A Genetic Programming Approach to Radiomic-Based Feature Construction for Survival Prediction in Non-Small Cell Lung Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. CT Dataset from NSCLC Patients
- More than one tumor exists in the volume.
- The post-operative survival time was lower than two years, but the patient remained alive.
- There are misalignments or reading errors between CT images and tumor contours.
2.2. Radiomic Feature Extraction
2.3. Proposed GP-Based Feature Construction Approach
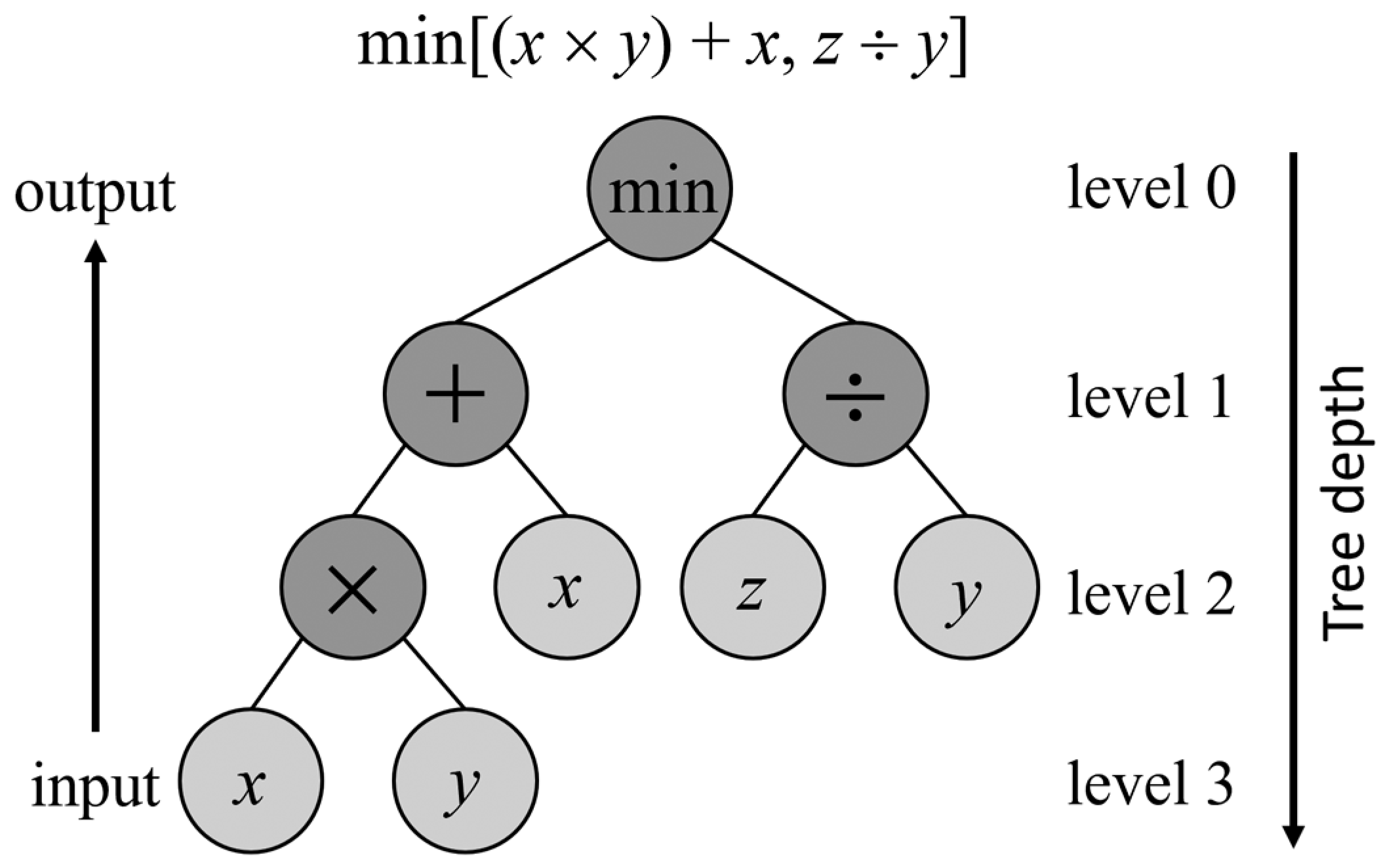
2.3.1. Genetic Programming Basics
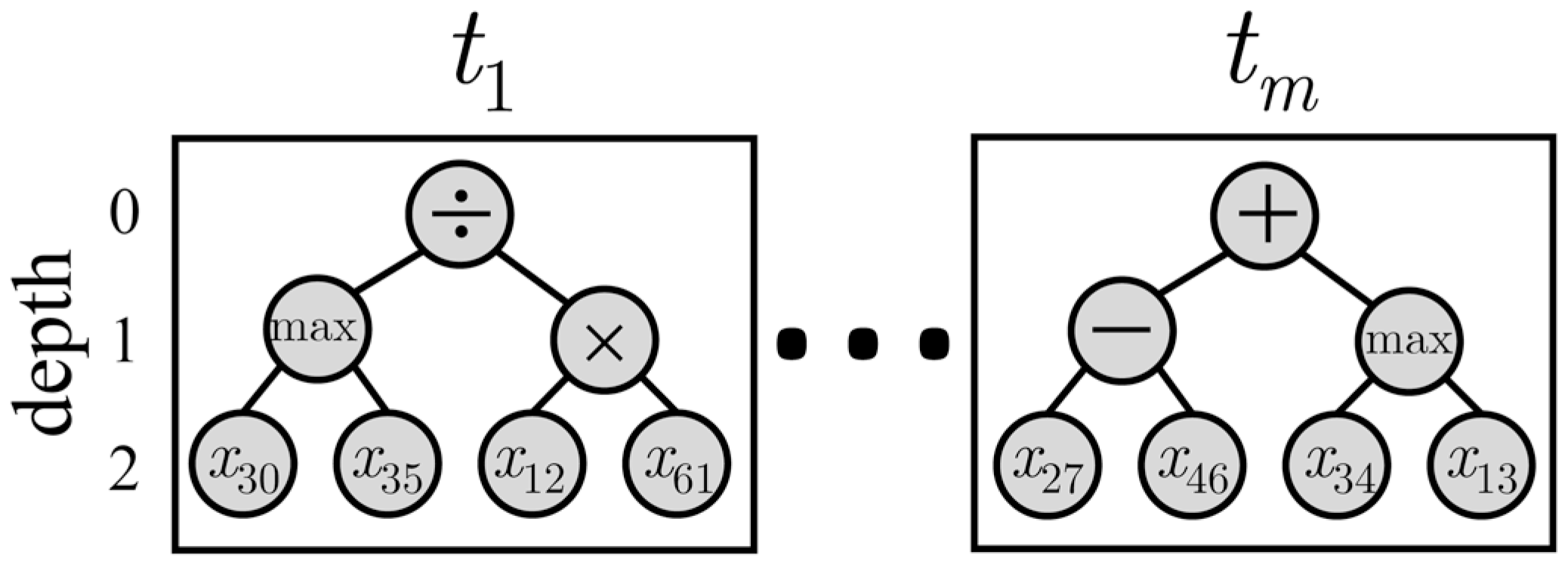
2.3.2. Multi-Tree Representation
2.3.3. Fitness Function
2.3.4. GP-Based Feature Construction Algorithm
| Algorithm 1 Feature construction using GP with multi-tree representation. |
|
| Algorithm 2 Crossover and mutation operators. |
|
2.4. Experimental Setup
2.4.1. Benchmark Techniques
- FS_FWD is a wrapper feature selection approach based on sequential forward selection, where features are sequentially added to an empty set of features until adding extra features does not improve the AUC value of a logistic regression classifier [44].
- FS_mRMR is a filter feature selection technique based on the minimal redundancy–maximal relevance (mRMR) criterion, where dependencies between variables and class labels are measured with mutual information [45].
- GP_AUC is a GP-based wrapper feature construction method in which the fitness function is the AUC value obtained from a logistic regression classifier trained with the constructed features [40].
- GP_CORR is a GP-based filter feature construction method, where the Spearman correlation measures the redundancy between constructed features, while the relevance between features and class labels is calculated in terms of AUC. The fitness function is the difference between the relevance and redundancy averages [46].
2.4.2. Experiments
3. Results
3.1. GP_GAIN Configurations
3.2. Internal Validation
3.3. External Validation
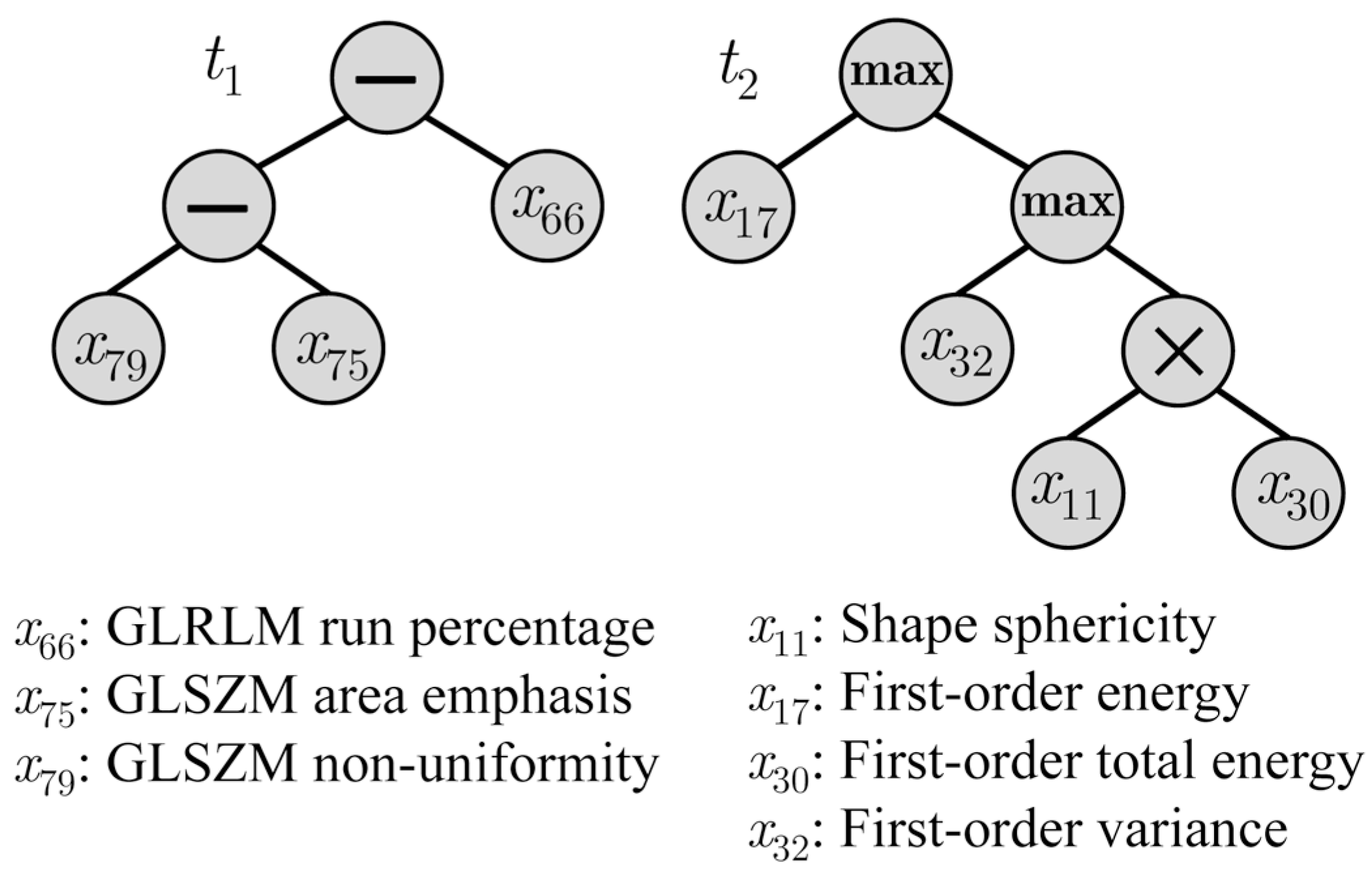
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AUC | Area under the ROC curve |
| CNN | Convolutional neural network |
| CT | Computer tomography |
| DL | Deep learning |
| GA | Genetic algorithm |
| GLCM | Gray-level co-occurrence matrix |
| GLDM | Gray-level dependence matrix |
| GLRLM | Gray-level run-length matrix |
| GLSZM | Gray-level size-zone matrix |
| GP | Genetic programming |
| LDA | Linear discriminant analysis |
| LR | Logistic regression |
| ML | Machine learning |
| MLP | Multi-layer perception |
| NB | Naive Bayes |
| NGTDM | Neighboring gray-tone difference matrix |
| NSCLC | Non-small cell lung cancer |
| RF | Random forest |
| ROI | Region of interest |
| SVM | Support vector machine |
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Vo, T.H.; Lee, G.S.; Yang, H.J.; Oh, I.J.; Kim, S.H.; Kang, S.R. Survival Prediction of Lung Cancer Using Small-Size Clinical Data with a Multiple Task Variational Autoencoder. Electronics 2021, 10, 1396. [Google Scholar] [CrossRef]
- Yang, Y.; Xu, L.; Sun, L.; Zhang, P.; Farid, S.S. Machine learning application in personalised lung cancer recurrence and survivability prediction. Comput. Struct. Biotechnol. J. 2022, 20, 1811–1820. [Google Scholar] [CrossRef] [PubMed]
- Ahsan, M.M.; Luna, S.A.; Siddique, Z. Machine-Learning-Based Disease Diagnosis: A Comprehensive Review. Healthcare 2022, 10, 541. [Google Scholar] [CrossRef] [PubMed]
- Scalco, E.; Rizzo, G.; Mastropietro, A. The stability of oncologic MRI radiomic features and the potential role of deep learning: A review. Phys. Med. Biol. 2022, 67, 09TR03. [Google Scholar] [CrossRef] [PubMed]
- Scalco, E.; Rizzo, G. Texture analysis of medical images for radiotherapy applications. Br. J. Radiol. 2017, 90, 20160642. [Google Scholar] [CrossRef] [PubMed]
- Zwanenburg, A.; Vallières, M.; Abdalah, M.A.; Aerts, H.J.; Andrearczyk, V.; Apte, A.; Ashrafinia, S.; Bakas, S.; Beukinga, R.J.; Boellaard, R.; et al. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-based Phenotyping. Radiology 2020, 295, 328–338. [Google Scholar] [CrossRef] [PubMed]
- Scrivener, M.; de Jong, E.E.; van Timmeren, J.E.; Pieters, T.; Ghaye, B.; Geets, X. Radiomics applied to lung cancer: A review. Transl. Cancer Res. 2016, 5, 398–409. [Google Scholar] [CrossRef]
- Parmar, C.; Grossmann, P.; Bussink, J.; Lambin, P.; Aerts, H.J.W.L. Machine Learning methods for Quantitative Radiomic Biomarkers. Sci. Rep. 2015, 5, 13087. [Google Scholar] [CrossRef]
- Zhang, Y.; Oikonomou, A.; Wong, A.; Haider, M.A.; Khalvati, F. Radiomics-based Prognosis Analysis for Non-Small Cell Lung Cancer. Sci. Rep. 2017, 7, 46349. [Google Scholar] [CrossRef]
- Braghetto, A.; Marturano, F.; Paiusco, M.; Baiesi, M.; Bettinelli, A. Radiomics and deep learning methods for the prediction of 2-year overall survival in LUNG1 dataset. Sci. Rep. 2022, 12, 14132. [Google Scholar] [CrossRef] [PubMed]
- Chaddad, A.; Desrosiers, C.; Toews, M.; Abdulkarim, B. Predicting survival time of lung cancer patients using radiomic analysis. Oncotarget 2017, 8, 104393. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.; Zhovannik, I.; Traverso, A.; Dankers, F.J.; Deist, T.M.; Kalendralis, P.; Monshouwer, R.; Bussink, J.; Fijten, R.; Aerts, H.J.; et al. Distributed radiomics as a signature validation study using the Personal Health Train infrastructure. Sci. Data 2019, 6, 218. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Yang, J.; Zhou, X.; Huang, L.; Zhao, W.; Wang, T.; Zhuang, J.; Tian, J. Development of a radiomics nomogram based on the 2D and 3D CT features to predict the survival of non-small cell lung cancer patients. Eur. Radiol. 2019, 29, 2196–2206. [Google Scholar] [CrossRef] [PubMed]
- Yadav, S.S.; Jadhav, S.M. Deep convolutional neural network based medical image classification for disease diagnosis. J. Big Data 2019, 6, 113. [Google Scholar] [CrossRef]
- Haarburger, C.; Weitz, P.; Rippel, O.; Merhof, D. Image-based survival prediction for lung cancer patients using CNNS. In Proceedings of the 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), Venice, Italy, 8–11 April 2019; pp. 1197–1201. [Google Scholar] [CrossRef]
- Hosny, A.; Parmar, C.; Coroller, T.P.; Grossmann, P.; Zeleznik, R.; Kumar, A.; Bussink, J.; Gillies, R.J.; Mak, R.H.; Aerts, H.J. Deep learning for lung cancer prognostication: A retrospective multi-cohort radiomics study. PLoS Med. 2018, 15, e1002711. [Google Scholar] [CrossRef] [PubMed]
- O’Mahony, N.; Campbell, S.; Carvalho, A.; Harapanahalli, S.; Hernandez, G.V.; Krpalkova, L.; Riordan, D.; Walsh, J. Deep Learning vs. Traditional Computer Vision. In Proceedings of the Advances in Computer Vision, Las Vegas, NV, USA, 2–3 May 2019; pp. 128–144. [Google Scholar] [CrossRef]
- Devarriya, D.; Gulati, C.; Mansharamani, V.; Sakalle, A.; Bhardwaj, A. Unbalanced breast cancer data classification using novel fitness functions in genetic programming. Expert Syst. Appl. 2020, 140, 112866. [Google Scholar] [CrossRef]
- Ain, Q.U.; Al-Sahaf, H.; Xue, B.; Zhang, M. A genetic programming approach to feature construction for ensemble learning in skin cancer detection. In Proceedings of the 2020 Genetic and Evolutionary Computation Conference. Association for Computing Machinery, 2020, GECCO ‘20, Cancún, Mexico, 8–12 July 2020; pp. 1186–1194. [Google Scholar] [CrossRef]
- Vanneschi, L. Machine Learning for Survival Prediction in Breast Cancer; NOVA IMS: Lisboa, Portugal, 2021. [Google Scholar]
- Ain, Q.U.; Al-Sahaf, H.; Xue, B.; Zhang, M. Genetic programming for automatic skin cancer image classification. Expert Syst. Appl. 2022, 197, 116680. [Google Scholar] [CrossRef]
- Sattar, M.; Majid, A.; Kausar, N.; Bilal, M.; Kashif, M. Lung cancer prediction using multi-gene genetic programming by selecting automatic features from amino acid sequences. Comput. Biol. Chem. 2022, 98, 107638. [Google Scholar] [CrossRef] [PubMed]
- Ochoa-Montiel, R.; Sossa, H.; Olague, G.; Sánchez-López, C. Machine Learning and Symbolic Learning for the Recognition of Leukemia L1, L2 and L3. In Proceedings of the Pattern Recognition; Vergara-Villegas, O.O., Cruz-Sánchez, V.G., Sossa-Azuela, J.H., Carrasco-Ochoa, J.A., Martínez-Trinidad, J.F., Olvera-López, J.A., Eds.; Springer: Cham, Switzerland, 2022; pp. 360–369. [Google Scholar] [CrossRef]
- Olson, R.S.; Urbanowicz, R.J.; Andrews, P.C.; Lavender, N.A.; Kidd, L.C.; Moore, J.H. Automating Biomedical Data Science Through Tree-Based Pipeline Optimization. In Proceedings of the Applications of Evolutionary Computation; Squillero, G., Burelli, P., Eds.; Springer: Cham, Switzerland, 2016; pp. 123–137. [Google Scholar] [CrossRef]
- Poli, R.; Koza, J.R. Genetic Programming. In Search Methodologies: Introductory Tutorials in Optimization and Decision Support Techniques; Burke, E.K., Kendall, G., Eds.; Springer: Boston, MA, USA, 2005; pp. 127–164. [Google Scholar] [CrossRef]
- Tran, B.; Xue, B.; Zhang, M. Genetic programming for multiple-feature construction on high-dimensional classification. Pattern Recognit. 2019, 93, 404–417. [Google Scholar] [CrossRef]
- Ain, Q.U.; Al-Sahaf, H.; Xue, B.; Zhang, M. A Multi-tree Genetic Programming Representation for Melanoma Detection Using Local and Global Features. In Proceedings of the AI 2018: Advances in Artificial Intelligence, Wellington, New Zealand, 11–14 December 2018; Springer: Cham, Switzerland, 2018; pp. 111–123. [Google Scholar] [CrossRef]
- Bhardwaj, H.; Sakalle, A.; Tiwari, A.; Verma, M.; Bhardwaj, A. Breast Cancer Diagnosis using Simultaneous Feature Selection and Classification: A Genetic Programming Approach. In Proceedings of the 2018 IEEE Symposium Series on Computational Intelligence (SSCI), Bangalore, India, 18–21 November 2018; pp. 2186–2192. [Google Scholar] [CrossRef]
- Scalco, E.; Rizzo, G.; Gómez-Flores, W. Automatic Feature Construction Based on Genetic Programming for Survival Prediction in Lung Cancer Using CT Images. In Proceedings of the 2022 44th Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Glasgow, UK, 11–15 July 2022; pp. 3797–3800. [Google Scholar] [CrossRef]
- Aerts, H.J.; Velazquez, E.R.; Leijenaar, R.T.; Parmar, C.; Grossmann, P.; Carvalho, S.; Bussink, J.; Monshouwer, R.; Haibe-Kains, B.; Rietveld, D.; et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 2014, 5, 4006. [Google Scholar] [CrossRef] [PubMed]
- Bakr, S.; Gevaert, O.; Echegaray, S.; Ayers, K.; Zhou, M.; Shafiq, M.; Zheng, H.; Benson, J.A.; Zhang, W.; Leung, A.N.; et al. A radiogenomic dataset of non-small cell lung cancer. Sci. Data 2018, 5, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Welch, M.L.; McIntosh, C.; Haibe-Kains, B.; Milosevic, M.F.; Wee, L.; Dekker, A.; Huang, S.H.; Purdie, T.G.; O’Sullivan, B.; Aerts, H.J.; et al. Vulnerabilities of radiomic signature development: The need for safeguards. Radiother. Oncol. 2019, 130, 2–9. [Google Scholar] [CrossRef] [PubMed]
- Clark, K.; Vendt, B.; Smith, K.; Freymann, J.; Kirby, J.; Koppel, P.; Moore, S.; Phillips, S.; Maffitt, D.; Pringle, M.; et al. The Cancer Imaging Archive (TCIA): Maintaining and operating a public information repository. J. Digit. Imaging 2013, 26, 1045–1057. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.; Liu, Z.; Guan, M.; Song, J.; Lian, Y.; Wang, S.; Tang, Z.; Dong, D.; Kong, L.; Wang, M.; et al. 2D and 3D CT Radiomics Features Prognostic Performance Comparison in Non-Small Cell Lung Cancer. Transl. Oncol. 2017, 10, 886–894. [Google Scholar] [CrossRef] [PubMed]
- Van Griethuysen, J.J.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.; Fillion-Robin, J.C.; Pieper, S.; Aerts, H.J.W.L. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef] [PubMed]
- Da-Ano, R.; Masson, I.; Lucia, F.; Doré, M.; Robin, P.; Alfieri, J.; Rousseau, C.; Mervoyer, A.; Reinhold, C.; Castelli, J.; et al. Performance comparison of modified ComBat for harmonization of radiomic features for multicenter studies. Sci. Rep. 2020, 10, 10248. [Google Scholar] [CrossRef]
- Fortin, J.P.; Parker, D.; Tunç, B.; Watanabe, T.; Elliott, M.A.; Ruparel, K.; Roalf, D.R.; Satterthwaite, T.D.; Gur, R.C.; Gur, R.E.; et al. Harmonization of multi-site diffusion tensor imaging data. Neuroimage 2017, 161, 149–170. [Google Scholar] [CrossRef] [PubMed]
- Engelbrecht, A.P. Computational Intelligence, 1st ed.; John Wiley & Sons: Hoboken, NJ, USA, 2007. [Google Scholar]
- Pei, W.; Xue, B.; Shang, L.; Zhang, M. Genetic programming for high-dimensional imbalanced classification with a new fitness function and program reuse mechanism. Soft Comput. 2020, 45, 18021–18038. [Google Scholar] [CrossRef]
- Hand, D.J.; Till, R.J. A Simple Generalisation of the Area Under the ROC Curve for Multiple Class Classification Problems. Mach. Learn. 2001, 45, 171–186. [Google Scholar] [CrossRef]
- Luke, S.; Panait, L. A Survey and Comparison of Tree Generation Algorithms. In Proceedings of the 3rd Annual Conference on Genetic and Evolutionary Computation, GECCO’01, San Francisco, CA, USA, 7–11 July 2001; pp. 81–88. [Google Scholar]
- Deb, K.; Pratap, A.; Agarwal, S.; Meyarivan, T. A fast and elitist multiobjective genetic algorithm: NSGA-II. IEEE Trans. Evol. Comput. 2002, 6, 182–197. [Google Scholar] [CrossRef]
- Massafra, R.; Bove, S.; Lorusso, V.; Biafora, A.; Comes, M.C.; Didonna, V.; Diotaiuti, S.; Fanizzi, A.; Nardone, A.; Nolasco, A.; et al. Radiomic Feature Reduction Approach to Predict Breast Cancer by Contrast-Enhanced Spectral Mammography Images. Diagnostics 2021, 11, 684. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Long, F.; Ding, C. Feature selection based on mutual information criteria of max-dependency, max-relevance, and min-redundancy. IEEE Trans. Pattern Anal. Mach. Intell. 2005, 27, 1226–1238. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Tang, K. Feature Selection for Maximizing the Area Under the ROC Curve. In Proceedings of the 2009 IEEE International Conference on Data Mining Workshops, Miami, FL, USA, 6 December 2009; pp. 400–405. [Google Scholar] [CrossRef]
- Duda, R.O.; Hart, P.E.; Stork, D.G. Pattern Classification, 2nd ed.; John Wiley & Sons: Hoboken, NJ, USA, 2001. [Google Scholar]
- Wu, J.; Chen, X.Y.; Zhang, H.; Xiong, L.D.; Lei, H.; Deng, S.H. Hyperparameter Optimization for Machine Learning Models Based on Bayesian Optimization. J. Electron. Sci. Technol. 2019, 17, 26–40. [Google Scholar] [CrossRef]
- Sokolova, M.; Lapalme, G. A systematic analysis of performance measures for classification tasks. Inform. Process. Manag. 2009, 45, 427–437. [Google Scholar] [CrossRef]
- Goel, M.K.; Khanna, P.; Kishore, J. Understanding survival analysis: Kaplan-Meier estimate. Int. J. Ayurveda Res. 2010, 1, 274–278. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Duan, H.; Li, X.; Ye, X.; Huang, G.; Nie, S. A prognostic analysis method for non-small cell lung cancer based on the computed tomography radiomics. Phys. Med. Biol. 2020, 65, 045006. [Google Scholar] [CrossRef] [PubMed]
- Miranda, I.M.; Ladeira, M.; de Castro Aranha, C. A Comparison Study Between Deep Learning and Genetic Programming Application in Cart Pole Balancing Problem. In Proceedings of the 2018 IEEE Congress on Evolutionary Computation (CEC), Rio de Janeiro, Brazil, 8–13 July 2018; pp. 1–7. [Google Scholar] [CrossRef]
- Sugai, Y.; Kadoya, N.; Tanaka, S.; Tanabe, S.; Umeda, M.; Yamamoto, T.; Takeda, K.; Dobashi, S.; Ohashi, H.; Takeda, K.; et al. Impact of feature selection methods and subgroup factors on prognostic analysis with CT-based radiomics in non-small cell lung cancer patients. Radiat. Oncol. 2021, 16, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Le, V.H.; Kha, Q.H.; Hung, T.N.K.; Le, N.Q.K. Risk score generated from CT-based radiomics signatures for overall survival prediction in non-small cell lung cancer. Cancers 2021, 13, 3616. [Google Scholar] [CrossRef] [PubMed]
- Shao, L.; Liu, L.; Li, X. Feature Learning for Image Classification Via Multiobjective Genetic Programming. IEEE Trans. Neural Netw. Learn. Syst. 2014, 25, 1359–1371. [Google Scholar] [CrossRef]
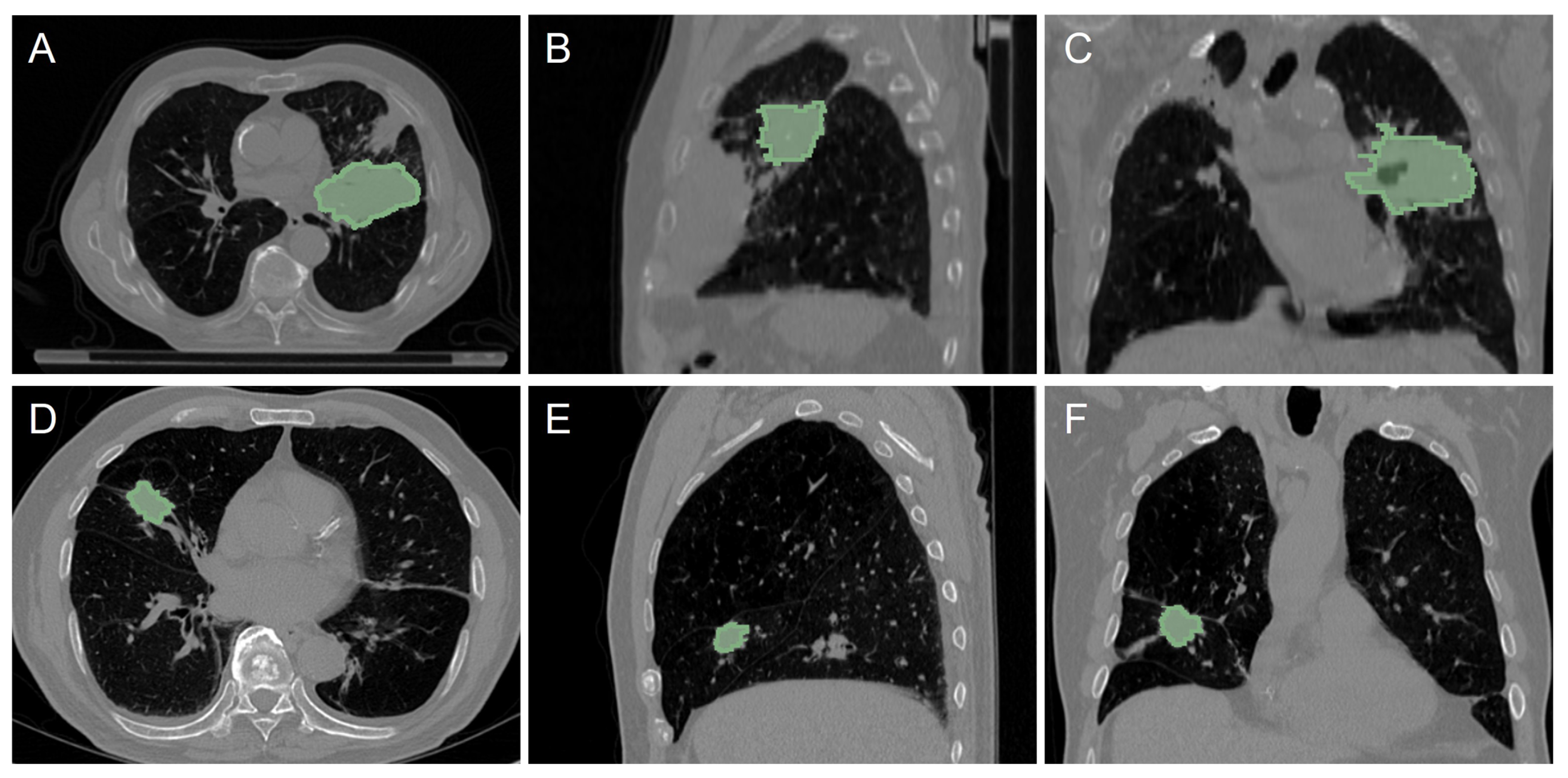
| LUNG1 | LUNG2 | |
|---|---|---|
| Image parameters | ||
| Pixel size | mm2 | – mm2 |
| Slice thickness | 3.0 mm | 0.8–3.0 mm |
| Patient information | ||
| Number of patients | 403 | 130 |
| Age (years) | 68.2 ± 10.1 | 69.7 ± 8.6 |
| Gender | ||
| Male | 279 (69.2%) | 98 (75.4%) |
| Female | 124 (30.8%) | 32 (24.6%) |
| Histology | ||
| Adenocarcinoma | 47 (11.7%) | 102 (78.5%) |
| Squamous Cell | 149 (37.0%) | 25 (19.2%) |
| Large Cell | 108 (26.8%) | 0 (0%) |
| Other | 99 (24.6%) | 3 (2.3%) |
| T stage | ||
| T1 | 88 (21.8%) | 68 (52.3%) |
| T2 | 150 (37.2%) | 44 (33.8%) |
| T3 | 49 (12.2%) | 13 (10.0%) |
| T4 | 116 (28.8%) | 5 (3.8%) |
| Median overall survival time (years) | 1.5 ± 2.2 | 3.9 ± 1.8 |
| Survival >2 years | 161 (40.0%) | 102 (78.5%) |
| Survival <2 years | 242 (60.0%) | 28 (21.5%) |
| Classifier | Hyperparameter | Value |
|---|---|---|
| RF | Features to sample | |
| Decision trees | ||
| Leaf node samples | ||
| MLP | Hidden units | |
| Learning rate | ||
| Momentum factor | ||
| SVM | Margin penalty | |
| Gaussian bandwidth |
| Parameter | Value |
|---|---|
| Population size | 50 |
| Number of generations () | 100 |
| Mutation probability () | 0.10 |
| Crossover probability () | 0.90 |
| Tournament size | 5 |
| Number of trees (m) | 1 to 5 |
| Maximum tree depth () | 2 to 7 |
| Function set | |
| Terminal set | 105 radiomic features |
| Index | Expression |
|---|---|
| Accuracy | |
| Recall | |
| Precision |
| Index | Tree Depth | Number of Trees | ||||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | ||
| AUC | 2 | 0.68 | 0.66 | 0.66 | 0.66 | 0.65 |
| 3 | 0.67 | 0.69 | 0.63 | 0.65 | 0.67 | |
| 4 | 0.65 | 0.67 | 0.68 | 0.66 | 0.65 | |
| 5 | 0.65 | 0.67 | 0.66 | 0.68 | 0.66 | |
| 6 | 0.69 | 0.65 | 0.66 | 0.63 | 0.67 | |
| 7 | 0.67 | 0.67 | 0.67 | 0.66 | 0.67 | |
| Accuracy | 2 | 0.59 | 0.62 | 0.58 | 0.61 | 0.59 |
| 3 | 0.61 | 0.65 | 0.59 | 0.59 | 0.61 | |
| 4 | 0.60 | 0.58 | 0.64 | 0.60 | 0.59 | |
| 5 | 0.61 | 0.60 | 0.62 | 0.61 | 0.60 | |
| 6 | 0.61 | 0.58 | 0.62 | 0.57 | 0.61 | |
| 7 | 0.61 | 0.59 | 0.60 | 0.62 | 0.62 | |
| Precision | 2 | 0.49 | 0.52 | 0.48 | 0.50 | 0.47 |
| 3 | 0.52 | 0.56 | 0.49 | 0.49 | 0.52 | |
| 4 | 0.50 | 0.48 | 0.54 | 0.50 | 0.49 | |
| 5 | 0.51 | 0.50 | 0.51 | 0.52 | 0.50 | |
| 6 | 0.51 | 0.46 | 0.51 | 0.46 | 0.51 | |
| 7 | 0.51 | 0.49 | 0.51 | 0.53 | 0.52 | |
| Recall | 2 | 0.70 | 0.62 | 0.59 | 0.56 | 0.57 |
| 3 | 0.69 | 0.61 | 0.60 | 0.65 | 0.50 | |
| 4 | 0.51 | 0.61 | 0.55 | 0.56 | 0.64 | |
| 5 | 0.61 | 0.61 | 0.58 | 0.52 | 0.66 | |
| 6 | 0.66 | 0.55 | 0.61 | 0.66 | 0.70 | |
| 7 | 0.59 | 0.65 | 0.62 | 0.61 | 0.67 | |
| Index | Method | LR | LDA | NB | RF | SVM | MLP |
|---|---|---|---|---|---|---|---|
| AUC | FS_mRMR | 0.63 | 0.62 | 0.61 | 0.64 | 0.65 | 0.62 |
| FS_FWD | 0.65 | 0.65 | 0.63 | 0.60 | 0.66 | 0.63 | |
| GP_AUC | 0.61 | 0.61 | 0.62 | 0.61 | 0.60 | 0.60 | |
| GP_CORR | 0.67 | 0.63 | 0.65 | 0.62 | 0.59 | 0.63 | |
| GP_GAIN | 0.69 | 0.68 | 0.70 | 0.64 | 0.68 | 0.67 | |
| Accuracy | FS_mRMR | 0.57 | 0.57 | 0.59 | 0.62 | 0.57 | 0.58 |
| FS_FWD | 0.62 | 0.60 | 0.60 | 0.56 | 0.60 | 0.58 | |
| GP_AUC | 0.56 | 0.55 | 0.57 | 0.56 | 0.54 | 0.55 | |
| GP_CORR | 0.60 | 0.57 | 0.58 | 0.58 | 0.56 | 0.58 | |
| GP_GAIN | 0.65 | 0.65 | 0.64 | 0.59 | 0.62 | 0.64 | |
| Precision | FS_mRMR | 0.47 | 0.46 | 0.48 | 0.53 | 0.49 | 0.48 |
| FS_FWD | 0.52 | 0.50 | 0.50 | 0.46 | 0.49 | 0.48 | |
| GP_AUC | 0.47 | 0.46 | 0.48 | 0.45 | 0.45 | 0.45 | |
| GP_CORR | 0.50 | 0.47 | 0.48 | 0.48 | 0.46 | 0.47 | |
| GP_GAIN | 0.56 | 0.55 | 0.55 | 0.39 | 0.52 | 0.54 | |
| Recall | FS_mRMR | 0.52 | 0.52 | 0.65 | 0.56 | 0.61 | 0.63 |
| FS_FWD | 0.65 | 0.61 | 0.52 | 0.56 | 0.63 | 0.51 | |
| GP_AUC | 0.62 | 0.64 | 0.61 | 0.63 | 0.71 | 0.63 | |
| GP_CORR | 0.60 | 0.55 | 0.63 | 0.62 | 0.52 | 0.55 | |
| GP_GAIN | 0.61 | 0.59 | 0.63 | 0.53 | 0.59 | 0.61 | |
| K–M () | FS_mRMR | 149.829 | 92.688 | 0.538 | 3.188 | 18.356 | 0.230 |
| FS_FWD | 0.001 | 0.311 | 2.041 | 33.303 | 0.020 | 4.625 | |
| GP_AUC | 1.834 | 2.291 | 0.267 | 2.247 | 5.895 | 19.451 | |
| GP_CORR | 6.176 | 136.507 | 28.744 | 28.731 | 277.792 | 83.912 | |
| GP_GAIN | 0.012 | 0.205 | 0.000 | 2.116 | 0.951 | 0.019 |
| Dataset | ||
|---|---|---|
| Method | LUNG1 | LUNG2 |
| FS-mRMR | 0.608 | 0.692 |
| FS-SFS-FWD | 0.606 | 0.643 |
| GP-AUC | 0.601 | 0.639 |
| GP-CORR | 0.579 | 0.659 |
| GP-GAIN | 0.586 | 0.644 |
| Index | Method | LR | LDA | NB | RF | SVM | MLP |
|---|---|---|---|---|---|---|---|
| AUC | FS_mRMR | 0.65 | 0.66 | 0.59 | 0.66 | 0.65 | 0.62 |
| FS_FWD | 0.65 | 0.65 | 0.65 | 0.58 | 0.67 | 0.62 | |
| GP_AUC | 0.65 | 0.65 | 0.65 | 0.65 | 0.65 | 0.65 | |
| GP_CORR | 0.69 | 0.69 | 0.69 | 0.65 | 0.71 | 0.67 | |
| GP_GAIN | 0.68 | 0.68 | 0.63 | 0.68 | 0.67 | 0.67 | |
| Accuracy | FS_mRMR | 0.63 | 0.64 | 0.52 | 0.39 | 0.64 | 0.69 |
| FS_FWD | 0.66 | 0.56 | 0.61 | 0.52 | 0.65 | 0.64 | |
| GP_AUC | 0.58 | 0.58 | 0.58 | 0.62 | 0.58 | 0.58 | |
| GP_CORR | 0.58 | 0.58 | 0.58 | 0.62 | 0.55 | 0.54 | |
| GP_GAIN | 0.74 | 0.68 | 0.65 | 0.74 | 0.54 | 0.74 | |
| Precision | FS_mRMR | 0.86 | 0.86 | 0.80 | 0.87 | 0.84 | 0.83 |
| FS_FWD | 0.87 | 0.87 | 0.86 | 0.83 | 0.85 | 0.83 | |
| GP_AUC | 0.86 | 0.86 | 0.86 | 0.87 | 0.88 | 0.88 | |
| GP_CORR | 0.89 | 0.89 | 0.89 | 0.83 | 0.89 | 0.92 | |
| GP_GAIN | 0.85 | 0.87 | 0.86 | 0.82 | 0.88 | 0.84 | |
| Recall | FS_mRMR | 0.63 | 0.65 | 0.52 | 0.26 | 0.67 | 0.76 |
| FS_FWD | 0.67 | 0.52 | 0.60 | 0.49 | 0.68 | 0.68 | |
| GP_AUC | 0.56 | 0.56 | 0.56 | 0.60 | 0.55 | 0.55 | |
| GP_CORR | 0.54 | 0.54 | 0.54 | 0.65 | 0.48 | 0.45 | |
| GP_GAIN | 0.81 | 0.70 | 0.66 | 0.85 | 0.48 | 0.82 | |
| K–M () | FS_mRMR | 1.67 | 1.77 | 7.46 | 42.23 | 5.88 | 6.73 |
| FS_FWD | 0.03 | 1.94 | 1.02 | 22.75 | 0.77 | 1.33 | |
| GP_AUC | 0.20 | 0.20 | 0.20 | 0.09 | 0.05 | 0.05 | |
| GP_CORR | 1.75 | 1.75 | 0.66 | 18.77 | 5.16 | 5.23 | |
| GP_GAIN | 2.65 | 1.93 | 0.61 | 8.58 | 13.28 | 0.84 |
| Reference | Approach | Dataset | Main Result |
|---|---|---|---|
| [9] | Radiomics | L1-T L2-V | AUC = 0.65–0.66 |
| [12] | Radiomics | L1-T, V | AUC = 0.76 for TNM stage I group |
| [16] | Radiomics and DL | L1-T, V | C-index = 0.623 for R + DL C-index = 0.585 for DL |
| [13] | Radiomics | L1-T L2-V | AUC = 0.61 |
| [33] | Radiomics | L1-T | C-index = 0.60 |
| [11] | Radiomics and DL | L1-T, V | AUC = 0.67 for R AUC = 0.63 for DL AUC = 0.67 for R + DL |
| [14] | Radiomics | L1-T Other-V | C-index = 0.62–0.73 |
| Ours | GP-based radiomics | L1-T L2-V | AUC = 0.71, C-index = 0.66 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Scalco, E.; Gómez-Flores, W.; Rizzo, G. A Genetic Programming Approach to Radiomic-Based Feature Construction for Survival Prediction in Non-Small Cell Lung Cancer. Appl. Sci. 2024, 14, 6923. https://doi.org/10.3390/app14166923
Scalco E, Gómez-Flores W, Rizzo G. A Genetic Programming Approach to Radiomic-Based Feature Construction for Survival Prediction in Non-Small Cell Lung Cancer. Applied Sciences. 2024; 14(16):6923. https://doi.org/10.3390/app14166923
Chicago/Turabian StyleScalco, Elisa, Wilfrido Gómez-Flores, and Giovanna Rizzo. 2024. "A Genetic Programming Approach to Radiomic-Based Feature Construction for Survival Prediction in Non-Small Cell Lung Cancer" Applied Sciences 14, no. 16: 6923. https://doi.org/10.3390/app14166923
APA StyleScalco, E., Gómez-Flores, W., & Rizzo, G. (2024). A Genetic Programming Approach to Radiomic-Based Feature Construction for Survival Prediction in Non-Small Cell Lung Cancer. Applied Sciences, 14(16), 6923. https://doi.org/10.3390/app14166923










