Machine Learning and Image Processing-Based System for Identifying Mushrooms Species in Malaysia
Abstract
:1. Introduction
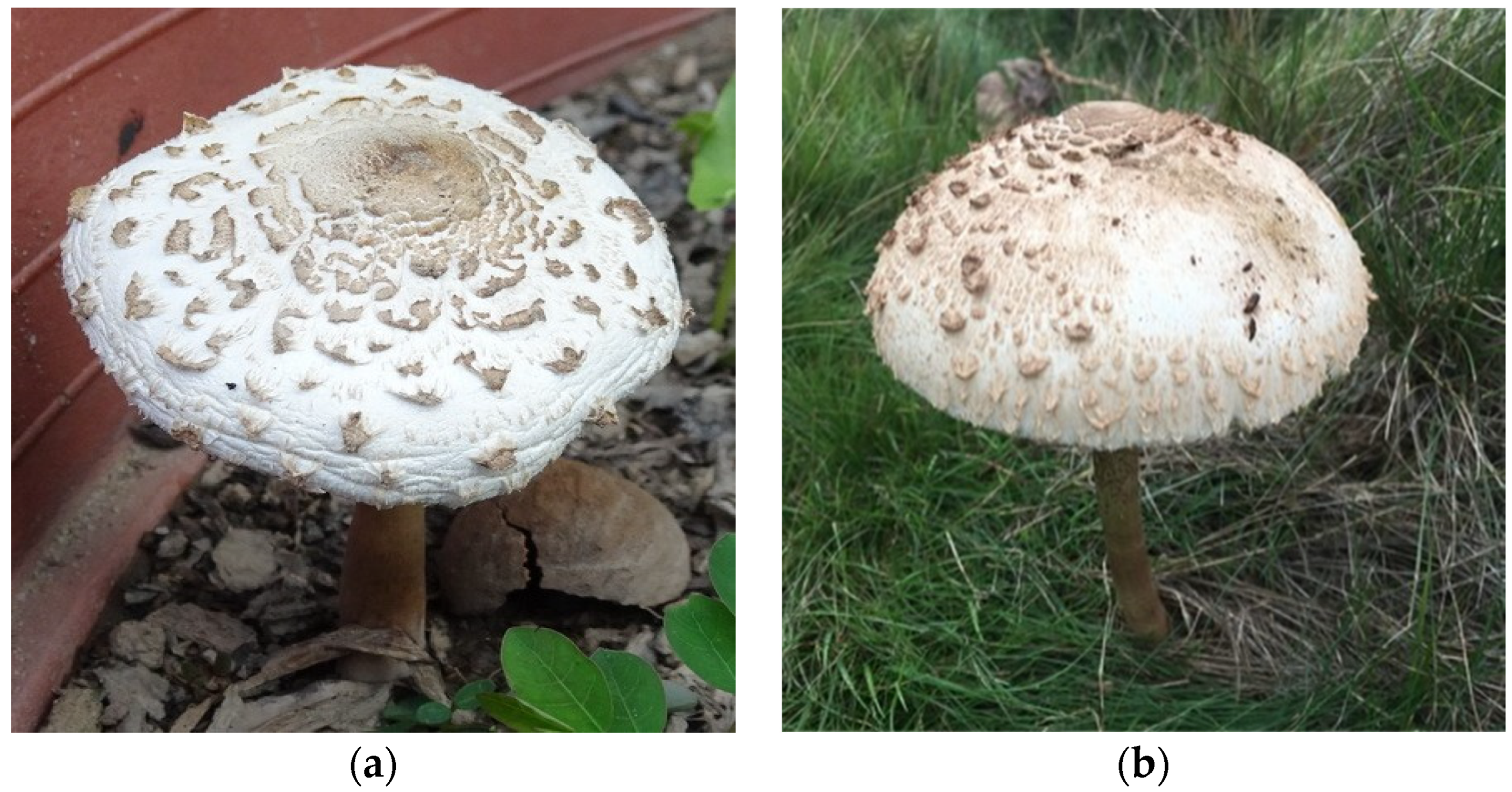
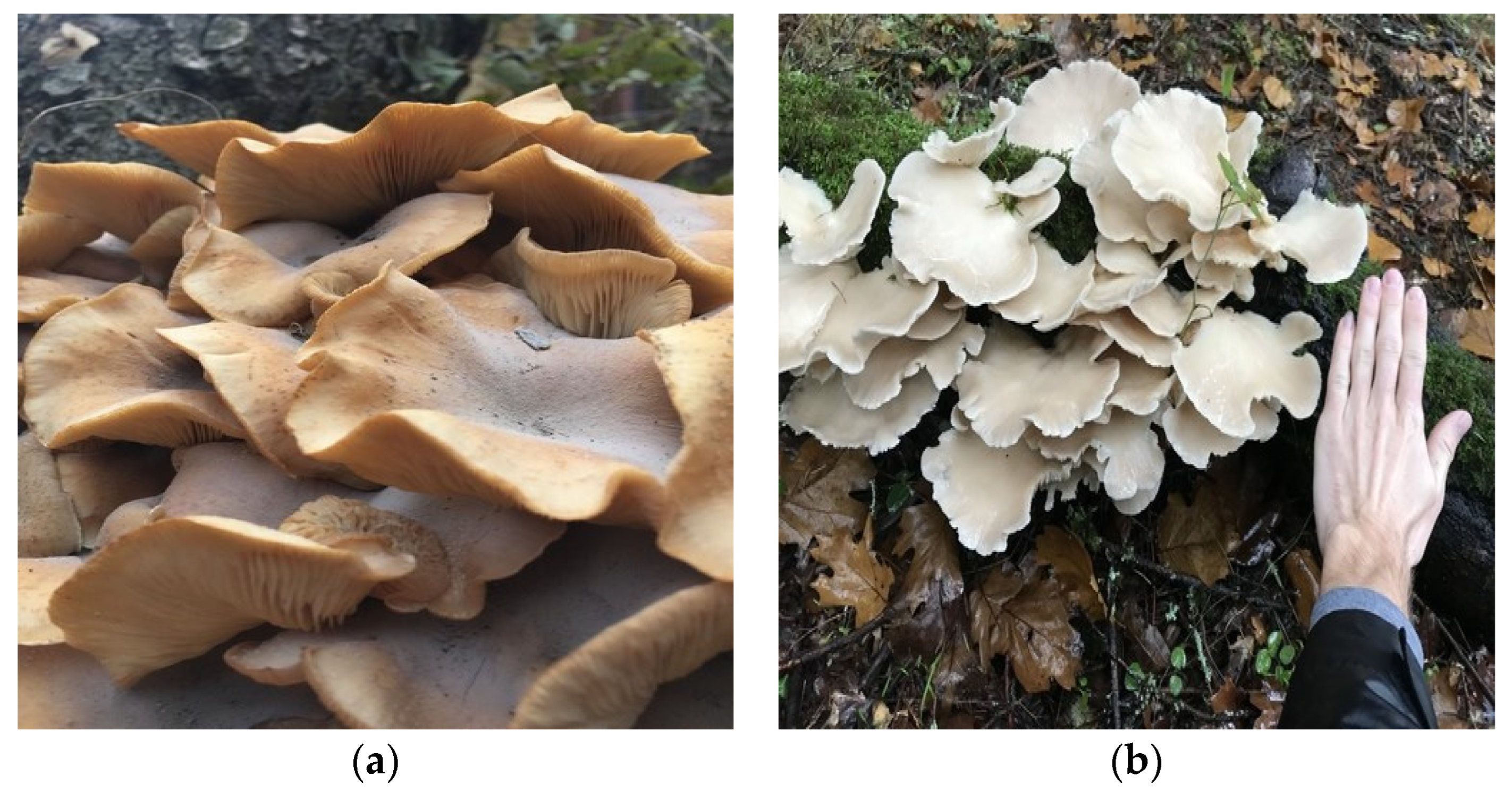
2. Dataset
3. Methods
3.1. Computer Vision for Image Classification
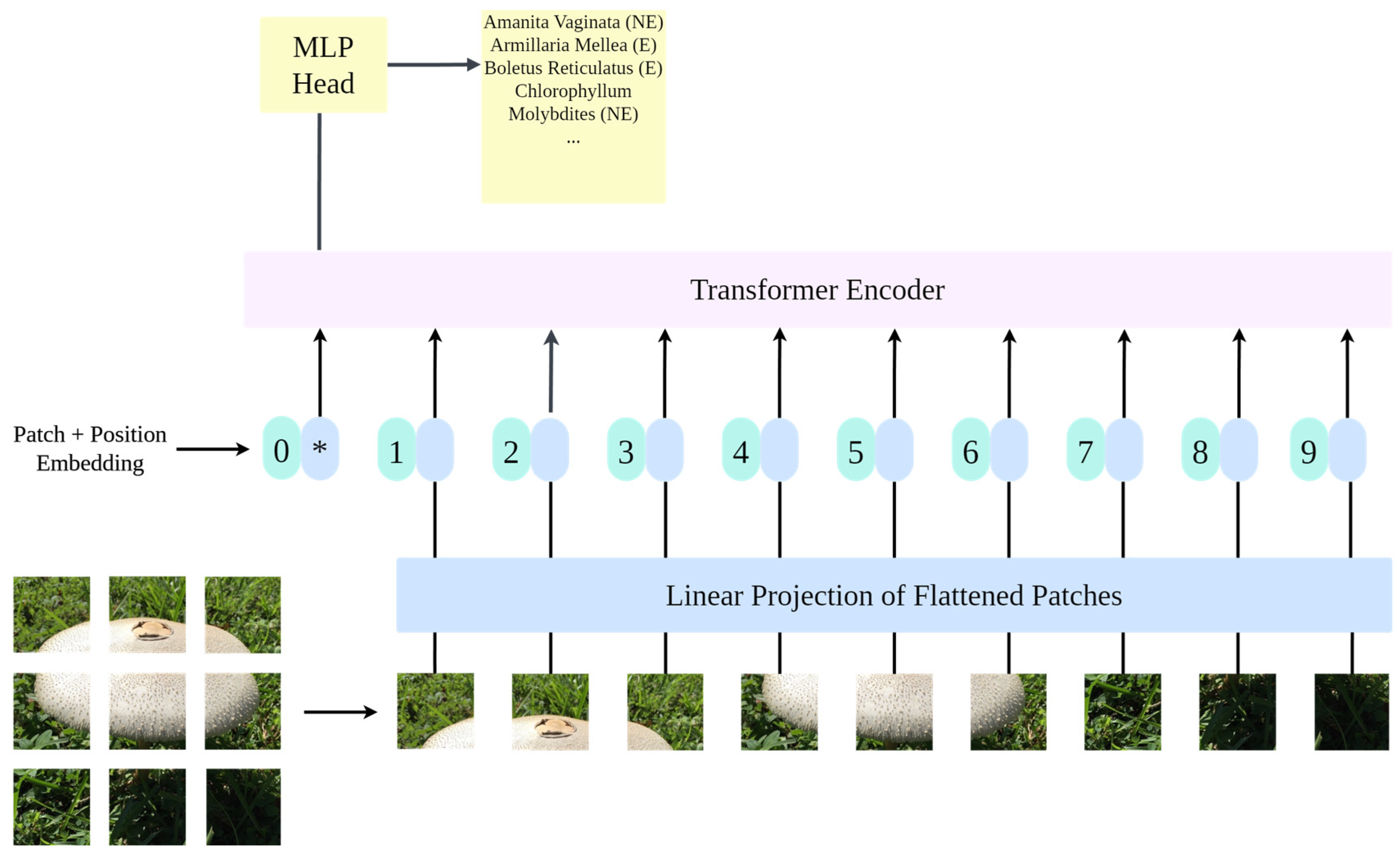
3.2. Vision Transformer
3.3. ResNet
4. Experiment
4.1. Experiment Settings
4.2. Data Augmentation
4.3. Performance Metrics
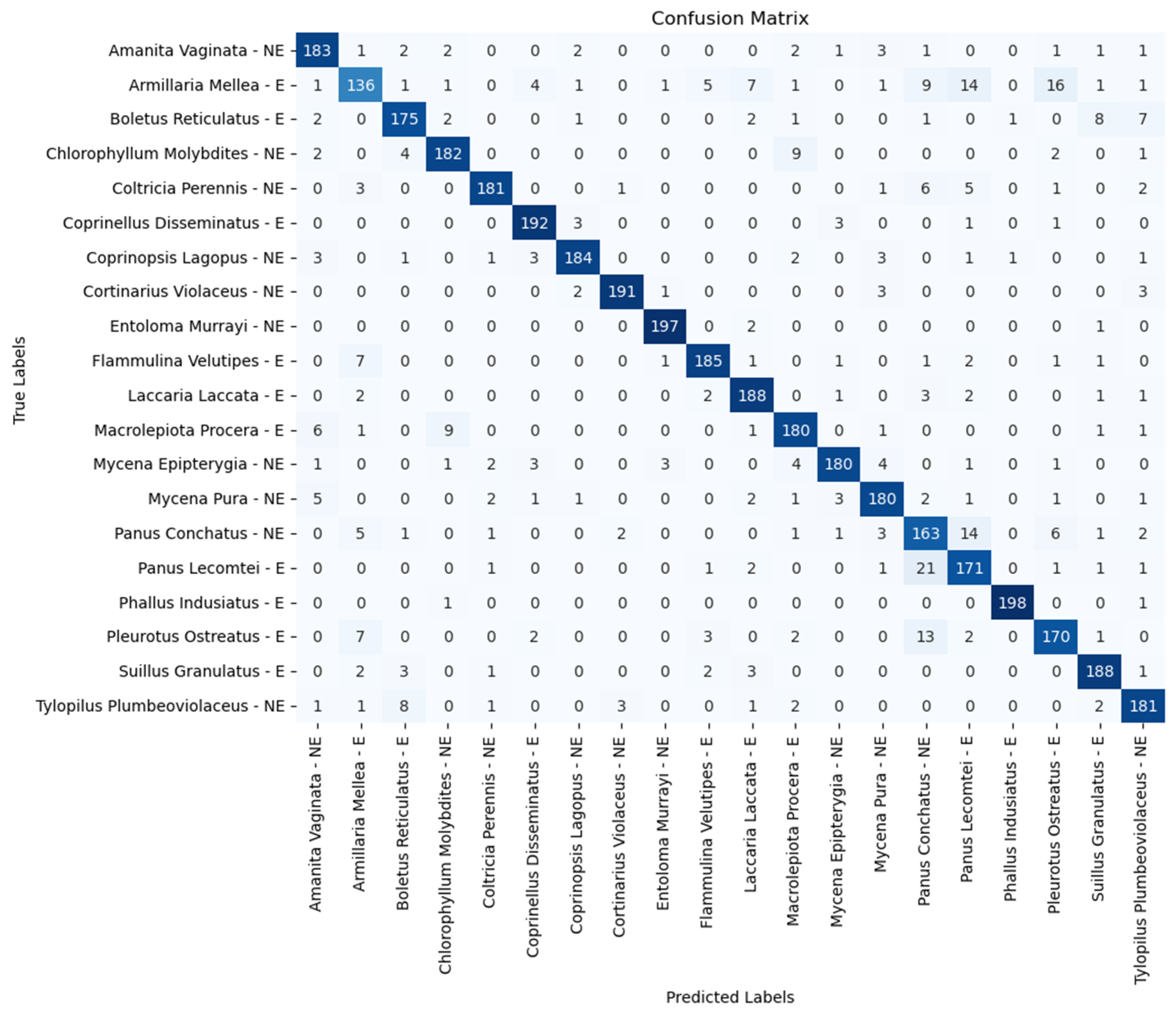
5. Results
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Roslan, F. Two Mushroom Poisoning Deaths Recorded in Sabah—Professor. The Borneo Post. 12 July 2018. Available online: https://www.theborneopost.com/2018/07/12/two-mushroom-poisoning-deaths-recorded-in-sabah-professor/ (accessed on 16 July 2024).
- O’Malley, G.F.; O’Malley, R. Mushroom Poisoning. MSD Manual Professional Edition. 13 June 2022. Available online: https://www.msdmanuals.com/professional/injuries-poisoning/poisoning/mushroom-poisoning (accessed on 16 July 2024).
- Hedman, H.; Holmdahl, J.; Mölne, J.; Ebefors, K.; Haraldsson, B.; Nyström, J. Long-term clinical outcome for patients poisoned by the fungal nephrotoxin orellanine. BMC Nephrol. 2017, 18, 121. [Google Scholar] [CrossRef] [PubMed]
- Anantharam, P.; Shao, D.; Imerman, P.M.; Burrough, E.; Schrunk, D.; Sedkhuu, T.; Tang, S.; Rumbeiha, W. Improved Tissue-Based analytical test methods for orellanine, a biomarker of cortinarius mushroom intoxication. Toxins 2016, 8, 158. [Google Scholar] [CrossRef] [PubMed]
- Wild Mushroom Poisoning—Fact Sheets. Available online: https://www.health.nsw.gov.au/environment/factsheets/Pages/wild-mushroom-poisoning.aspx (accessed on 16 July 2024).
- Leong, A. Family Ends in Hospital after Ingesting Poisonous Mushrooms. TRP. 15 July 2023. Available online: https://www.therakyatpost.com/news/2023/07/15/watch-family-ends-in-hospital-after-ingesting-poisonous-mushrooms/ (accessed on 16 July 2024).
- Chlorophyllum molybdites—Mushroom World. Available online: https://www.mushroom.world/show?n=Chlorophyllum-molybdites (accessed on 16 July 2024).
- Hamonangan, R.; Bagus, M.; Bagus, C.; Dinata, S.; Atmaja, K. Accuracy of classification poisonous or edible of mushroom using naïve bayes and k-nearest neighbors. J. Soft Comput. Explor. 2021, 2, 53–60. [Google Scholar]
- Ottom, M. Classification of mushroom fungi using machine learning techniques. Int. J. Adv. Trends Comput. Sci. Eng. 2019, 8, 2378–2385. [Google Scholar] [CrossRef]
- Rimi, I.F.; Habib, M.T.; Supriya, S.; Khan, M.A.A.; Hossain, S.A. Traditional machine learning and deep learning modeling for legume species recognition. SN Comput. Sci. 2022, 3, 430. [Google Scholar] [CrossRef]
- Wibowo, A.; Rahayu, Y.; Riyanto, A.; Hidayatulloh, T. Classification algorithm for edible mushroom identification. In Proceedings of the 2018 International Conference on Information and Communications Technology (ICOIACT), Yogyakarta, Indonesia, 6–7 March 2018. [Google Scholar]
- Demirel, Y.A.; Demirel, G. Deep learning based approach for classification of mushrooms. Gazi Univ. J. Sci. Part A Eng. Innov. 2023, 10, 487–498. [Google Scholar] [CrossRef]
- Zahan, N.; Hasan, M.Z.; Malek, M.A.; Reya, S.S. A Deep Learning-Based Approach for Edible, Inedible and Poisonous Mushroom Classification. In Proceedings of the 2021 International Conference on Information and Communication Technology for Sustainable Development (ICICT4SD), Dhaka, Bangladesh, 27–28 February 2021. [Google Scholar]
- INaturalist. (n.d.) Available online: https://www.inaturalist.org/ (accessed on 16 July 2024).
- Lee, S.S.; Alias, S.A.; Jones, E.G.B.; Zainuddin, N.; Chan, H.T. Checklist of Fungi of Malaysia. J. Trop. For. Sci. 2023, 25, 442. [Google Scholar]
- Help iNaturalist. iNaturalist. Available online: https://help.inaturalist.org/en/support/home (accessed on 16 July 2024).
- Gao, X.; Qian, Y.; Gao, A. COVID-VIT: Classification of COVID-19 from CT chest images based on vision transformer models. arXiv 2021, arXiv:2107.01682. [Google Scholar]
- Testagrose, C.; Shabbir, M.; Weaver, B.; Liu, X. Comparative study between Vision Transformer and EfficientNet on Marsh grass classification. In Proceedings of the International Florida Artificial Intelligence Research Society Conference, Clearwater Beach, FL, USA, 14–17 May 2023. [Google Scholar]
- Isiaka, F. Performance metrics of an intrusion detection system through Window-Based Deep Learning models. J. Data Sci. Intell. Syst. 2023, 2, 174–180. [Google Scholar] [CrossRef]
- Shang, L.; Zhang, Z.; Tang, F.; Cao, Q.; Pan, H.; Lin, Z. Signal process of ultrasonic guided wave for damage detection of localized defects in plates: From shallow learning to deep learning. J. Data Sci. Intell. Syst. 2023. [Google Scholar] [CrossRef]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An Image is Worth 16x16 Words: Transformers for Image Recognition at Scale. arXiv 2021, arXiv:2010.11929. [Google Scholar]
- Devlin, J.; Chang, M.; Lee, K.; Toutanova, K. BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding. arXiv 2019, arXiv:1810.04805. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar]
- Zhuang, F.; Qi, Z.; Duan, K.; Xi, D.; Zhu, Y.; Zhu, H.; Xiong, H.; He, Q. A Comprehensive Survey on Transfer Learning. Proc. IEEE 2021, 109, 43–76. [Google Scholar] [CrossRef]





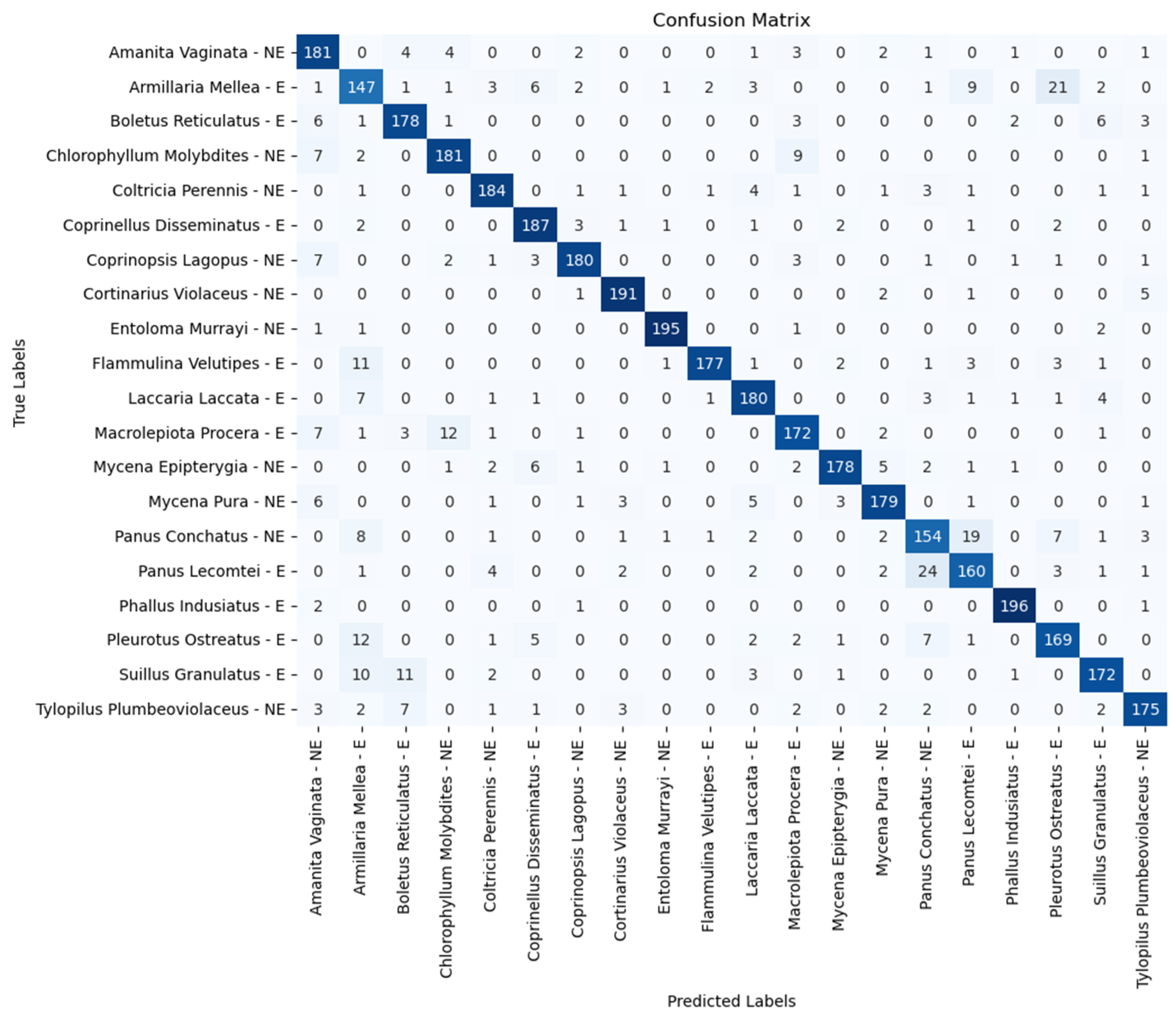
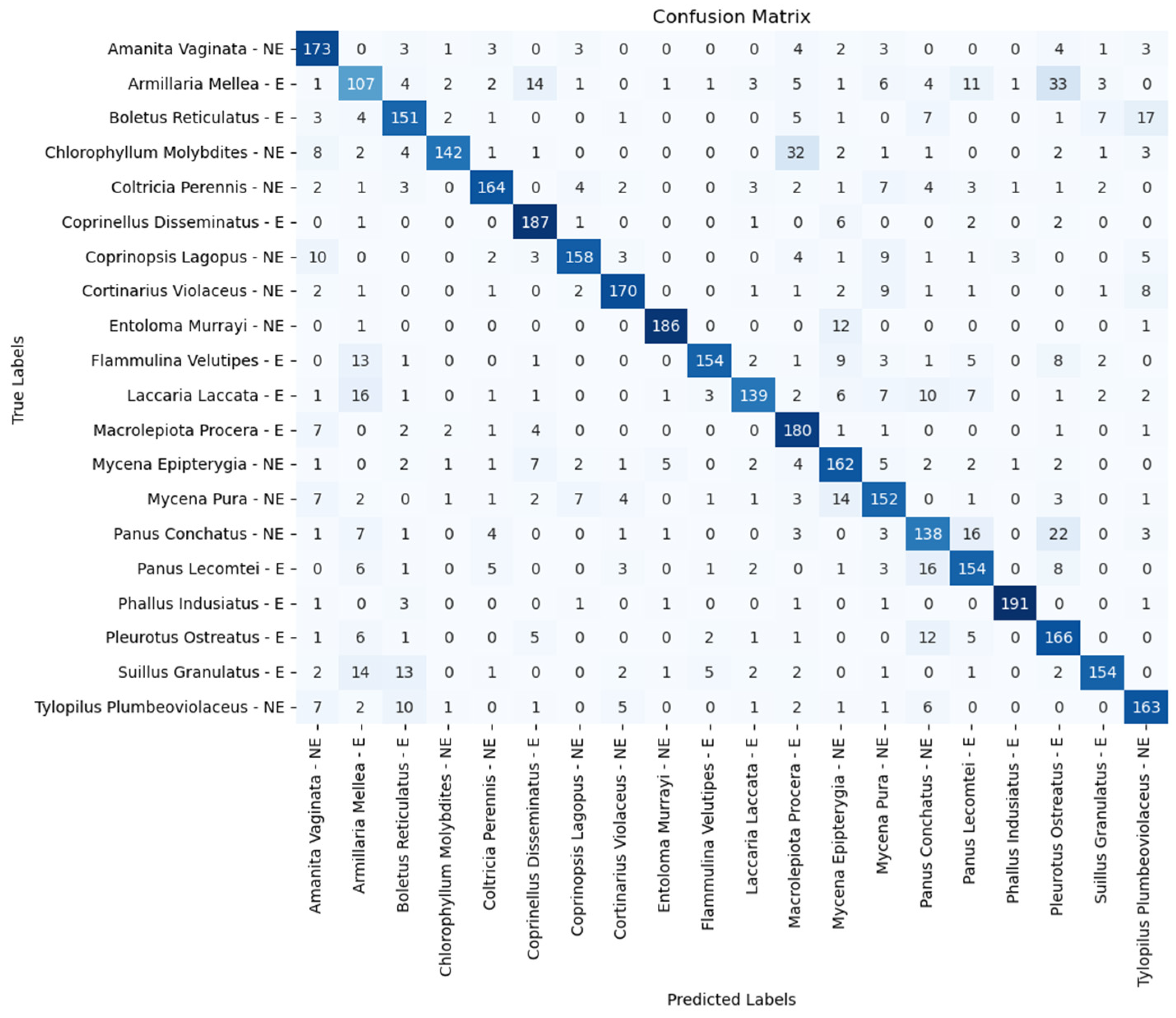


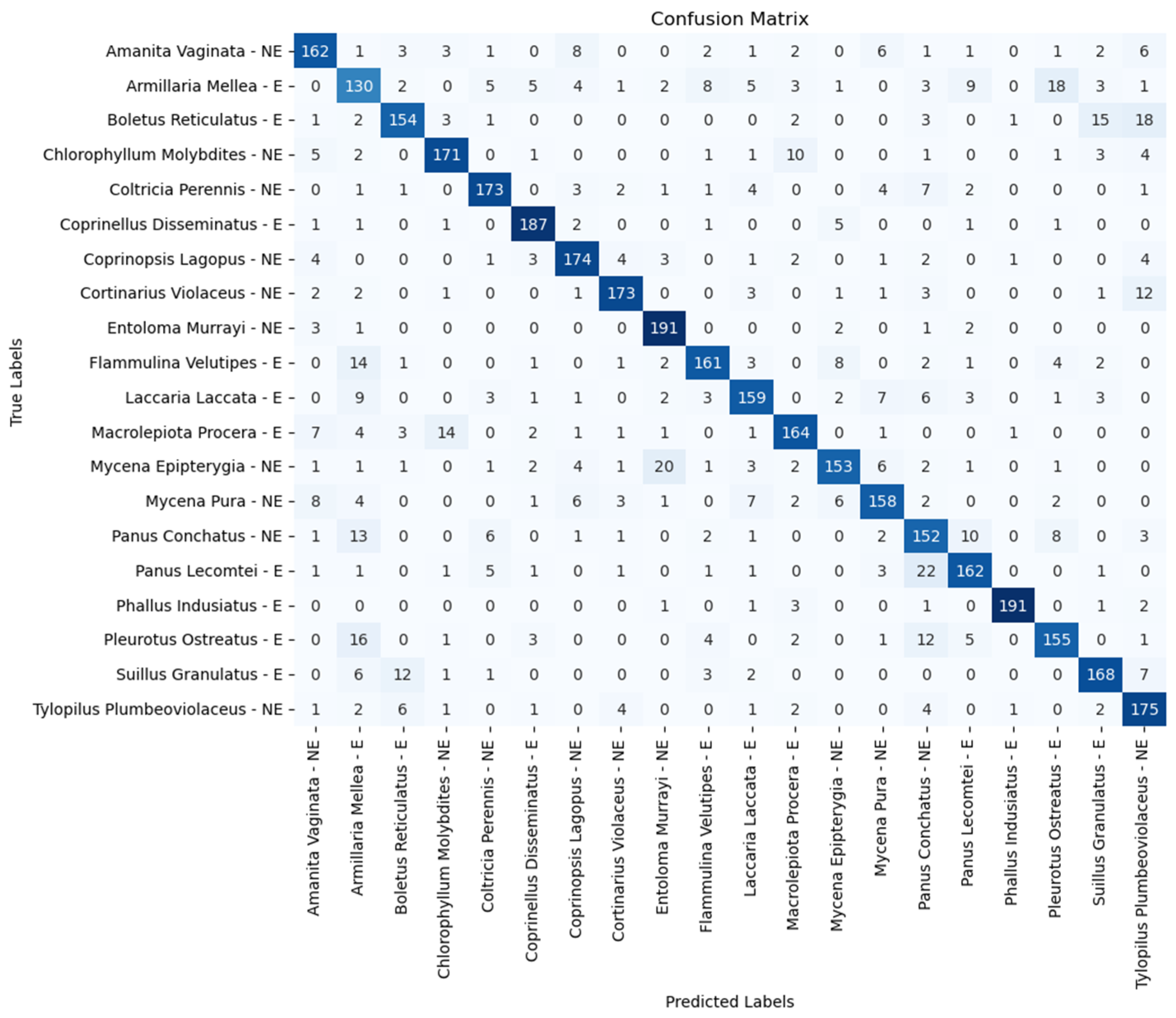
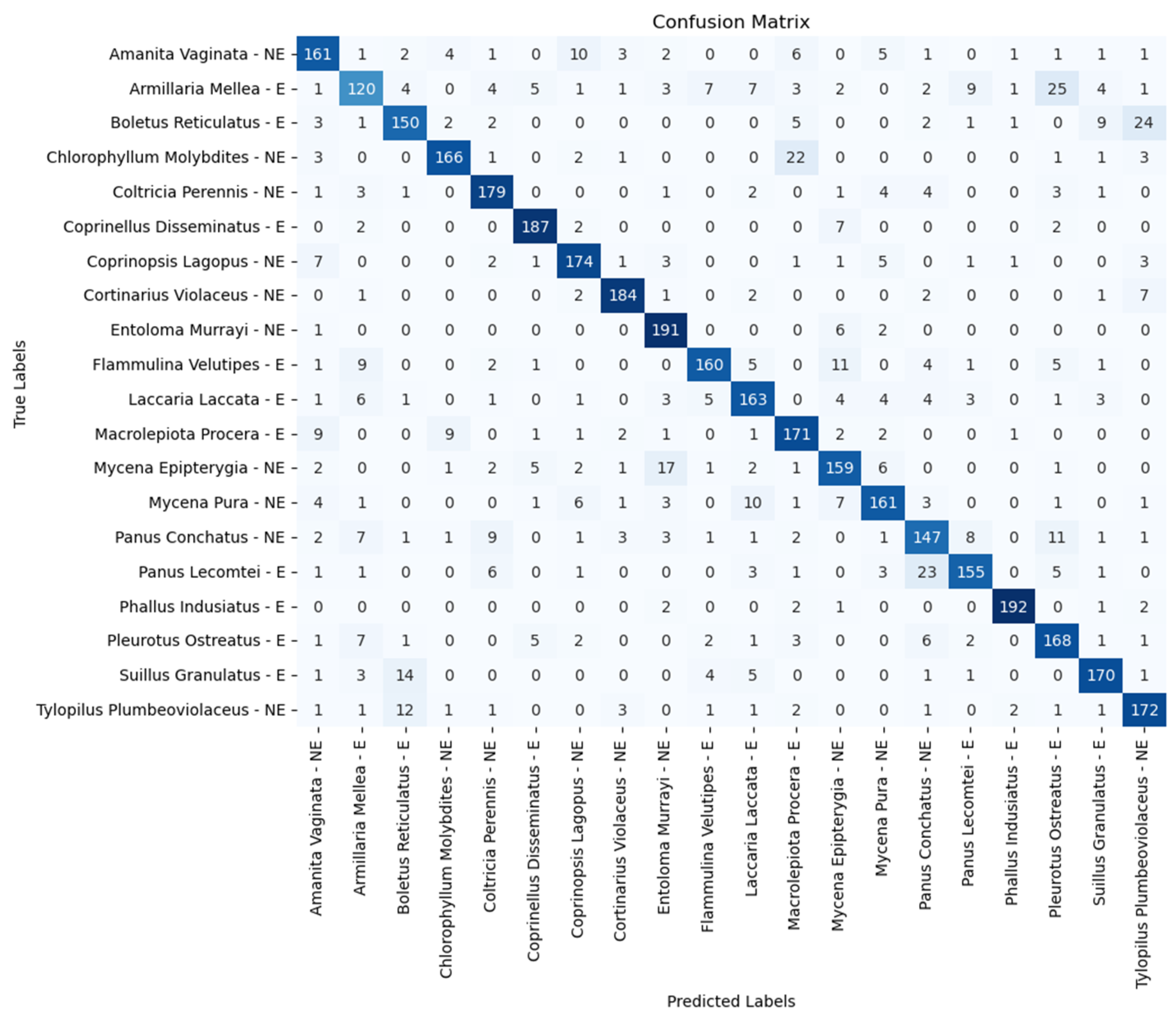
| Model | Accuracy (%) | Precision (%) | Recall (%) | F1 Score (%) |
|---|---|---|---|---|
| ViT-B/16 | 90.12 | 90.20 | 90.12 | 90.10 |
| ViT-B/32 | 85.87 | 85.98 | 85.87 | 85.88 |
| ViT-L/16 | 90.47 | 90.52 | 90.47 | 90.46 |
| ViT-L/32 | 88.40 | 88.52 | 88.40 | 88.42 |
| ResNet-18 | 79.77 | 80.60 | 79.77 | 79.82 |
| ResNet-34 | 81.67 | 82.08 | 81.67 | 81.63 |
| Resnet-50 | 84.00 | 84.00 | 84.00 | 83.95 |
| ResNet-101 | 82.82 | 83.07 | 82.82 | 82.85 |
| ResNet-152 | 83.25 | 83.30 | 83.25 | 83.15 |
| Mushroom Species | Accuracy (%) | |||
|---|---|---|---|---|
| ViT-B/16 | ViT-B/32 | ViT-L/16 | ViT-L/32 | |
| Amanita vaginata—E | 91.51 | 86.00 | 88.50 | 90.50 |
| Armillaria mellea—E | 68.00 | 68.00 | 74.50 | 73.50 |
| Boletus reticulatus—E | 87.50 | 84.00 | 89.50 | 89.00 |
| Chlorophyllum molybdites—NE | 91.00 | 85.00 | 89.50 | 90.50 |
| Coltricia perennis—NE | 90.50 | 87.00 | 93.00 | 92.00 |
| Coprinellus disseminatus—E | 96.00 | 92.00 | 97.50 | 93.50 |
| Coprinopsis lagopus—NE | 92.00 | 91.50 | 92.50 | 90.00 |
| Cortinarius violaceus—NE | 95.50 | 90.50 | 95.00 | 95.50 |
| Entoloma murrayi—NE | 98.50 | 96.50 | 97.50 | 97.50 |
| Flammulina velutipes—E | 92.50 | 84.50 | 90.50 | 88.50 |
| Laccaria laccata—E | 94.00 | 85.00 | 93.50 | 90.00 |
| Macrolepiota procera—E | 90.00 | 89.00 | 88.50 | 86.00 |
| Mycena epipterygia—NE | 90.00 | 89.00 | 92.50 | 89.00 |
| Mycena pura—NE | 90.00 | 85.00 | 95.00 | 89.50 |
| Panus conchatus—NE | 81.50 | 76.00 | 78.00 | 77.00 |
| Panus lecomtei—E | 85.50 | 78.50 | 87.50 | 80.00 |
| Phallus indusiatus—E | 99.00 | 97.50 | 99.50 | 98.00 |
| Pleurotus ostreatus—E | 85.00 | 77.00 | 94.00 | 84.50 |
| Suillus granulatus—E | 94.00 | 86.00 | 93.50 | 86.00 |
| Tylopilus plumbeoviolaceus—NE | 90.50 | 89.50 | 89.50 | 87.50 |
| Mushroom Species | Accuracy (%) | ||||
|---|---|---|---|---|---|
| ResNet-18 | ResNet-34 | ResNet-50 | ResNet-101 | ResNet-152 | |
| Amanita vaginata—E | 86.50 | 82.50 | 80.50 | 81.00 | 80.50 |
| Armillaria mellea—E | 53.50 | 57.50 | 61.50 | 65.00 | 60.00 |
| Boletus reticulatus—E | 75.50 | 81.00 | 76.50 | 77.00 | 75.00 |
| Chlorophyllum molybdites—NE | 71.00 | 82.00 | 84.50 | 85.50 | 83.00 |
| Coltricia perennis—NE | 82.00 | 84.00 | 88.50 | 86.50 | 89.50 |
| Coprinellus disseminatus—E | 93.50 | 93.50 | 92.50 | 93.50 | 93.50 |
| Coprinopsis lagopus—NE | 79.00 | 84.00 | 85.00 | 87.00 | 87.00 |
| Cortinarius violaceus—NE | 85.00 | 92.50 | 89.00 | 86.50 | 92.00 |
| Entoloma murrayi—NE | 93.00 | 89.50 | 93.50 | 95.50 | 95.50 |
| Flammulina velutipes—E | 77.00 | 68.50 | 85.50 | 80.50 | 80.00 |
| Laccaria laccata—E | 69.50 | 79.50 | 80.50 | 79.50 | 81.50 |
| Macrolepiota procera—E | 90.00 | 88.00 | 88.50 | 82.00 | 85.50 |
| Mycena epipterygia—NE | 81.00 | 81.50 | 83.50 | 76.50 | 79.50 |
| Mycena pura—NE | 76.00 | 74.00 | 76.50 | 79.00 | 80.50 |
| Panus conchatus—NE | 69.00 | 70.00 | 77.00 | 76.00 | 73.50 |
| Panus lecomtei—E | 77.00 | 82.50 | 82.50 | 81.00 | 77.50 |
| Phallus indusiatus—E | 95.50 | 97.50 | 98.50 | 95.50 | 96.00 |
| Pleurotus ostreatus—E | 83.00 | 85.50 | 82.50 | 77.50 | 84.00 |
| Suillus granulatus—E | 77.00 | 80.00 | 85.00 | 84.00 | 85.00 |
| Tylopilus plumbeoviolaceus—NE | 81.50 | 80.00 | 88.50 | 87.50 | 86.00 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lim, J.Y.; Wee, Y.Y.; Wee, K. Machine Learning and Image Processing-Based System for Identifying Mushrooms Species in Malaysia. Appl. Sci. 2024, 14, 6794. https://doi.org/10.3390/app14156794
Lim JY, Wee YY, Wee K. Machine Learning and Image Processing-Based System for Identifying Mushrooms Species in Malaysia. Applied Sciences. 2024; 14(15):6794. https://doi.org/10.3390/app14156794
Chicago/Turabian StyleLim, Jia Yi, Yit Yin Wee, and KuokKwee Wee. 2024. "Machine Learning and Image Processing-Based System for Identifying Mushrooms Species in Malaysia" Applied Sciences 14, no. 15: 6794. https://doi.org/10.3390/app14156794





