Effect of Genotype × Environment Interactions on the Yield and Stability of Sugarcane Varieties in Ecuador: GGE Biplot Analysis by Location and Year
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sources of Genotypes and Test Sites
2.2. Yield
2.3. Statistical Analysis
3. Results
3.1. Analysis of Average Yield
3.2. Relevance of Genotypic, Environmental, and Genotype-Environmental Effects
3.3. Effect of Location on Yield Components
3.4. Comparison of Sources of Variation
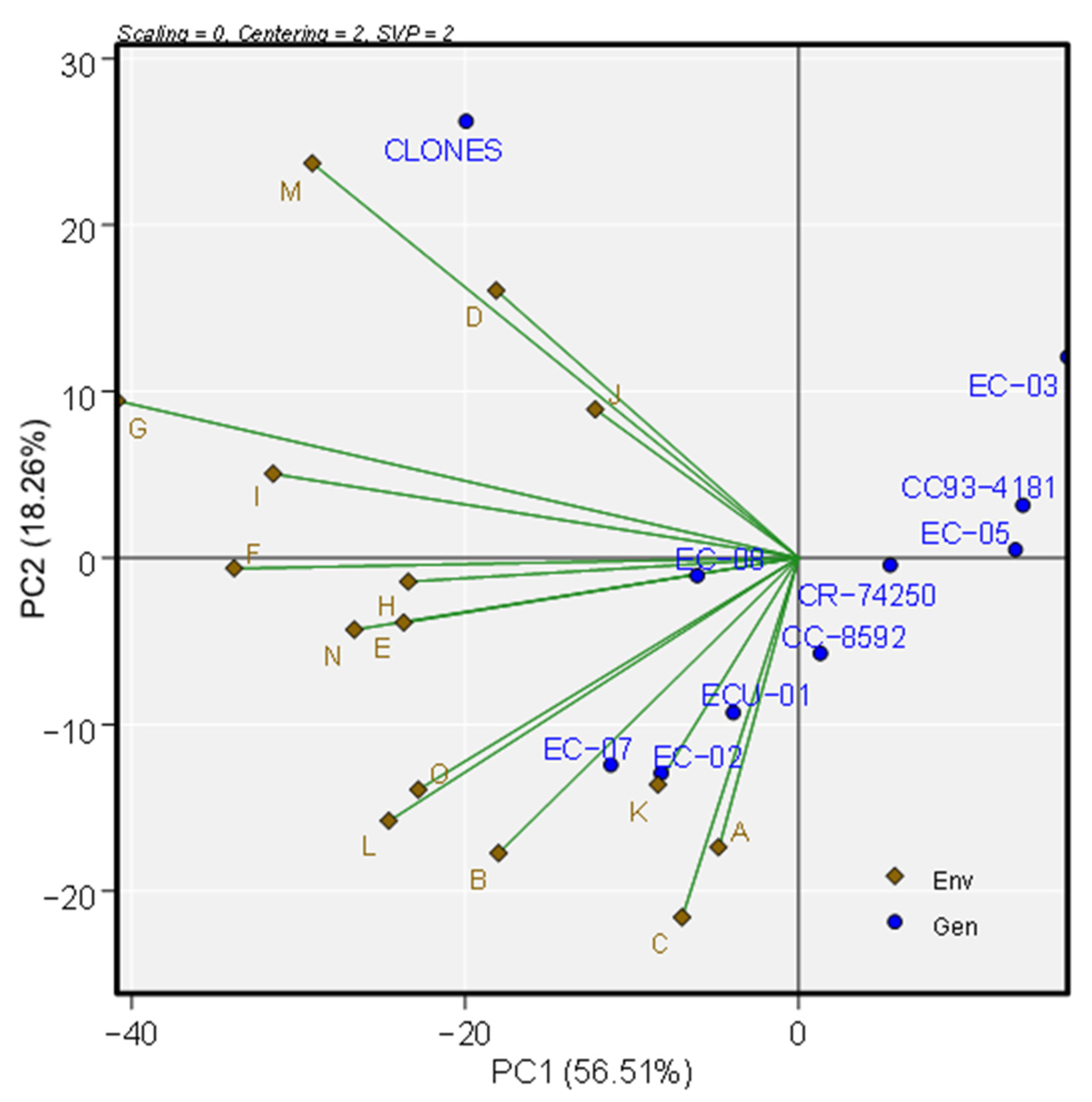
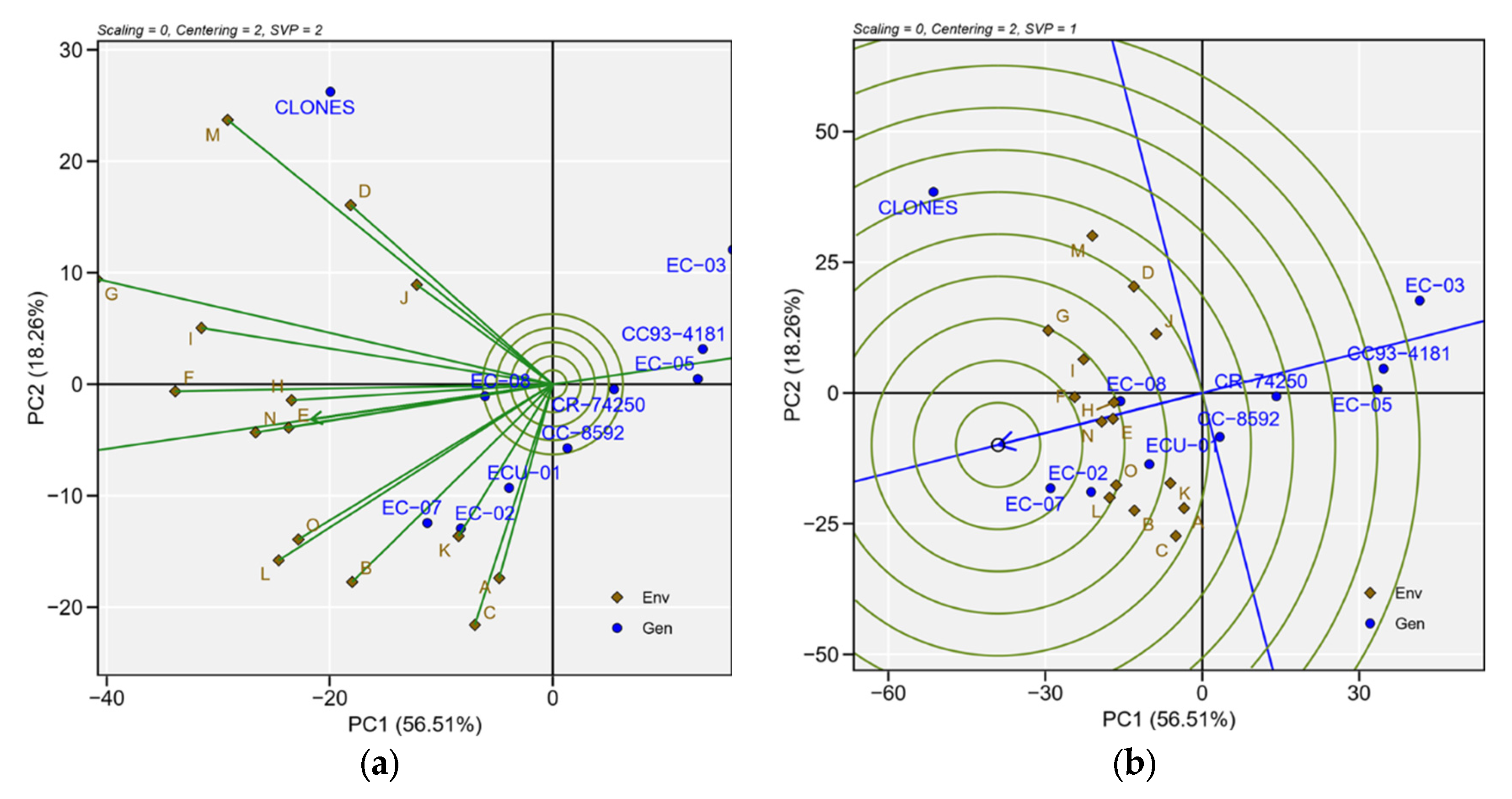
3.5. Biplot Patterns for Multivariate Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lee, H.; Sohn, Y.J.; Jeon, S.; Yang, H.; Son, J.; Kim, Y.J.; Park, S.J. Sugarcane wastes as microbial feedstocks: A review of the biorefinery framework from resource recovery to production of value-added products. Bioresour. Technol. 2023, 376, 128879. [Google Scholar] [CrossRef]
- Ministerio de Agricultura y Ganadería (MAGAP). Luego de Siete años Aumenta el Precio de la Tonelada de Caña de Azúcar. Available online: https://www.agricultura.gob.ec/luego-de-siete-anos-aumenta-el-precio-de-la-tonelada-de-cana-de-azucar (accessed on 8 July 2024).
- INEC (Instituto Nacional de Estadística y Censo). Encuesta de Superficie y Producción Agropecuaria Continua. ESPAC. 2022. Available online: https://www.ecuadorencifras.gob.ec/encuesta-de-superficie-y-produccion-agropecuaria-continua-2022/ (accessed on 8 July 2024).
- Silva, E.; Martínez, F.; Madrid, C.; León, T.; Castillo, R.; Mendoza, J.; Garcés, F.; Salazar, M.; Aucatoma, B.; Fiallos, B.; et al. EC-07 y EC-08, nuevas variedades mejoradas de caña de azúcar. Boletín Divulg. 2016, 10, 1–9. [Google Scholar]
- Yadawad, A.; Patil, S.B.; Kongawad, B.Y.; Kadlag, A.D.; Hemaprabha, G. Multi environmental evaluation for selection of stable and high yielding sugarcane (Saccharum officinarum L.) clones based on AMMI and GGE biplot models. Indian. J. Genet. Plant Breed. 2023, 83, 389–397. [Google Scholar] [CrossRef]
- Natarajan, S.; Basnayake, J.; Lakshmanan, P.; Fukai, S. Limited contribution of water availability in genotype-by-environment interaction in sugarcane yield and yield components. J. Agron. Crop Sci. 2020, 206, 665–678. [Google Scholar] [CrossRef]
- Dang, X.; Hu, X.; Ma, Y.; Li, Y.; Kan, W.; Dong, X. AMMI and GGE biplot analysis for genotype × environment interactions affecting the yield and quality characteristics of sugar beet. PeerJ 2024, 12, e16882. [Google Scholar] [CrossRef]
- Oladosu, Y.; Rafii, M.Y.; Abdullah, N.; Magaji, U.; Miah, G.; Hussin, G.; Ramli, A. Genotype × Environment interaction and stability analyses of yield and yield components of established and mutant rice genotypes tested in multiple locations in Malaysia. Acta Agric. Scand. B Soil Plant Sci. 2017, 67, 590–606. [Google Scholar] [CrossRef]
- Liu, Q.; Huang, L.; Fu, C.; Zhang, T.; Ding, W.; Yang, C. Genotype–environment interaction of crocin in Gardenia jasminoides by AMMI and GGE biplot analysis. Food Sci. Nutr. 2022, 10, 4080–4087. [Google Scholar] [CrossRef]
- Yan, W.; Cornelius, P.L.; Crossa, J.; Hunt, L.A.; Yan, W. Two Types of GGE Biplots for Analyzing Multi-Environment Trial Data. Crop Sci. 2001, 41, 656–663. [Google Scholar] [CrossRef]
- Yan, W.; Hunt, L.A.; Sheng, Q.; Szlavnics, Z. Cultivar Evaluation and Mega-Environment Investigation Based on the GGE Biplot. Crop Sci. 2000, 40, 597–605. [Google Scholar] [CrossRef]
- Frutos, E.; Galindo, M.P.; Leiva, V. An interactive biplot implementation in R for modeling genotype-by-environment interaction. Stoch. Env. Res. Risk Assess. 2014, 28, 1629–1641. [Google Scholar] [CrossRef]
- Gabriel, K.R. The biplot graphic display of matrices with application to principal component analysis. Biometrika 1971, 58, 453–467. [Google Scholar] [CrossRef]
- Galindo-Villardón, P. Una Alternativa de Representación Simultánea: HJ-Biplot. Qüestiió 1986, 10, 13–23. [Google Scholar]
- Mehareb, E.M.; Osman, M.A.M.; Attia, A.E.; Bekheet, M.A.; Fouz, A.E.F.M. Stability assessment for selection of elite sugarcane clones across multi-environment based on AMMI and GGE-biplot models. Euphytica 2022, 218, 95. [Google Scholar] [CrossRef]
- Maulana, H.; Solihin, E.; Trimo, L.; Hidayat, S.; Wijaya, A.A.; Hariadi, H.; Amien, S.; Ruswandi, D.; Karuniawan, A. Genotype-by-environment interactions (GEIs) and evaluate superior sweet potato (Ipomoea batatas [L.] Lam) using combined analysis and GGE biplot. Heliyon 2023, 9, e20203. [Google Scholar] [CrossRef]
- Esan, V.I.; Oke, G.O.; Ogunbode, T.O.; Obisesan, I.A. AMMI and GGE biplot analyses of Bambara groundnut [Vigna subterranea (L.) Verdc.] for agronomic performances under three environmental conditions. Front. Plant Sci. 2023, 13, 997429. [Google Scholar] [CrossRef]
- Khan, M.M.H.; Rafii, M.Y.; Ramlee, S.I.; Jusoh, M.; Al Mamun, M. AMMI and GGE biplot analysis for yield performance and stability assessment of selected Bambara groundnut (Vigna subterranea L. Verdc.) genotypes under the multi-environmental trials (METs). Sci. Rep. 2021, 11, 22791. [Google Scholar]
- Yan, W. Singular-Value Partitioning in Biplot Analysis of Multienvironment Trial Data. Agron. J. 2002, 94, 990–996. [Google Scholar]
- Akan, K.; Cat, A.; Hocaoglu, O.; Tekin, M. Evaluating Scald Reactions of Some Turkish Barley (Hordeum vulgare L.) Varieties Using GGE Biplot Analysis. Agronomy 2023, 13, 2975. [Google Scholar] [CrossRef]
- Zhao, F.; Li, Y.; Cui, T.; Bai, J. GGE Biplot-Based Transcriptional Analysis of 7 Genes Involved in Steroidal Glycoalkaloid Biosynthesis in Potato (Solanum tuberosum L.). Agronomy 2023, 13, 2127. [Google Scholar] [CrossRef]
- Valenzuela-Cobos, J.D.; Guevara-Viejó, F.; Vicente-Galindo, P.; Galindo-Villardón, P. Eco-Friendly Biocontrol of Moniliasis in Ecuadorian Cocoa Using Biplot Techniques. Sustainability 2023, 15, 4223. [Google Scholar] [CrossRef]
- Mehdi, F.; Cao, Z.; Zhang, S.; Gan, Y.; Cai, W.; Peng, L.; Wu, Y.; Wang, W.; Yang, B. Factors affecting the production of sugarcane yield and sucrose accumulation: Suggested potential biological solutions. Front. Plant Sci. 2024, 15, 1374228. [Google Scholar] [CrossRef]
- Ellis, R.N.; Basford, K.E.; Cooper, M.; Leslie, J.K.; Byth, D.E. A methodology for analysis of sugarcane productivity trends. I. Analysis across districts. Aust. J. Agric. Res. 2001, 52, 1001–1009. [Google Scholar] [CrossRef]
- Zhang, H.; Feng, Z.; Wang, J.; Yun, X.; Qu, F.; Sun, C.; Wang, Q. Genotype by environment interaction for grain yield in foxtail millet (Setaria italica) using AMMI model and GGE Biplot. Plant Growth Regul. 2022, 99, 101–121. [Google Scholar] [CrossRef]
- Yan, W.; Kang, M.S. GGE Biplot Analysis: A Graphical Tool for Breeders, Geneticists, and Agronomists. GGE Biplot Analysis [Internet]. 28 August 2002. Available online: https://www.taylorfrancis.com/books/mono/10.1201/9781420040371/gge-biplot-analysis-weikai-yan-manjit-kang (accessed on 9 July 2024).
- Yan, W.; Tinker, N.A. Biplot analysis of multi-environment trial data: Principles and applications. Can. J. Plant Sci. 2006, 86, 623–645. [Google Scholar] [CrossRef]
- Klomsa-ard, P.; Jaisil, P.; Patanothai, A. Performance and Stability for Yield and Component Traits of Elite Sugarcane Genotypes Across Production Environments in Thailand. Sugar Tech 2013, 15, 354–364. [Google Scholar] [CrossRef]
- Reddy, P.S.; Reddy, B.V.S.; Rao, P.S. Genotype by sowing date interaction effects on sugar yield components in sweet sorghum (Sorghum bicolor L. moench). SABRAO J. Breed. Genet. 2014, 46, 305–312. [Google Scholar]
- Shojaei, S.H.; Mostafavi, K.; Ghasemi, S.H.; Bihamta, M.R.; Illés, Á.; Bojtor, C.; Nagy, J.; Harsányi, E.; Vad, A.; Széles, A.; et al. Sustainability on Different Canola (Brassica napus L.) Cultivars by GGE Biplot Graphical Technique in Multi-Environment. Sustainability 2023, 15, 8945. [Google Scholar] [CrossRef]
- Orbe, D.; Cuichán, M. Encuesta de Superficie y Producción Agropecuaria Continua; INEC (Instituto Nacional de Estadística y Censos): Loja, Ecuador, 2022; pp. 2–14. [Google Scholar]
- de Guenni, L.B.; García, M.; Muñoz, G.; Santos, J.L.; Cedeño, A.; Perugachi, C.; Castillo, J. Predicting monthly precipitation along coastal Ecuador: ENSO and transfer function models. Theor. Appl. Climatol. 2017, 129, 1059–1073. [Google Scholar] [CrossRef]
- Silva, E.; Madrid, C.; Martínez, F.; León, T. Programa de variedades. In Centro de Investigación de la Caña de Azúcar (CINCAE), Informe Anual 2021; El Triunfo, Ecuador, 2022; pp. 2–10. Available online: https://cincae.org/wp-content/uploads/2022/06/Informe-Anual-2021.pdf (accessed on 2 June 2024).
- Melios, S.; Ninou, E.; Irakli, M.; Tsivelika, N.; Sistanis, I.; Papathanasiou, F.; Didos, S.; Zinoviadou, K.; Karantonis, H.C.; Argiriou, A.; et al. Effect of Genotype, Environment, and Their Interaction on the Antioxidant Properties of Durum Wheat: Impact of Nitrogen Fertilization and Sowing Time. Agriculture 2024, 14, 328. [Google Scholar] [CrossRef]
- Bucarám Ortíz, J.; Garcia, Y.; Vega, A.; Amador Sacoto, C.; Borodulina, T.; Mancero-Castillo, D. Dynamics of co2 emission and dispersion from two sugarcane mills in the Guayas basin. Chilean J. Agric. Anim. Sci. 2023, 39, 296–304. [Google Scholar] [CrossRef]
- Amador-Sacoto, C.; Helfgott-Lerner, S. Sustainability of Sugarcane Farms in the Milagro Canton, Ecuador. Int. J. Adv. Sci. Eng. Inf. Technol. 2023, 13, 837–843. [Google Scholar] [CrossRef]


| Sector | Sugarcane Farmer | Year | Hectares | Total Harvested Material | Average Yields (Ton/Ha) | Enviroment Code |
|---|---|---|---|---|---|---|
| 01 | Ingenio | 2018 | 15.88 | 1141.15 | 71.85 | A |
| 2019 | 16.54 | 1300.73 | 78.62 | B | ||
| 2020 | 16.15 | 1268.12 | 78.53 | C | ||
| 02 | Ingenio and Isabel maria | 2018 | 15.23 | 1130.96 | 73.43 | D |
| 2019 | 16.39 | 1407.28 | 85.93 | E | ||
| 2020 | 16.42 | 1398.54 | 85.22 | F | ||
| 03 | Ingenio | 2018 | 17.36 | 1312.59 | 75.6 | G |
| 2019 | 17.33 | 1666.7 | 96.19 | H | ||
| 2020 | 18.18 | 1773.76 | 97.58 | I | ||
| 04 | Ingenio | 2018 | 17.08 | 1059.5 | 62.03 | J |
| 2019 | 16.76 | 1246.57 | 74.39 | K | ||
| 2020 | 16.48 | 1244.68 | 75.55 | L | ||
| 05 | Ingenio | 2018 | 16.25 | 1048.75 | 64.55 | M |
| 2019 | 16.17 | 1265.4 | 78.25 | N | ||
| 2020 | 16.63 | 1309.51 | 78.77 | O |
| Average Total Yield (t. ha−1) | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotypes | Sector 01 | Sector 02 | Sector 03 | Sector 04 | Sector 05 | ||||||||||
| 2018 | 2019 | 2020 | 2018 | 2019 | 2020 | 2018 | 2019 | 2020 | 2018 | 2019 | 2020 | 2018 | 2019 | 2020 | |
| CC-8592 | 70.45 | 69.22 | 72.71 | 57.85 | 63.34 | 77.1 | 83.68 | 92.09 | 72.42 | 79.14 | 79.52 | 84.64 | 96.63 | 75.83 | 78.51 |
| CC93-4181 | 66.97 | 60.62 | 66.69 | 71.91 | 59.03 | 65.25 | 78.71 | 91.52 | 73.66 | 54.41 | 64.28 | 69.98 | 78.74 | 67.14 | 72.01 |
| CLONES | 63.69 | 88.24 | 111.68 | 77.85 | 102.28 | 72.99 | 97 | 107.61 | 72.95 | 88.94 | 59.08 | 101.45 | 116.37 | 72.81 | 75.53 |
| CR-74250 | 70.03 | 64.76 | 74.85 | 65.75 | 61.79 | 72.06 | 81.56 | 95.09 | 78.37 | 72.76 | 64.98 | 74.68 | 88.36 | 76.24 | 70.86 |
| EC-02 | 70.95 | 67.81 | 91.03 | 53.39 | 60.01 | 86.95 | 88.09 | 103.06 | 81.79 | 86.95 | 81.75 | 91.55 | 106.18 | 82.44 | 85.62 |
| EC-03 | 58.14 | 56.02 | 64.13 | 60.57 | 62.72 | 59.09 | 71.96 | 87.97 | 59.94 | 67.88 | 67.9 | 69.28 | 89.98 | 48.56 | 64.67 |
| EC-05 | 86.66 | 60.73 | 74.19 | 57.68 | 61.76 | 72.56 | 77.64 | 83.56 | 81.93 | 71.47 | 72.17 | 70.92 | 84.35 | 49.74 | 50.85 |
| EC-07 | 87.77 | 47.94 | 97.28 | 75.77 | 72.03 | 83.32 | 99.32 | 109.72 | 81.68 | 82.09 | 83.82 | 93.21 | 106.31 | 74.91 | 85.74 |
| EC-08 | 77.63 | 78.32 | 84.42 | 75.72 | 75.14 | 78.92 | 82.3 | 102.06 | 78 | 77.05 | 85.9 | 90.51 | 99.36 | 73.17 | 79.82 |
| ECU-01 | 77.42 | 70.06 | 73.36 | 67.94 | 71.13 | 84.28 | 92.24 | 95.3 | 84.83 | 85.79 | 75.58 | 87.62 | 91.37 | 80.87 | 82.89 |
| Source | Degrees of Freedom (Df) | Sum of Square (SS) | Mean of Square (MS) | Proportion in the Total SS (%SS) | F-Value (F) |
|---|---|---|---|---|---|
| G | 9 | 82,664 | 9185 | 7.96 | 30.65 *** |
| Sector | 4 | 77,022 | 19,256 | 7.42 | 64.26 *** |
| Year | 2 | 102,203 | 51,101 | 9.84 | 170.53 *** |
| G × Sector | 36 | 19,543 | 543 | 1.88 | 1.81 ** |
| G × Year | 18 | 10,569 | 587 | 1.02 | 1.96 ** |
| Sector × Year | 8 | 11,363 | 1420 | 1.10 | 4.74 *** |
| G × Sector × Year | 72 | 14,212 | 197 | 1.37 | 0.66 (ns) |
| Location | Genotypes | G SS% | Year | Y SS% | G × Year | G × Y SS% | Residuals | R SS% |
|---|---|---|---|---|---|---|---|---|
| Sector 01 | 12,562 *** | 9.42 | 3225 ** | 2.41 | 4102 (ns) | 3.08 | 113,475 | 85.08 |
| Sector 02 | 28,260 *** | 10.17 | 39,638 *** | 14.27 | 4910 (ns) | 1.77 | 205,058 | 73.8 |
| Sector 03 | 26,614 *** | 10.75 | 36,735 *** | 14.84 | 4046 (ns) | 1.63 | 180,116 | 72.77 |
| Sector 04 | 10,265 *** | 7.29 | 16,017 *** | 11.38 | 7614 * | 5.41 | 106,797 | 75.91 |
| Sector 05 | 14,688 *** | 9.84 | 14,670 *** | 9.83 | 4610 (ns) | 3.08 | 115,227 | 77.23 |
| Source of Variation | Yield (t. ha−1) | SD | Rank by Grand Mean | Rank by GGE |
|---|---|---|---|---|
| Genotypes | ||||
| CC-8592 | 76.54 ± 0.511 c | 19.63 | 6 | 4 |
| CC93-4181 | 68.67 ± 1.845 d,e | 17.58 | 9 | 6 |
| CLONES | 92.64 ± 2.334 a | 22.4 | 1 | 5 |
| CR-74250 | 72.52 ± 1.065 d | 16.13 | 7 | 5 |
| EC-02 | 85.27 ± 1.301 a,b | 23.83 | 2 | 2 |
| EC-03 | 63.98 ± 1.636 e | 15.06 | 10 | 7 |
| EC-05 | 72.63 ± 2.026 c,d | 16.13 | 8 | 6 |
| EC-07 | 86.1 ± 1.912 a,b | 20.18 | 3 | 1 |
| EC-08 | 83.29 ± 1.697 b | 19.47 | 4 | 3 |
| ECU-01 | 82.95 ± 0.813 b | 19.72 | 5 | 3 |
| Sector | ||||
| sector 01 | 75.17 ± 0.813 c | 17.17 | 3 | 3 |
| sector 02 | 79.84 ± 0.647 b | 19.71 | 2 | 2 |
| sector 03 | 88.44 ± 0.799 a | 22.99 | 1 | 1 |
| sector 04 | 70.77 ± 0.81 c,d | 17.56 | 5 | 3 |
| sector 05 | 73.68 ± 0.807 d | 18.03 | 4 | 2 |
| Years | ||||
| 2018 | 68.45 ± 0.58 b | 18.42 | 3 | 3 |
| 2019 | 82.31 ± 0.58 a | 18.95 | 2 | 2 |
| 2020 | 83.61 ± 0.624 a | 19.52 | 1 | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Torres-Ordoñez, L.H.; Valenzuela-Cobos, J.D.; Guevara-Viejó, F.; Galindo-Villardón, P.; Vicente-Galindo, P. Effect of Genotype × Environment Interactions on the Yield and Stability of Sugarcane Varieties in Ecuador: GGE Biplot Analysis by Location and Year. Appl. Sci. 2024, 14, 6665. https://doi.org/10.3390/app14156665
Torres-Ordoñez LH, Valenzuela-Cobos JD, Guevara-Viejó F, Galindo-Villardón P, Vicente-Galindo P. Effect of Genotype × Environment Interactions on the Yield and Stability of Sugarcane Varieties in Ecuador: GGE Biplot Analysis by Location and Year. Applied Sciences. 2024; 14(15):6665. https://doi.org/10.3390/app14156665
Chicago/Turabian StyleTorres-Ordoñez, Luis Henry, Juan Diego Valenzuela-Cobos, Fabricio Guevara-Viejó, Purificación Galindo-Villardón, and Purificación Vicente-Galindo. 2024. "Effect of Genotype × Environment Interactions on the Yield and Stability of Sugarcane Varieties in Ecuador: GGE Biplot Analysis by Location and Year" Applied Sciences 14, no. 15: 6665. https://doi.org/10.3390/app14156665







