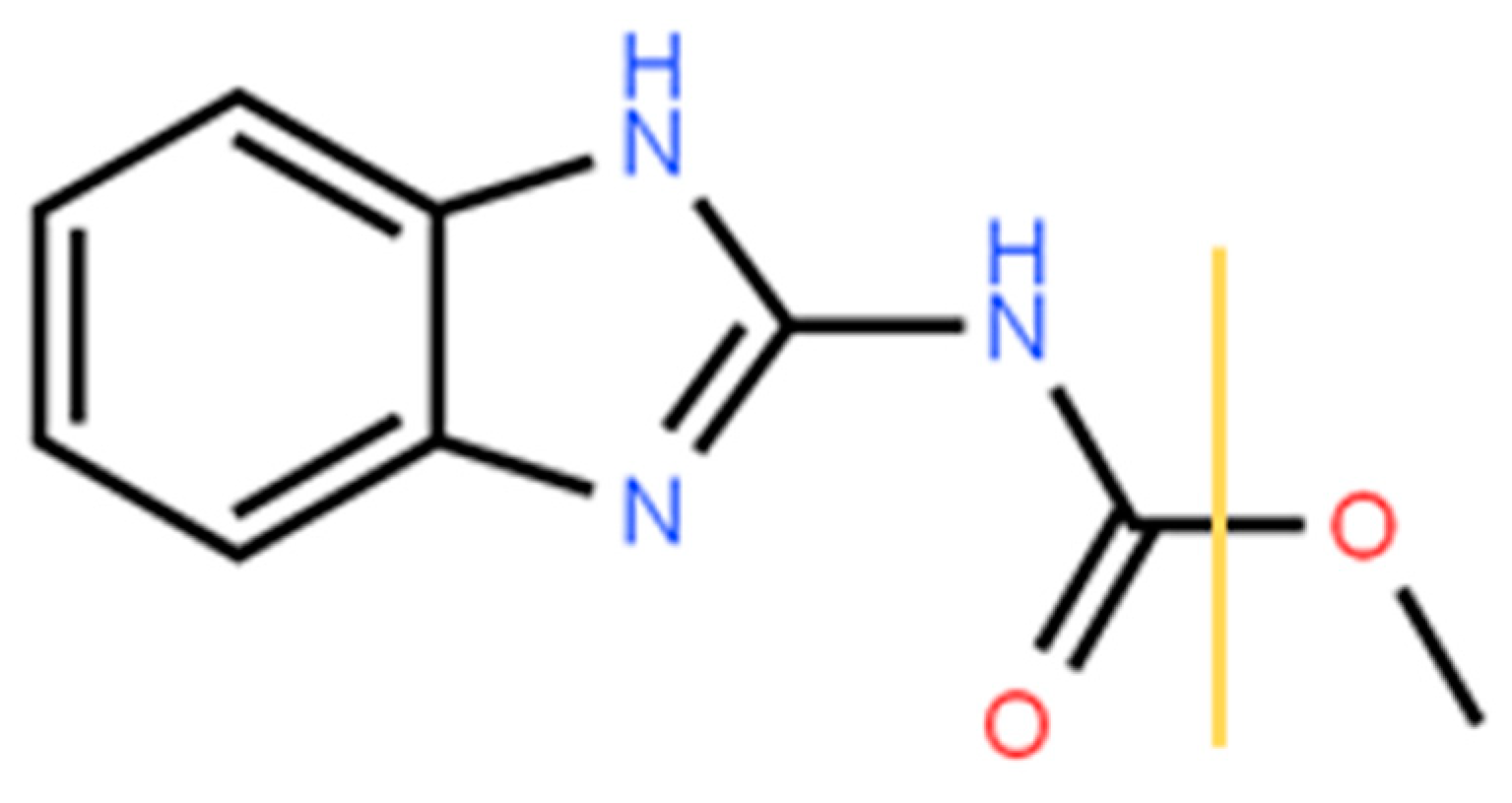
2.2. Material and Reagents
2.3. Experimental Procedure
3. Results
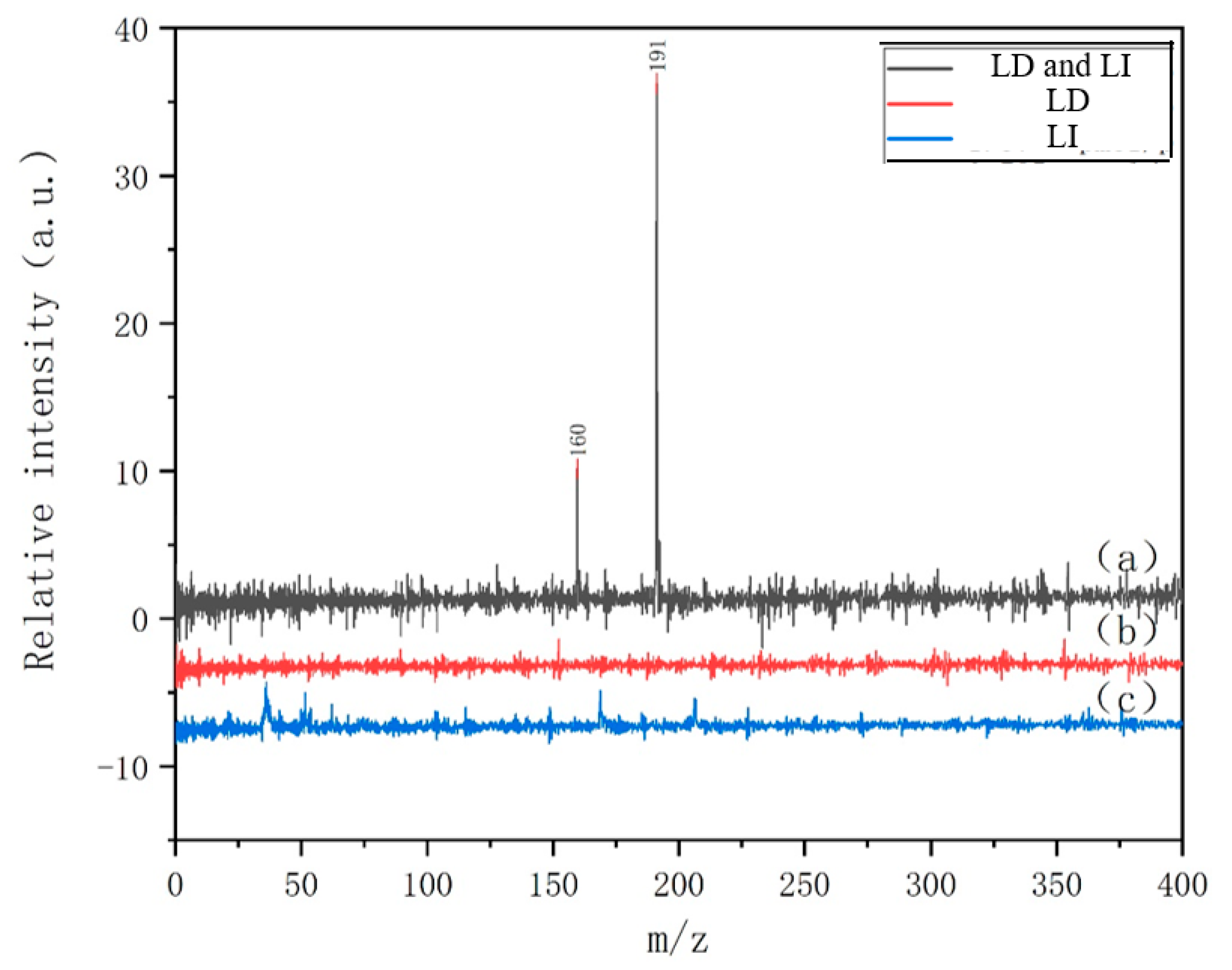
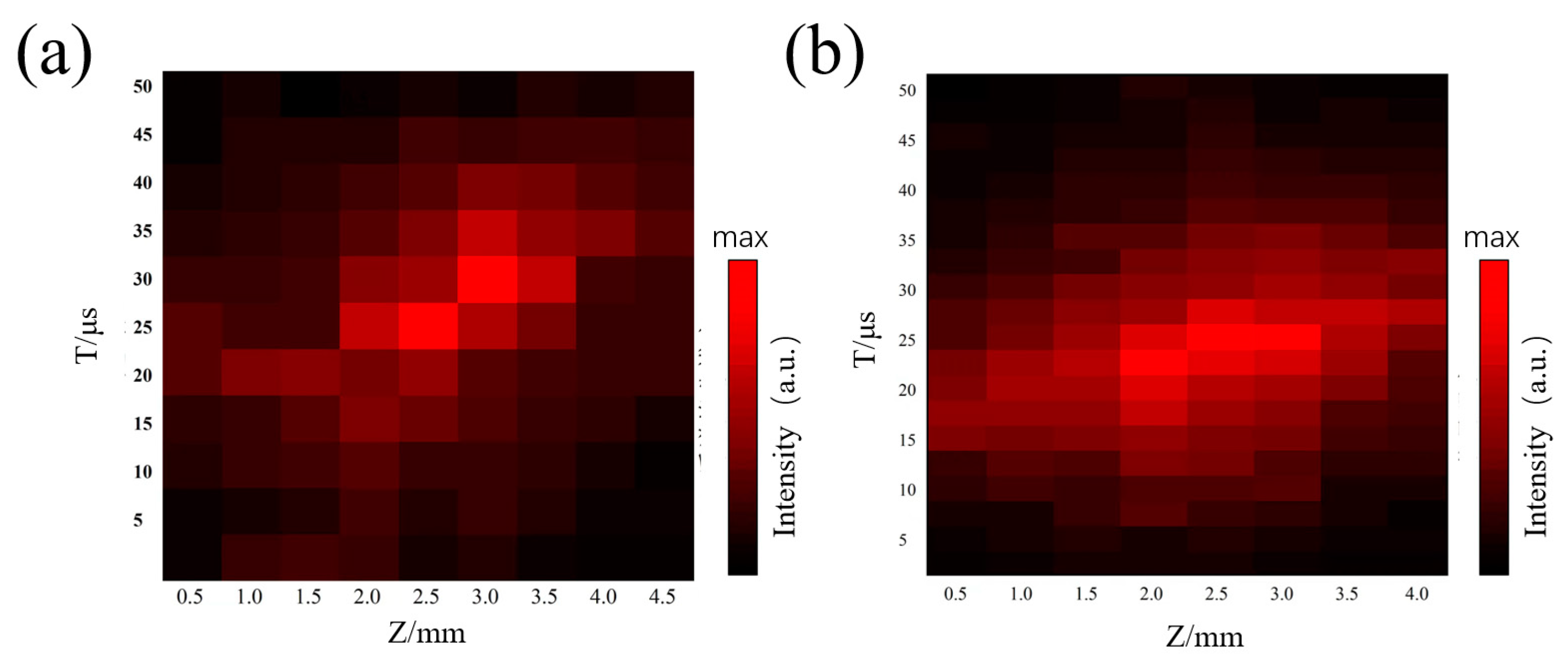
3.1. LDPI-MS Analysis of the Pure Sample of Carbendazim
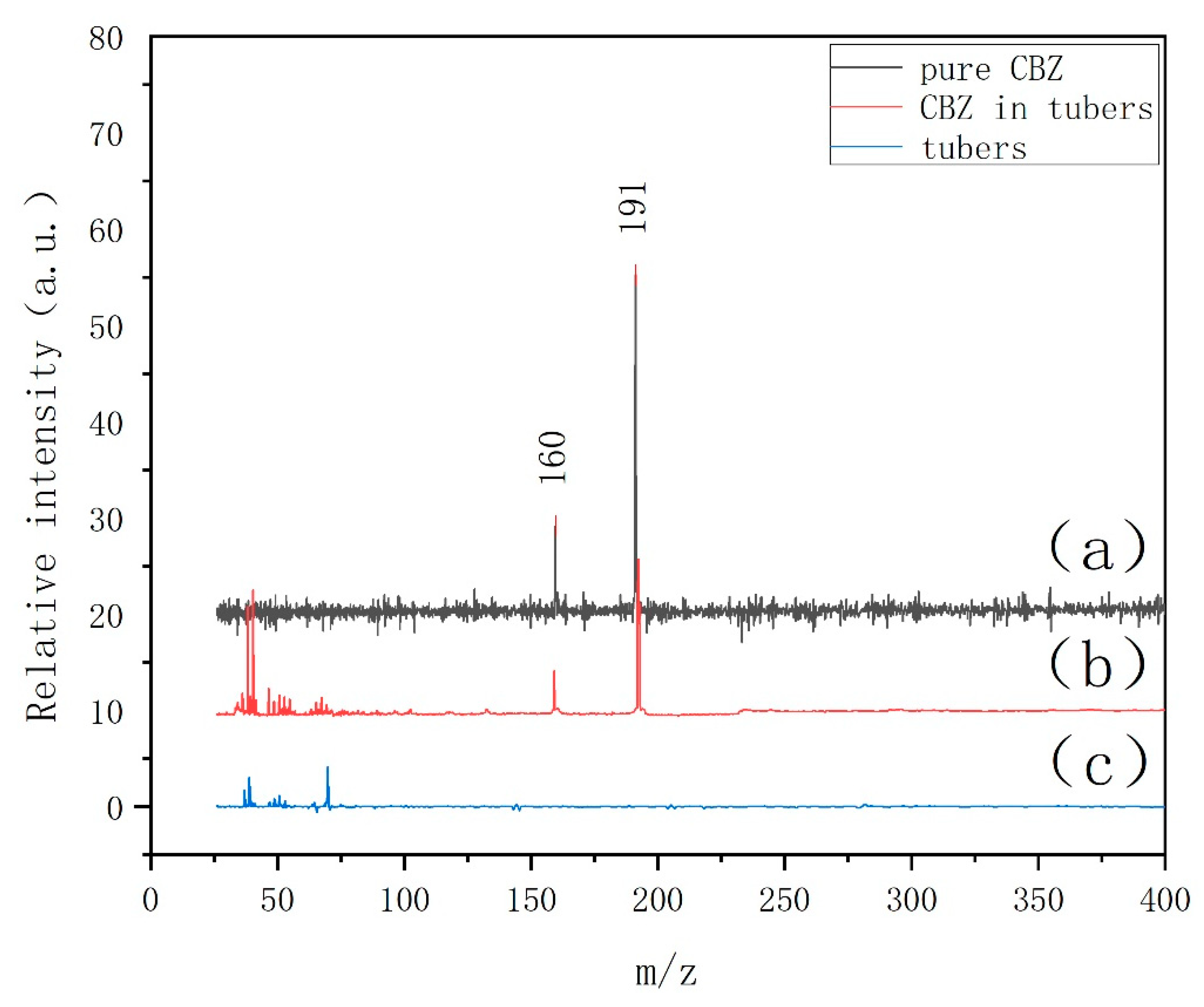
3.2. LDPI-MS Analysis of Carbendazim in Carrot Tissue
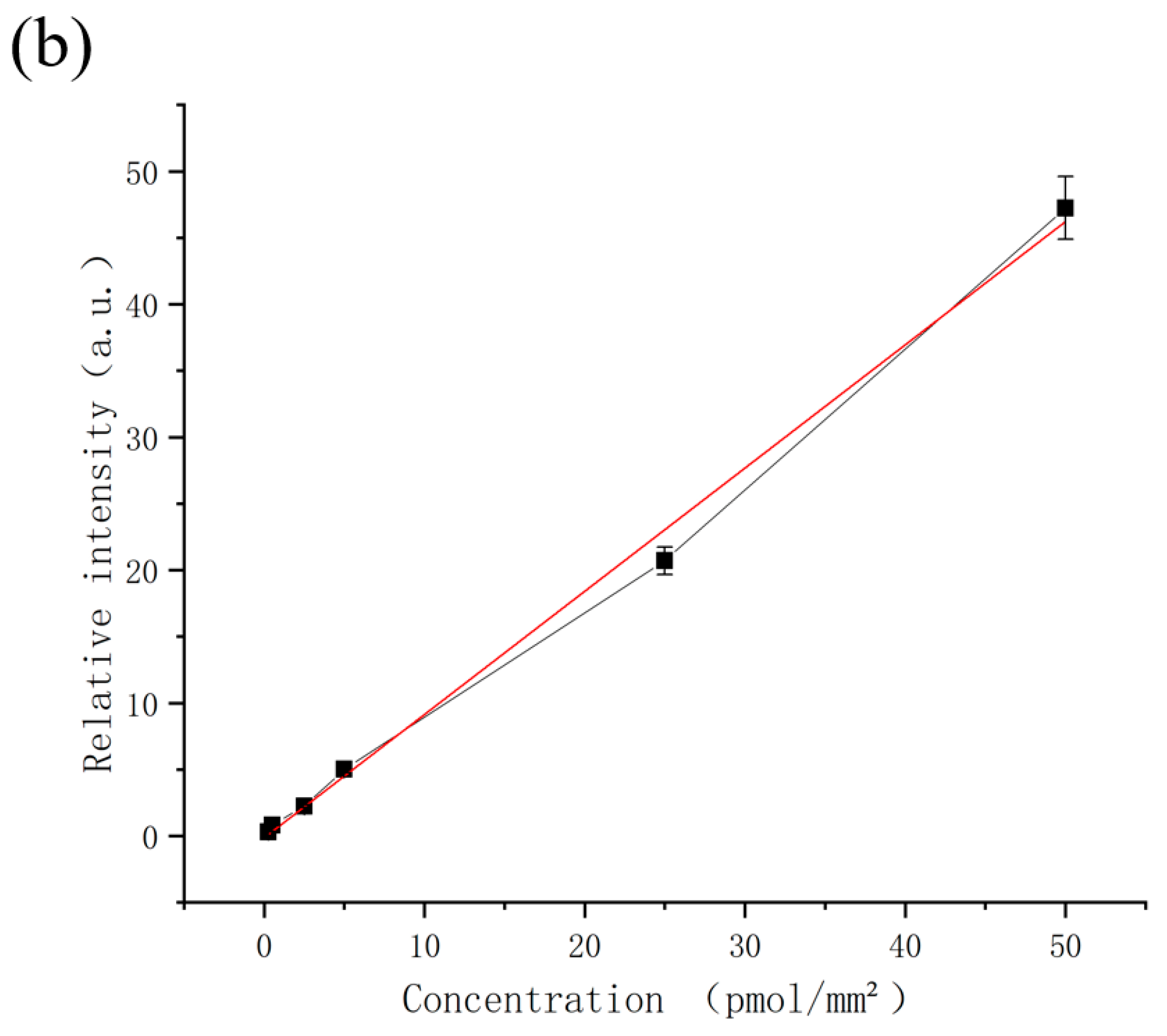
3.3. Signal Linearity and Limit of Detection
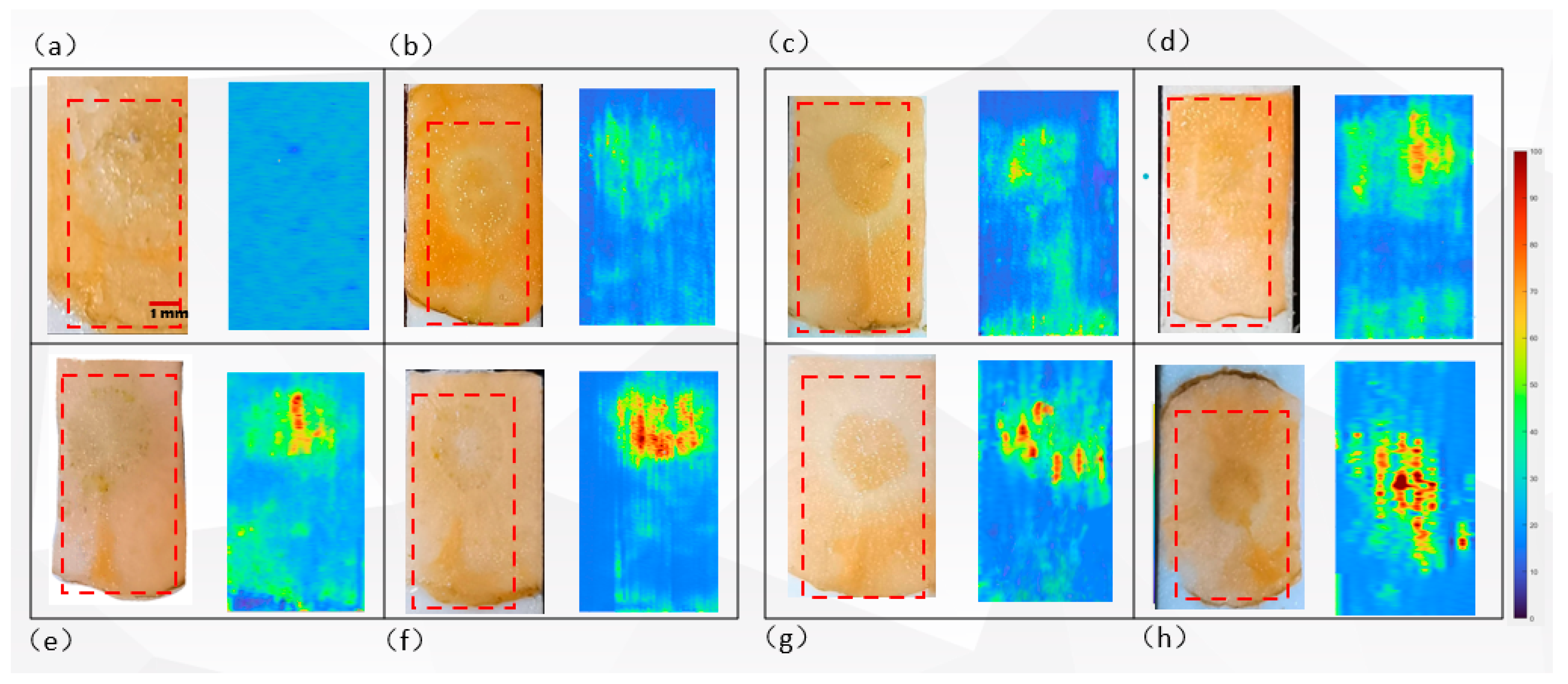
3.4. LDPI-MS Imaging of Carbendazim Residue in Carrot Tubers
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rozell, D. Using population projections in climate change analysis. Clim. Chang. 2017, 142, 521–529. [Google Scholar] [CrossRef]
- Zhang, J.J.; Yang, H. Metabolism and detoxification of pesticides in plants. Sci. Total Environ. 2021, 790, 148034. [Google Scholar] [CrossRef] [PubMed]
- Ucker, F.E.; Kalaoun, T.C.; Matias, R.K.; Gerônimo, F.H.C.; Ucker, L.C.F.; Silva, D.L. Consequences of Excessive Application of Pesticides and Impacts on the Environment and Human Healt. Int. J. Adv. Eng. Res. Sci. 2023, 10, 39–50. [Google Scholar] [CrossRef]
- Chakwiya, A.; Van der Linde, E.J.; Korsten, L. In vitro sensitivity testing of Cladobotryum mycophilum to carbendazim and prochloraz manganese. S. Afr. J. Sci. 2015, 111, 7. [Google Scholar] [CrossRef]
- Brauer, V.S.; Rezende, C.P.; Pessoni, A.M.; De Paula, R.G.; Rangappa, K.S.; Nayaka, S.C.; Gupta, V.K.; Almeida, F. Antifungal Agents in Agriculture: Friends and Foes of Public Health. Biomolecules 2019, 9, 521. [Google Scholar] [CrossRef] [PubMed]
- Ben Mordechay, E.; Mordehay, V.; Tarchitzky, J.; Chefetz, B. Fate of contaminants of emerging concern in the reclaimed wastewater-soil-plant continuum. Sci. Total Environ. 2022, 822, 153574. [Google Scholar] [CrossRef]
- Zhou, T.; Guo, T.; Wang, Y.; Wang, A.; Zhang, M. Carbendazim: Ecological risks, toxicities, degradation pathways and potential risks to human health. Chemosphere 2023, 314, 137723. [Google Scholar] [CrossRef]
- Martins, T.S.; Machado, S.A.S.; Oliveira, O.N.; Bott-Neto, J.L. Optimized paper-based electrochemical sensors treated in acidic media to detect carbendazim on the skin of apple and cabbage. Food Chem. 2023, 410, 135429. [Google Scholar] [CrossRef]
- Watanabe, E.; Kobara, Y.; Yogo, Y. Rapid and Simple Analysis of Pesticides Persisting on Green Pepper Surfaces Swabbing with Solvent-Moistened Cotton. J. Agric. Food Chem. 2012, 60, 9000–9005. [Google Scholar] [CrossRef]
- Michalik, B.; Simon, P.W.; Gabelman, W.H. Assessing Susceptibility of Carrot Roots to Bacterial Soft Rot. HortScience 1992, 27, 1020–1022. [Google Scholar] [CrossRef]
- da Silva, C.L.; de Lima, E.C.; Tavares, M.F.M. Investigation of preconcentration strategies for the trace analysis of multi-residue pesticides in real samples by capillary electrophoresis. J. Chromatogr. A 2003, 1014, 109–116. [Google Scholar] [CrossRef]
- Zhou, L.; Jiang, Y.; Lin, Q.; Wang, X.; Zhang, X.; Xu, J.; Chen, Z. Residue transfer and risk assessment of carbendazim in tea. J. Sci. Food Agric. 2018, 98, 5329–5334. [Google Scholar] [CrossRef]
- Singh, S.; Singh, N.; Kumar, V.; Datta, S.; Wani, A.B.; Singh, D.; Singh, K.; Singh, J. Toxicity, monitoring and biodegradation of the fungicide carbendazim. Environ. Chem. Lett. 2016, 14, 317–329. [Google Scholar] [CrossRef]
- Crapnell, R.D.; Adarakatti, P.S.; Banks, C.E. Electroanalytical overview: The sensing of carbendazim. Anal. Methods 2023, 15, 4811–4826. [Google Scholar] [CrossRef]
- Suresh, I.; Selvaraj, S.; Nesakumar, N.; Rayappan, J.B.B.; Kulandaiswamy, A.J. Nanomaterials based non-enzymatic electrochemical and optical sensors for the detection of carbendazim: A review. Trends Environ. Anal. Chem. 2021, 31, e00137. [Google Scholar] [CrossRef]
- Wang, S.-Y.; Shi, X.-C.; Liu, F.-Q.; Laborda, P. Chromatographic Methods for Detection and Quantification of Carbendazim in Food. J. Agric. Food Chem. 2020, 68, 11880–11894. [Google Scholar] [CrossRef]
- Khosropour, H.; Keramat, M.; Laiwattanapaisal, W. A dual action electrochemical molecularly imprinted aptasensor for ultra-trace detection of carbendazim. Biosens. Bioelectron. 2024, 243, 115754. [Google Scholar] [CrossRef]
- Peacock, P.M.; Zhang, W.-J.; Trimpin, S. Advances in Ionization for Mass Spectrometry. Anal. Chem. 2016, 89, 372–388. [Google Scholar] [CrossRef]
- Fan, J.; Ma, W.; Yu, Y.; Li, Y.; Nie, Z. Recent advances in entirely hand-held ionization sources for mass spectrometry. Anal. Bioanal. Chem. 2023, 416, 2057–2063. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.H. Basic principle from electrospray ionization to soft ionization mass spectrometry and development of ion source. ChemRxiv 2022, 10, 2432–2434. [Google Scholar] [CrossRef]
- OMirgorodskaya, O.A.; Shevchenko, A.A.; Chernushevich, I.V.; Dodonov, A.F.; Miroshnikov, A.I. Electrospray-ionization-time-of-flight-mass-spectrometry-in-protein-chemistry. Anal. Chem. 2002, 66, 99–107. [Google Scholar] [CrossRef]
- Wang, C.; Qin, L.-Y.; Li, D.-M.; Hu, L.-G.; Xue, J.-J.; Zhai, X.-P.; Wang, Q.; Guo, L.; Tang, L.; Xie, J.-W. Doped nanomaterial facilitates 3D printing target plate for rapid detection of alkaloids in laser desorption/ionization mass spectrometry. Anal. Bioanal. Chem. 2023, 415, 6825–6838. [Google Scholar] [CrossRef]
- Yang, Z.; Chang, Z.; Deng, K.; Gu, J.; Wu, Y.; Sun, Q.; Luo, Q. Reactive Matrices for MALDI-MS of Cholesterol. Anal. Chem. 2023, 95, 16786–16790. [Google Scholar] [CrossRef]
- Drzeżdżon, J.; Jacewicz, D.; Sielicka, A.; Chmurzyński, L. MALDI-MS for polymer characterization—Recent developments and future prospects. TrAC Trends Anal. Chem. 2019, 115, 121–128. [Google Scholar] [CrossRef]
- Xiang, L.; Wang, F.; Bian, Y.; Harindintwali, J.D.; Wang, Z.; Wang, Y.; Dong, J.; Chen, H.; Schaeffer, A.; Jiang, X.; et al. Visualizing the Distribution of Phthalate Esters and Plant Metabolites in Carrot by Matrix-Assisted Laser Desorption/Ionization Imaging Mass Spectrometry. J. Agric. Food Chem. 2022, 70, 15311–15320. [Google Scholar] [CrossRef]
- Lu, Q.; Xu, Z.; You, X.; Ma, S.; Zenobi, R. Atmospheric Pressure Mass Spectrometry Imaging Using Laser Ablation, Followed by Dielectric Barrier Discharge Ionization. Anal. Chem. 2021, 93, 6232–6238. [Google Scholar] [CrossRef]
- Shariatgorji, M.; Källback, P.; Gustavsson, L.; Schintu, N.; Svenningsson, P.; Goodwin, R.J.A.; Andren, P.E. Controlled-pH Tissue Cleanup Protocol for Signal Enhancement of Small Molecule Drugs Analyzed by MALDI-MS Imaging. Anal. Chem. 2012, 84, 4603–4607. [Google Scholar] [CrossRef]
- Wang, P.; Giese, R.W. Recommendations for quantitative analysis of small molecules by matrix-assisted laser desorption ionization mass spectrometry. J. Chromatogr. A 2017, 1486, 35–41. [Google Scholar] [CrossRef]
- Peterson, D.S. Matrix-free methods for laser desorption/ionization mass spectrometry. Mass Spectrom. Rev. 2006, 26, 19–34. [Google Scholar] [CrossRef]
- Li, X.; Hang, L.; Wang, T.; Leng, Y.; Zhang, H.; Meng, Y.; Yin, Z.; Hang, W. Nanoscale Three-Dimensional Imaging of Drug Distributions in Single Cells via Laser Desorption Post-Ionization Mass Spectrometry. J. Am. Chem. Soc. 2021, 143, 21648–21656. [Google Scholar] [CrossRef] [PubMed]
- Trevor, J.L.; Lykke, K.R.; Pellin, M.J.; Hanley, L. Two-Laser Mass Spectrometry of Thiolate, Disulfide, and Sulfide Self-Assembled Monolayers. Langmuir 1998, 14, 1664–1673. [Google Scholar] [CrossRef]
- Cui, Y.; Veryovkin, I.V.; Majeski, M.W.; Cavazos, D.R.; Hanley, L. High Lateral Resolution vs Molecular Preservation in near-IR fs-Laser Desorption Postionization Mass Spectrometry. Anal. Chem. 2014, 87, 367–371. [Google Scholar] [CrossRef]
- Soltwisch, J.; Kettling, H.; VENS-Cappell, S.; Wiegelmann, M.; Muthing, J.; Dreisewerd, K. Mass spectrometry imaging with laser-induced postionization. Science 2015, 348, 211–215. [Google Scholar] [CrossRef]
- Gasper, G.L.; Takahashi, L.K.; Zhou, J.; Ahmed, M.; Moore, J.F.; Hanley, L. Laser Desorption Postionization Mass Spectrometry of Antibiotic-Treated Bacterial Biofilms Using Tunable Vacuum Ultraviolet Radiation. Anal. Chem. 2010, 82, 7422–7478. [Google Scholar] [CrossRef]
- Soltwisch, J. Mass spectrometry imaging with MALDI-2. Nat. Methods 2015, 12, 387. [Google Scholar] [CrossRef]
- Akhmetov, A.; Moore, J.F.; Gasper, G.L.; Koin, P.J.; Hanley, L. Laser desorption postionization for imaging MS of biological material. J. Mass Spectrom. 2010, 45, 137–145. [Google Scholar] [CrossRef]
- Holbrook, J.H.; Kemper, G.; Hummon, A. Quantitative Mass Spectrometry Imaging: Therapeutics & Biomolecules. Chem. Commun. 2024, 60, 2137–2151. [Google Scholar] [CrossRef]
- Lu, Q.; Hu, Y.; Chen, J.; Jin, S. Laser Desorption Postionization Mass Spectrometry Imaging of Folic Acid Molecules in Tumor Tissue. Anal. Chem. 2017, 89, 8238–8243. [Google Scholar] [CrossRef]
- Nie, W.; Lu, Q.; Hu, T.; Xie, M.; Hu, Y. Visualizing the distribution of curcumin in the root of Curcuma longa via VUV-postionization mass spectrometric imaging. Analyst 2022, 148, 175–181. [Google Scholar] [CrossRef] [PubMed]
- Hanley, L.; Zimmermann, R. Light and Molecular Ions: The Emergence of Vacuum UV Single-Photon Ionization in MS. Anal. Chem. 2009, 11, 4174–4182. [Google Scholar] [CrossRef] [PubMed]
- Que, F.; Wang, G.-L.; Feng, K.; Xu, Z.-S.; Wang, F.; Xiong, A.-S. Hypoxia enhances lignification and affects the anatomical structure in hydroponic cultivation of carrot taproot. Plant Cell Rep. 2018, 37, 1021–1032. [Google Scholar] [CrossRef]
- Panadés, R.; Ibarz, A.; Esplugas, S. Photodecomposition of carbendazim in aqueous solutions. Water Res. 2000, 34, 2951–2954. [Google Scholar] [CrossRef]
- Dettenmaier, E.M.; Doucette, W.J.; Bugbee, B. Chemical Hydrophobicity and Uptake by Plant Roots. Environ. Sci. Technol. 2009, 43, 324–329. [Google Scholar] [CrossRef]
- Ahn, S.; Lee, J.-Y.; Kim, B. Accurate Determination of Carbaryl, Carbofuran and Carbendazim in Vegetables by Isotope Dilution Liquid Chromatography/Tandem Mass Spectrometry. Chromatographia 2020, 84, 27–35. [Google Scholar] [CrossRef]
- Roman, M.; Dobrowolski, J.C.; Baranska, M.; Baranski, R. Spectroscopic Studies on Bioactive Polyacetylenes and Other Plant Components in Wild Carrot Root. J. Nat. Prod. 2011, 74, 1757–1763. [Google Scholar] [CrossRef] [PubMed]
- Létondor, C.; Pascal-Lorber, S.; Laurent, F. Uptake and distribution of chlordecone in radish: Different contamination routes in edible roots. Chemosphere 2015, 118, 20–28. [Google Scholar] [CrossRef] [PubMed]



| Condition | CBZ (m/z 191) Signal Intensity (a.u.) | ||||||
|---|---|---|---|---|---|---|---|
| Variate | Duplicate Experimental Data | Average | |||||
| Temperature (°C) | 15 | 46.3 | 45.8 | 46.6 | 47.1 | 45.5 | 46.3 |
| 20 | 46.8 | 49.2 | 48.5 | 46.1 | 50.2 | 48.2 | |
| 25 | 47.3 | 46.7 | 48.0 | 51.1 | 54.2 | 49.4 | |
| Shaking frequency (/min) | 0 | 38.2 | 41.0 | 38.5 | 37.6 | 38.1 | 38.7 |
| 5 | 46.8 | 49.2 | 48.5 | 46.1 | 50.2 | 48.2 | |
| 10 | 45.8 | 47.3 | 50.0 | 48.9 | 46.3 | 47.7 | |
| Thickness (μm) | 50 | 25.3 | 24.8 | 23.9 | 24.1 | 23.9 | 24.4 |
| 100 | 46.8 | 49.2 | 48.5 | 46.1 | 50.2 | 48.2 | |
| 150 | 55.2 | 51.6 | 54.5 | 55.3 | 54.0 | 54.1 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, T.; Zhang, H.; Hu, Y. Visual Analysis of Carbendazim Residues in Carrot Tubers via Postionization Mass Spectrometry Imaging. Appl. Sci. 2024, 14, 5431. https://doi.org/10.3390/app14135431
Wang T, Zhang H, Hu Y. Visual Analysis of Carbendazim Residues in Carrot Tubers via Postionization Mass Spectrometry Imaging. Applied Sciences. 2024; 14(13):5431. https://doi.org/10.3390/app14135431
Chicago/Turabian StyleWang, Tianyu, He Zhang, and Yongjun Hu. 2024. "Visual Analysis of Carbendazim Residues in Carrot Tubers via Postionization Mass Spectrometry Imaging" Applied Sciences 14, no. 13: 5431. https://doi.org/10.3390/app14135431




