Abstract
A significant part of our stone heritage is made of limestone. Researchers are increasingly concerned about the risk of biodeterioration of these important objects. In this article, we present an up-to-date review of the microbial diversity of biodeteriorated limestone cultural heritage (CHL). This is based on an extensive bibliographic search of the literature investigating biodiversity using culture-dependent (CD) and culture-independent (CI) techniques. In the case of the former, only articles in which microorganisms were identified using molecular tools that generate DNA sequences were selected, with the aim of providing traceable identification based on the sequences submitted to public databases. The literature search resulted in the selection of 50 articles published between 2004 and 2023. The biodiversity data obtained from the CHL were organized into the following groups: fungi (626 records), bacteria and cyanobacteria (786 and 103 records, respectively), algae (51 records), and archaea (27 records). Within each group, the microbial diversity studied was compared according to results obtained using CD and CI techniques. Of all the articles selected, 12 used both approaches, demonstrating the growing effort to discover the total microbiome of biodeteriorated cultural heritage assets.
1. Introduction
The Mediterranean Basin countries are recognized to accommodate some of the world’s most important cultural heritage sites, including stone monuments and lithic works of art (Figure 1). Due to their cultural, artistic, and religious meaning, the safeguarding of this precious cultural heritage is highly important. The wide availability of tertiary sedimentary rocks in Southern European countries, especially Spain, Portugal, and Italy, has made limestone the main source of creating stone monuments [1]. Limestone has undoubtedly been used since the Stone Age and even earlier, and the first records of its usage relate to the Egyptian Second Dynasty, which was about 5800 years ago, when it was used for the construction of the Giza pyramids [2].

Figure 1.
Biodeteriorated limestone outdoor monument of Aristotle, located at Dornava mansion (Slovenia) from the middle of 18th century. The statue shows discoloration phenomena and superficial disintegration of limestone. This photo was taken by Maja Gutman Levstek.
Limestone is a sedimentary rock composed of two main calcium carbonate (CaCO3) minerals, calcite and aragonite, assembled in trigonal and orthorhombic systems, respectively. It may also contain materials such as quartz, sand, chert, and clay. The bioreceptivity of limestone is determined by a number of factors, including its porosity, pH, texture, roughness, chemical composition, density, and moisture [3]. There is a growing concern that microbial biofilms on cultural heritage buildings and monuments, especially on carboniferous stones such as limestone, impact their aesthetic and physical effects, since limestone is soluble in weak acid solutions and, therefore, particularly vulnerable to biocorrosion [1].
The locations from which the microbial samples discussed in this review originate range from the Giza pyramid complex [3], the archaeological site of the Herculaneum’s House of the Bicentenary in Italy (1 CE) [4], the archaeological site of Copan, one of the most remarkable Classic Maya (250–900 CE) centers in Mesoamerica [5], the ancient Chinese Buddhist culture sites from West Lake Cultural Landscape dating back to the Northern Song (960–1219 CE) and Qing dynasties (1636–1912 CE) [6], the 15th century French Notre Dame Gothic Cathedral in Rheims [7], and some other selected examples, including prominent monuments, buildings, and landscapes of historical significance. Studied caves of CH significance from France [8,9,10] and Spain [11] are included as well. Carbonate rocks (limestone in particular) are easily biodeteriorated, yet it is the vulnerability of the substratum that made the limestone attractive for creating stone monuments and buildings, as it is soft and easy to carve [1]. Microorganisms, including bacteria, fungi, archaea, algae, and lichens, are major causes of damage to stone monuments and have, therefore, received significant attention from conservators, restorers, and conservation scientists. The microbial colonization of stones depends on environmental factors such as water availability, pH, exposure to wind and rain, nutrient sources, and petrologic parameters such as mineral composition, type of cement, and the porosity and permeability of the rock material [12]. Culture-dependent techniques have been adopted for many years for investigating microbial communities or for assessing specific patterns of degradation caused by certain microbial taxa. This strategy has the advantage of testing specific degradation problems, yet it fails us when a comprehensive view of the complexity of the microbial communities is needed, which is mainly due to the fact that approximately 99% of viable microorganisms cannot be cultured [13]. In this review, we refer to the deterioration of cultural heritage objects/items made of limestone (CHL) colonized by microbes that are identified and/or detected using molecular methods. Culture-dependent (CD) methods refer to all methods in which the identification of pure microbial cultures is based on selected target DNA regions, mostly obtained using Sanger sequencing, while culture-independent (CI) methods include Illumina short-fragment and PacBio or Nanopore long-fragment sequencing on extracted DNA samples obtained directly from the CHL biodeteriorated sites.
2. Materials and Methods
Google Scholar, Scopus, and Web of Science engines were used for screening the taxonomic literature relating to the microbes involved in biodeterioration. Keywords such as “biodeterioration”, “microorganism”, “limestone”, either “cultural heritage” or “historical buildings”, and “molecular methods” were used. The bibliographic search was performed by the three authors independently.
The bibliography was selected if (i) the microbial samples were detected/isolated from CHL assets, either epi- or endolithically; (ii) identification of microorganisms was based on molecular marker genes; and/or (iii) the microbiomes were determined using culture-independent methods. Articles that included cultural methods with phenotypic identification only, or those describing microbial detection by microscopy only, or those that employed other physico-chemical analyses without molecular genetic approaches, were not included in order to make the taxonomic information precise. The only exception were lichens, since they are not culturable as such, and their identification mainly relies on morphology and some biochemical characteristics. In addition, the studies on samples taken from limestone mortar or wall paintings were not included because the chemical properties of these substrates differ from pure limestone.
After the bibliographic search, the names of all of the microorganisms mentioned in 50 eligible articles were entered into Supplemental Tables S1–S5. Since different articles presented results at different taxonomic levels, we used the NCBI (National Center for Biotechnology Information) Taxonomy database to standardize the taxonomic entries. After the literature review, the data on organisms obtained from CHL were organized into the following taxonomic/functional groups: fungi, bacteria, cyanobacteria, algae, and archaea, each being represented in its own table (Supplementary Material Tables S1–S5). The reason for this organization is that the vast majority of articles on cultural heritage asset biodeterioration separate organisms into these groups. However, according to NCBI taxonomy, archaea, and bacteria present independent monophyletic superkingdoms, and cyanobacteria are a monophyletic phylum within the superkingdom of bacteria. The reason for placing cyanobacteria in a separate table in our study is the general belief that these photoautotrophs, along with algae, are the primary colonizers of stone [14]. Fungi and algae belong to the monophyletic eukarya superkingdom. In contrast to the monophyletic kingdom of fungi, algae present a polyphyletic group. The data on unidentified microorganisms recognized by culture-independent techniques based on DNA sequences, which were not classified to the genus level, were not entered into the tables. In the 1st column of Tables S1–S5, the CHL objects are listed. The 2nd to 7th columns show the taxonomic classification according to the NCBI Taxonomy database. The 8th and 9th columns specify whether culture-dependent or culture-independent approaches were used. The 10th column indicates whether a taxon was derived from an indoor or outdoor environment or from both of these. The 11th column lists the countries where the specimens were collected, the 12th column contains the information about the aesthetic changes, e.g., color changes or other damages, and the 13th column lists the references.
3. Results
3.1. The Origin of Studies
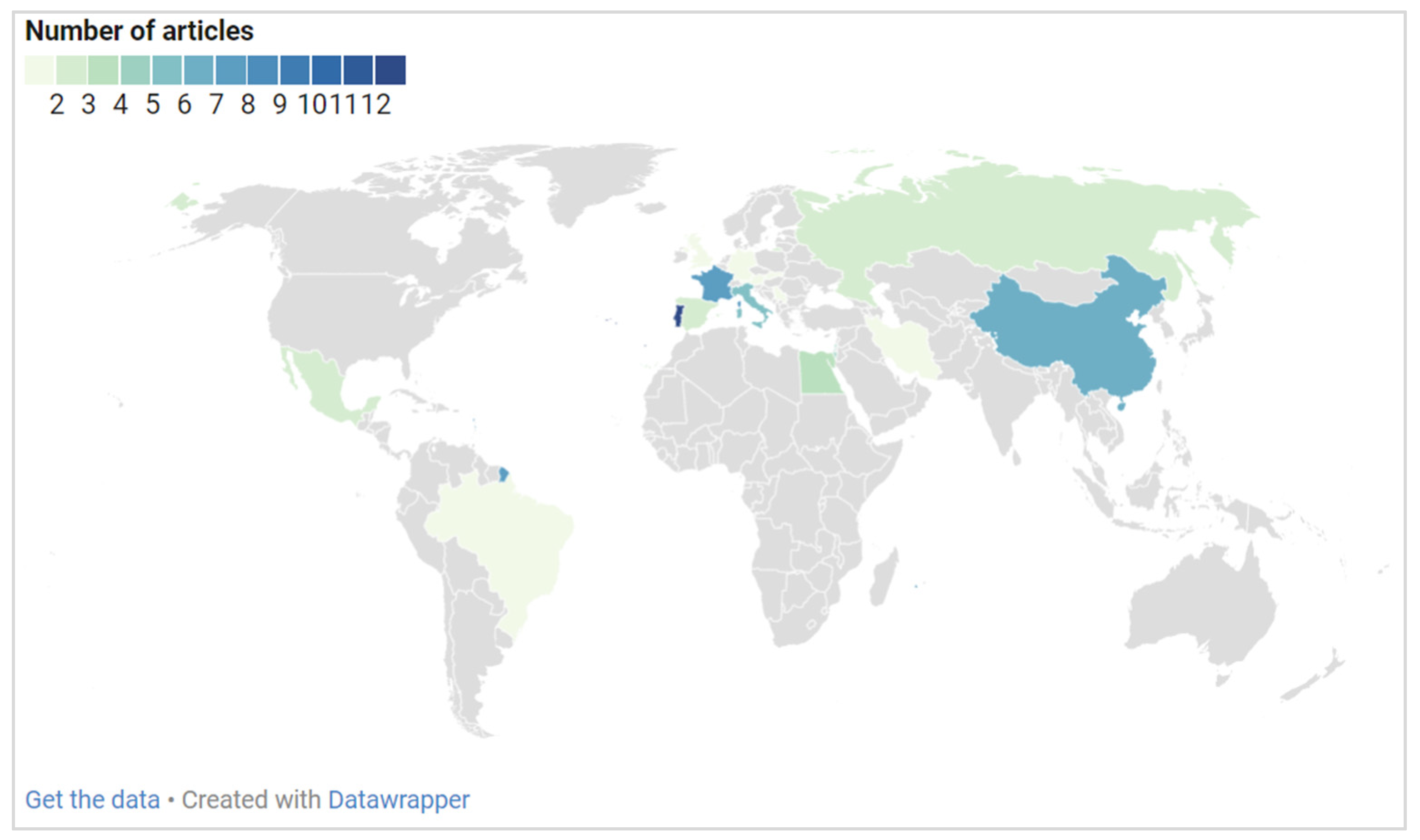
Figure 2 provides information on the geographical distribution of the included studies. Most of the studies were performed in European countries. As evident, Portugal is the leading country in this field, with 13 studies having been carried out. It is followed by France with seven, China with six, Italy with five, and Israel with four studies. The microbiome of biodeteriorated limestone was also investigated in Egypt in three studies. For Mexico, Russia, and Spain, two studies were included, and, for Austria, Brazil, Germany, Iran, Serbia, Slovakia, and the United Kingdom (UK), a single research article was included.
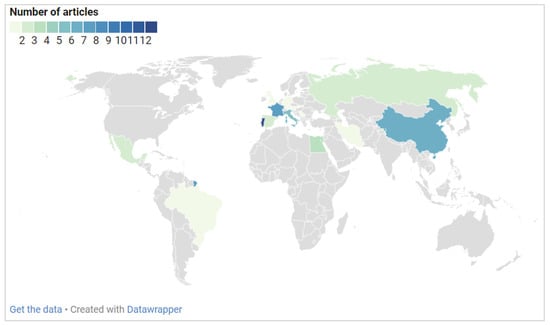
Figure 2.
The number of studies on biodeteriorated limestone using molecular identification approaches by country marked up on the world map in different colors according to the number of articles.
3.2. Biodiveristy
3.2.1. Overall Biodiversity
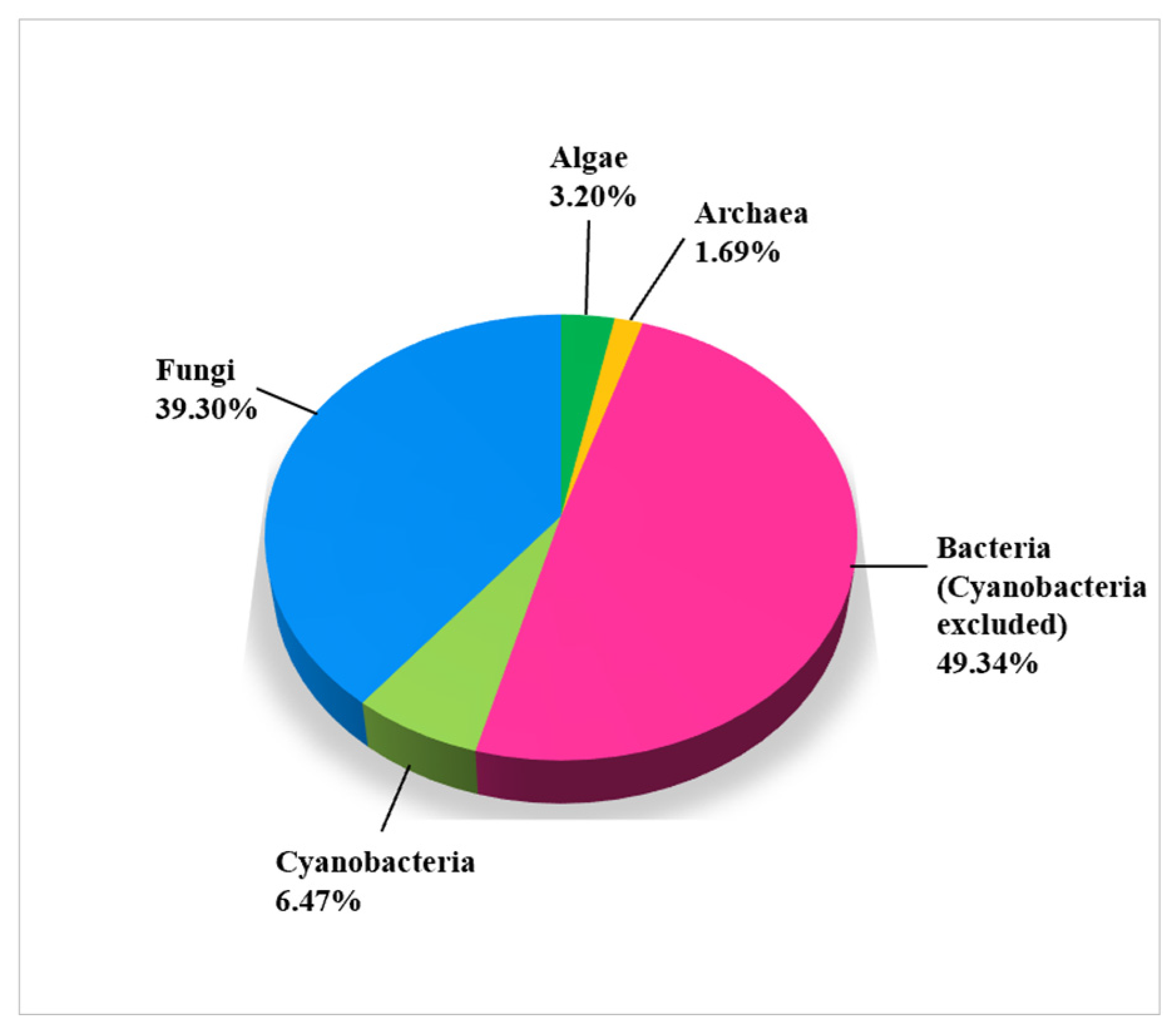
As seen in Figure 3, a generous number of microbial isolates from limestone cultural heritage assets belong to bacteria (55.81%, of which 6.47% belong to cyanobacteria). The second most frequently isolated group identified by the molecular markers is fungi (39.30%). There are few research articles on the use of molecular methods identifying algae, so they represent only 3.20% of all microbes. Archaea are the least studied microbial group, representing only 1.69% of all microbial records.
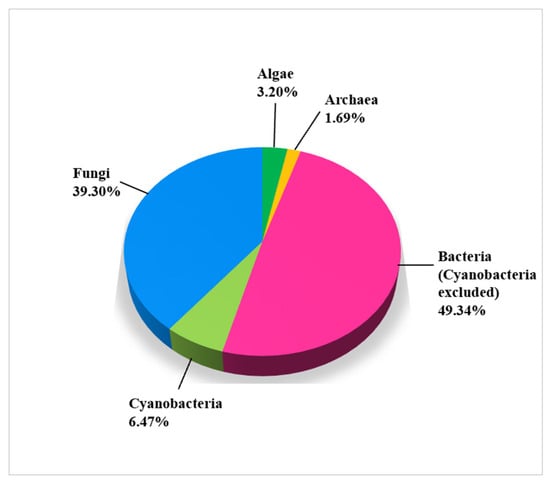
Figure 3.
Biodiversity of microbial groups on damaged CHL with molecular identification (1403 records).
3.2.2. Fungi
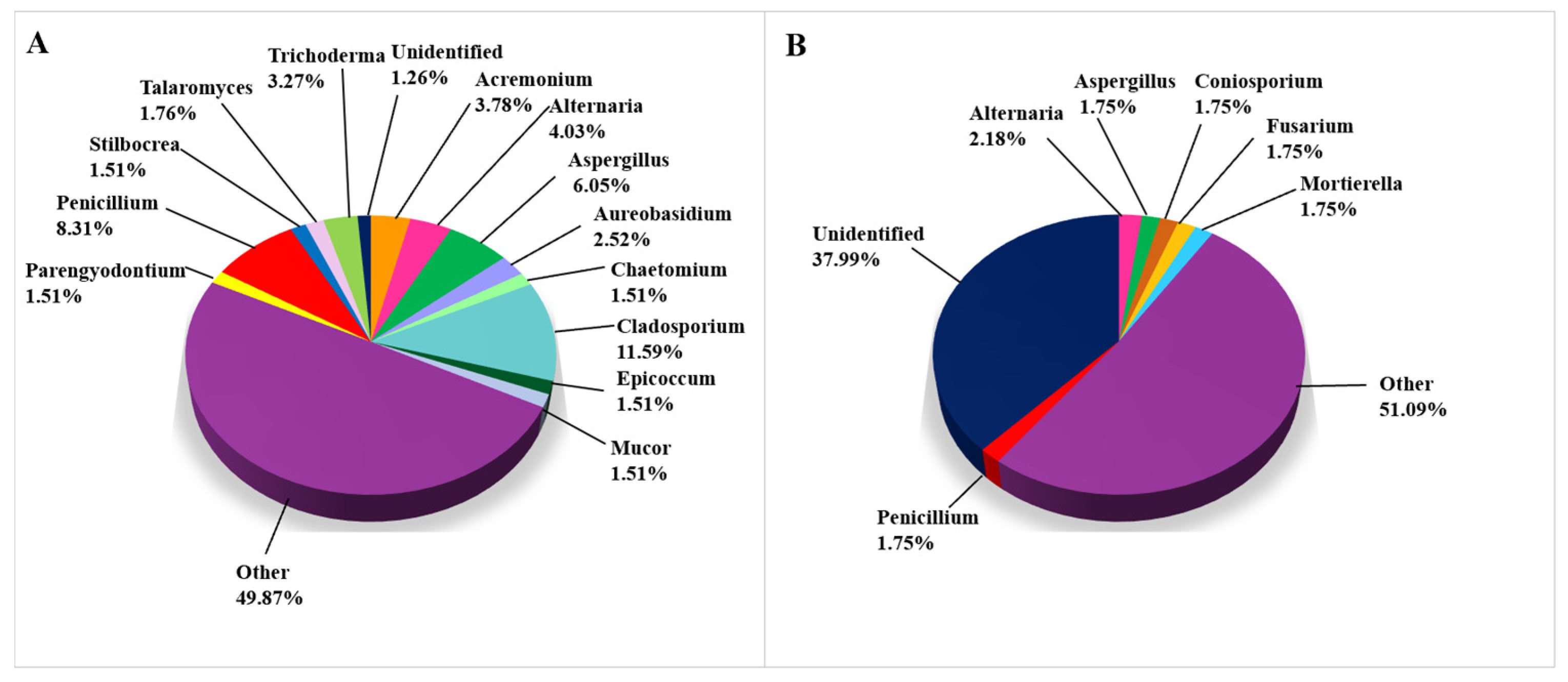
Fungal taxa on biodeteriorated limestone (Figure 4) were extensively studied using CD methods (Figure 4A), as 397 out of 626 records were obtained this way, while the remaining 229 taxa were detected using CI approaches, as shown in Figure 4B, where we list all of the genera occurring in more than 1.5% of studies. All genera occurring in less than 1.5% were grouped in the category “Other”. Among them, the genera Clonostachys, Cyphelophora, Emericellopsis, Fusarium, Geosmithia, Mortierella, Ovicillium, and Stachybotrys were recorded with the frequency of 1.0–1.5%, while the same frequency was noticed for the genera Coniosporium, Exophiala, and Rhodotorula using CI techniques. In the CI studies, 38% of fungal taxa remained unidentified at the genus level (Figure 4B), indicating that these taxa most likely have not yet been cultured, if sequences do not report “artifact taxa” due to the possibility of sequencing errors. Of the isolated cultures, only 1.26% of the taxa recorded remained unidentified and were assigned only to a higher taxonomic level.
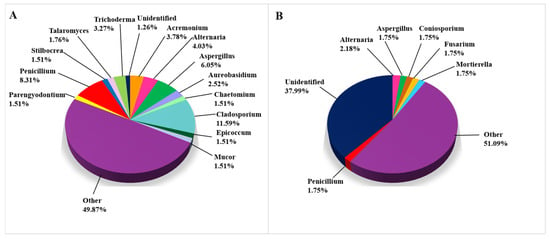
Figure 4.
Fungi on damaged limestone cultural heritage. (A) Fungal genera identified using CD approaches (397 records obtained from 24 cultural heritage sites); (B) Fungal genera identified using CI approaches (229 records obtained from 13 cultural heritage sites).
Although Penicillium is the most often detected genus among fungi (33 records), Cladosporium cladosporioides is the most common fungal species on biodeteriorated CHL, having been detected in 10 studies. Alternaria alternata and Penicillium chrysogenum were detected in six articles, Acremonium charticola and Epicoccum nigrum in five, while Penicillium brevicompactum and Stachybotrys chartarum were detected in in four articles. All of the following fungal species occurred rather frequently (in three articles): Alternaria infectoria, Aspergillus flavus, Aspergillus niger, Aureobasidium melanogenum, Aureobasidium pullulans, Cladosporium halotolerans, C. ramotenellum, C. sphaerospermum, Penicillium crustosum, P. glabrum, Stilbocrea macrostoma, and Talaromyces funiculosus (Supplemental Table S1).
3.2.3. Bacteria
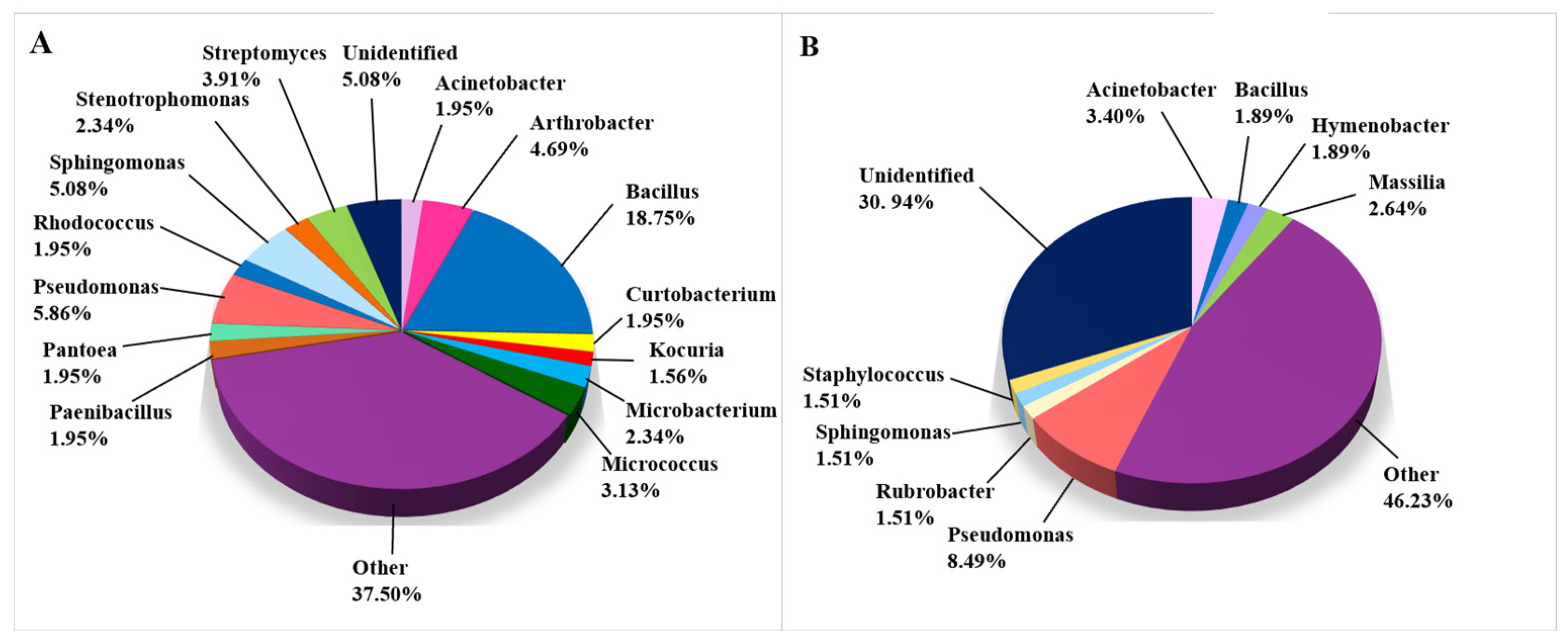
The bacterial taxa that were isolated and identified on biodeteriorated CHL are depicted in Figure 5. In contrast to the fungal taxa, the bacterial taxa were detected mainly using CI methods (530 of 786 records, Figure 5B), while the detection of 256 bacterial taxa relied on CD methods (Figure 5A). The percentage of genera determined is shown in Figure 5 for those with a frequency of more than 1.5%, while genera occurring in less than 1.5% are joined in the category “Other” for the taxa determined using CD approaches. For the taxa determined with CI approaches, the occurrence of genera specified under “Other” was less than 3%. Only 5.08% of cultivated bacterial taxa remained unidentified, while 30.0% of all taxa obtained using CI methods remained unclassified, possibly representing not-yet-cultivated organisms. The genus Bacillus was most often isolated from CHL (48 records), while Pseudomonas was most often detected in CI assets (45 records). Arthrobacter agilis, which was isolated and determined in six articles, is the most frequently occurring species among bacteria in biodeteriorated limestone of cultural heritage significance. Frequent taxa include the members of Bacillus cereus and B. subtilis species complexes, mentioned in five different articles, and B. licheniformis, B. safensis, Pseudomonas oryzihabitans, and Pseudomonas stutzeri, which were mentioned in four articles. The following bacterial species were isolated in three articles: B. aryabhattai, B. megaterium, B. muralis, B. simplex, and Rubrobacter radiotolerans (Supplemental Table S2).
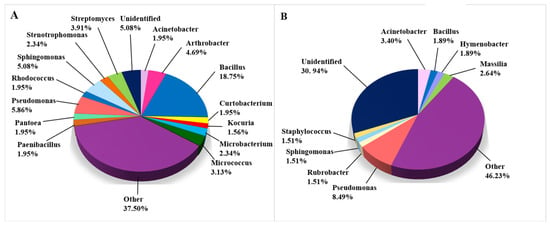
Figure 5.
Bacteria on damaged limestone cultural heritage. (A) Bacterial genera identified using CD approaches (256 records obtained from 19 cultural heritage sites); (B) Bacterial genera identified using CI approaches (530 records obtained from 21 cultural heritage sites).
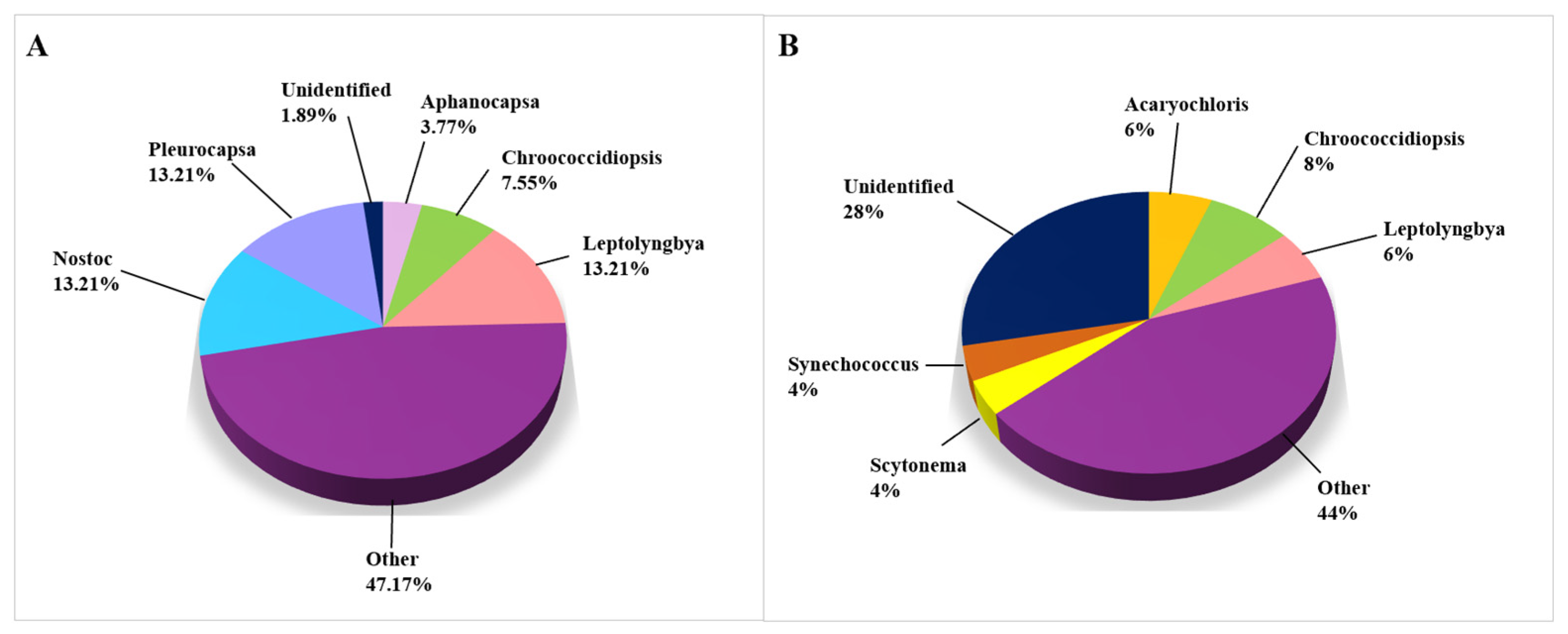
3.2.4. Cyanobacteria
Figure 6 represents the cyanobacterial taxa that were isolated and identified on biodeteriorated CHL. The genera that occurred in less than 3% of studies are categorized under “Other” (purple fraction). Research on the cyanobacterial microbiome was carried out very evenly using both CD methods (53 taxa, Figure 6A) and CI methods (50 taxa, Figure 6B). The trend of a higher percentage of unidentified taxa on the genus level obtained using CI methods persists, since there are 28.00% of unidentified cyanobacterial taxa, while the same in the CD-based approaches research represents only 1.89%. The percentage of determined genera is shown in Figure 6. The most often encountered cyanobacterial genera were Leptolyngbia, Nostoc, and Pleurocapsa (each genus mentioned in 13.2% of studies), while the species Acaryochloris marina, Anabaena cylindrica, Loriellopsis cavernicola, and Nostoc punctiforme occurred in two articles (Supplemental Table S3).
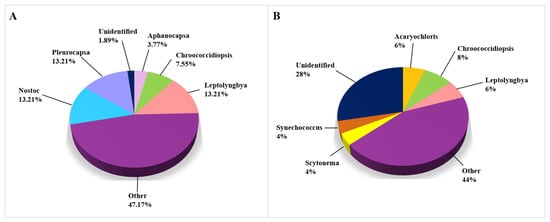
Figure 6.
Cyanobacteria on damaged limestone cultural heritage. (A) Cyanobacterial genera identified using CD approaches (53 records obtained from 12 cultural heritage sites); (B) Cyanobacterial genera identified using CI approaches (50 records obtained from 14 cultural heritage sites).
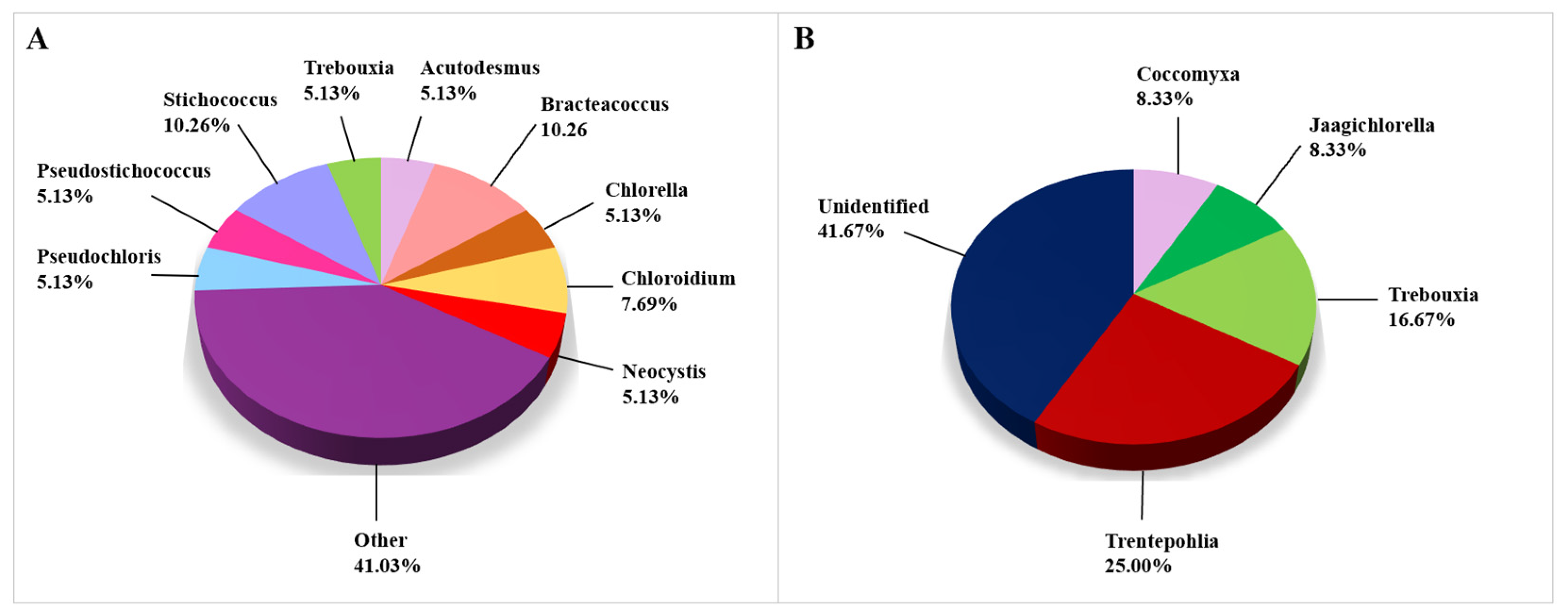
3.2.5. Algae
Algal genera that were obtained from biodeteriorated CHL using CD approaches are depicted in Figure 7A, in which the genera that appeared in less than 5% are presented in the category named “Other”. Since the number of genera detected using the CI approach was low, we have presented all of them in one graph (Figure 7B). Most taxonomic records on algae were obtained by the use of CD approaches, namely 39 of the 51 algal taxa, while 12 taxa were identified using the CI approaches. All algae were identified to the genus level in the CD studies. However, there were 41.67% of unidentified algal genera in studies conducted using CI approaches. The most frequently observed algal genus was Bracteacoccus, while the most frequently encountered species were Acutodesmus bajacalifornicus and Pseudostichococcus monallantoides. Both species were discovered in two articles (Supplemental Table S4).
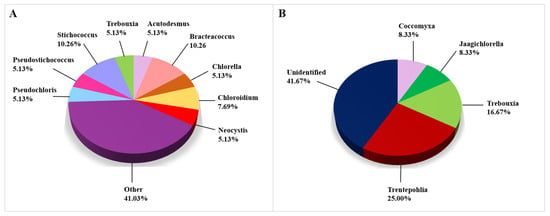
Figure 7.
Algae identified on damaged limestone cultural heritage. (A) Algal genera identified using CD approaches (39 records obtained from 3 cultural heritage sites); (B) Genera of algae identified using CI approaches (12 records obtained from 5 cultural heritage sites).
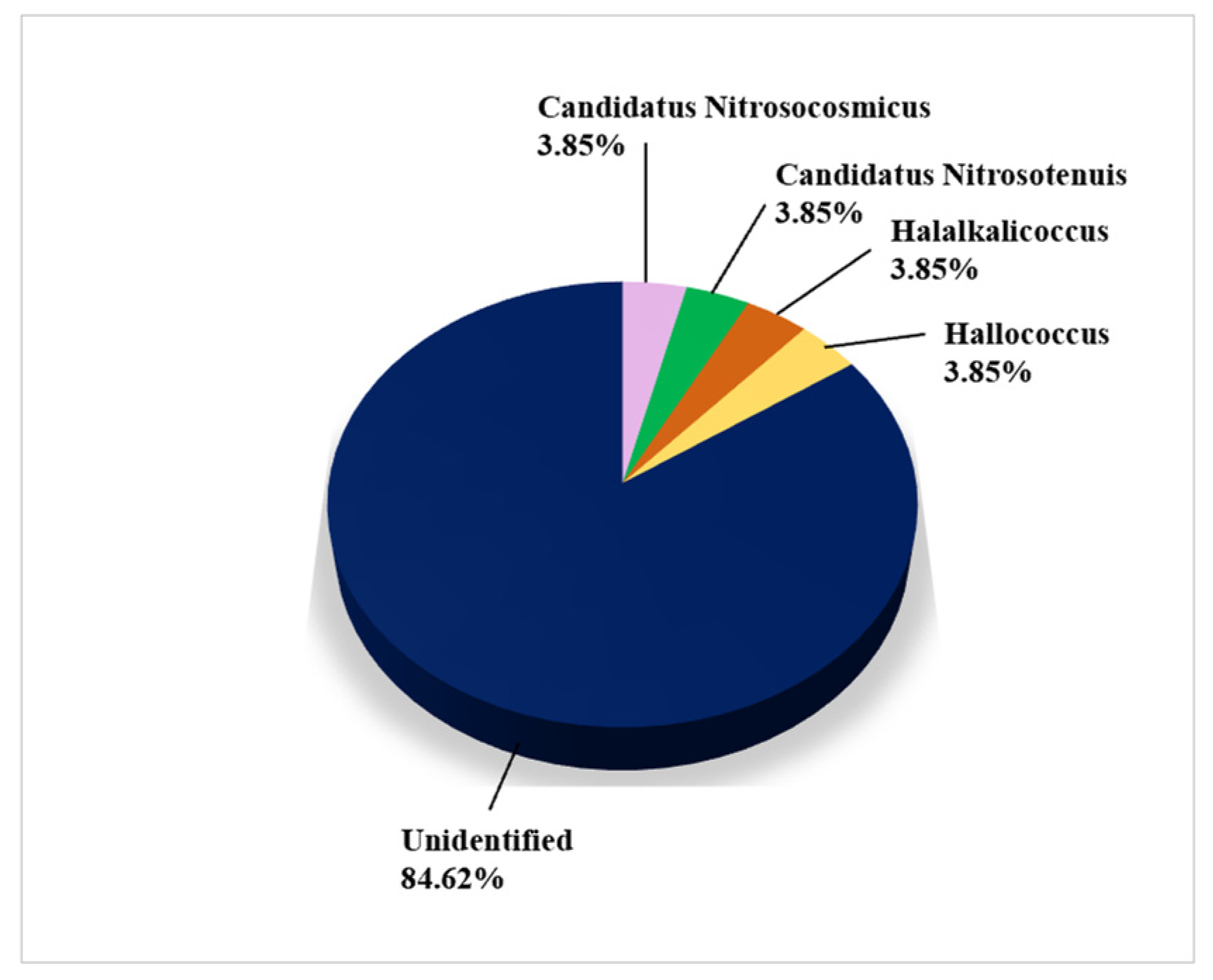
3.2.6. Archaea
Figure 8 shows the genera of archaea isolated and identified on biodeteriorated CHL using CI approaches. Only a single species, Haloferax mediterannei, isolated from the Herculaneum’s House of the Bicentenary, was identified on the basis of a culture (Supplemental Table S5).
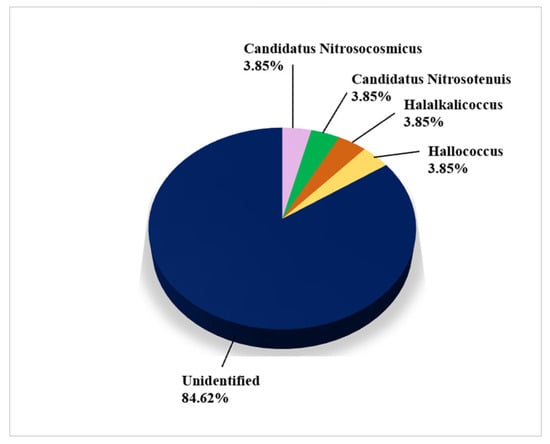
Figure 8.
Archaea on damaged limestone cultural heritage identified using CI approaches (26 records obtained from 4 cultural heritage sites).
3.2.7. Lichens
Taxonomically, lichens, with a mutualistic association of fungi (mycobionts) and cyanobacteria or algae (photobionts), are classified according to the fungal partner. In this review, like in the biodeterioration studies, they are presented separately from fungi. Only 6 of the 50 selected articles present lichen identifications, while, in another 8 articles, there were mere observations of lichens using microscopy without any further identification being made [7,15,16,17,18,19,20,21]. Kirchhoff et al. [22] identified lichens at the species level using Sanger sequencing of PCR products obtained from DNA extracted from lichens, revealing Xanthoria elegans, Caloplaca demissa, Texosporium sancti-jacobi, Caloplaca holocarpa, and Xanthomendoza hasseana. Zhu et al. [6] recognized the genera Leprocaulon, Botryolepraria, Pseudostichococcus, Lepraria, and Leprocaulon as the dominating lichens on the studied statues and monuments. In the remaining four articles [23,24,25,26], the authors identified the lichens by their morphological characters, as follows: Gholipour-Shahraki and Mohammadi [23] identified the genera Calogaya, Acarospora, Lecanora, Polysporina, and Verrucaria; Nir et al. [24] identified Caloplaca; Ding et al. [26] recognized Aspicilia, Caloplaca, Lecanora, Verrucaria, Dirina massiliensis, and Xanthoria; and Pinheiro [25] mentioned Aspicilia, Caloplaca, Caloplaca aurantia, Lecanora, Squamarina crassa, Thyrea, Verrucaria, and Xanthoria parietina.
3.3. Culture-Dependent Methods
3.3.1. Isolation of Microorganisms
A variety of culture media was employed for the isolation of the different groups of microorganisms (Supplemental Table S6). These were either general enumeration media (e.g., DRBC or DG18 for fungi) or general cultivation media (e.g., MEA, OA, PDA, or CzA for fungi; NA, TSA, or BHI for bacteria; BG-11 for cyanobacteria and algae; and media with a high salt concentration for isolation of archaea). The majority of studies used at least two or even more culture media. For the isolation of fungi, the media were supplemented with antibiotics, such as chloramphenicol (100 mg/L) or streptomycin (0.5 g/L). In the media utilized for the isolation of bacteria, fungi were suppressed with cycloheximide (50 mg/L). Fluorescent illumination and light:dark cycles were applied when culturing phototrophic microbes, like algae and cyanobacteria. The incubation temperatures ranged from 15 to 30 °C.
Culture methods were also applied on communities. Kirchoff et al. [22], for example, took samples scratched from the stone surface and grew them in culture media (3N BBM+V medium and Z medium) prior to total DNA isolation, PCR amplification, and sequencing.
3.3.2. Identification of Microorganisms
The selection and diversity of isolated cultures was studied prior to sequencing in some studies in order to assess the diversity of the obtained cultures. For bacteria, repetitive extragenic palindrome PCR amplifications were used as fingerprint methods, while, for fungi, random amplified microsatellite polymorphism (RAMP) was utilized [27].
The identification of all microorganisms obtained in pure cultures, or even lichens as unculturable organisms growing on original substrata, was performed by employing Sanger sequencing (Table 1) as a standard technique for individual sequencing reactions using a specific DNA primer set for targeting the microorganisms on a specific template of DNA isolated from a pure culture [28]. The identification level depends on the specificity of the region defined by the PCR primer pair. For fungal identification, the ITS rDNA region was most often used as the specific barcode for identification [21]. Some older studies also employed 18S sequencing [27], which is less specific and may have resulted in a higher taxa identification level. Prokaryotes (bacteria, cyanobacteria, and archaea) were mostly identified by 16S rDNA sequences, while algae by 18S rDNA.

Table 1.
Table with collected information on all included studies, including the microbial groups studied and the sequencing methods and other molecular tools used.
All of the sequences were deposited to public databases (the National Center for Biotechnology Information or the China National Center for Bioinformation database) and are available for identification verification and sequence comparison.
3.3.3. Biodeterioration Studies on Pure Cultures
The usage of cultures extends beyond the isolation of specific microbes. Studies have also been conducted on microbial pure cultures to test their stone deterioration potential, such as the excretion of weak acids, the ability to solubilize CaCO3, and the ability to precipitate CaCO3. The former was tested on the Bruni medium, utilized to test CaCO3 dissolution resulting in a clear cone around the colonies [54], and the ability to precipitate CaCO3 on B4, where the positive strains produced crystals [27]. All of the cultures that were used in the 50 selected articles are listed in Supplemental Table S6.
3.4. Culture-Independent Methods
3.4.1. DNA Isolation
Prior to the application of these methods, total DNA was extracted from biodeteriorated CHL using different commercial kits. One of the most often cited was Power Soil DNA Isolation Kit (MO BIO Laboratories, Inc., Carlsbad, USA) [6,20,21,24,34,39,41], followed by DNeasy PowerSoil (Qiagen, Hilden, Germany) [11,22,40,46,47], sometimes aiming at the epilithic microbiome, and sometimes also on the endolithic microbiomes [22].
3.4.2. Assessing Abundance and Diversity of Taxa by DNA Sequencing
In early studies, the total community of the biodeteriorated CHL was assessed after polymerase chain reaction (PCR) amplification from total DNA, and the sequence was obtained via cloning PCR products into E. coli (clone library preparation) and subsequential Sanger sequencing [18,22,29]. There was a wide variety of next-generation sequencing (NGS) methods used (second, third, and fourth generation), by which the outcome sequences were defined by the specificity of the used primer pairs in the PCR amplification reactions. The most often analyzed in the case of bacterial diversity assessment were amplicons of variable V3 and V4 regions of 16S rDNA, for fungi ITS 1 or ITS2 rDNA, and for algae the hypervariable regions V2-V4 of the 18S rRNA gene. In the second-generation sequencing, 454 pyrosequencing using GS FLX++ technology was used in a single article reviewed here [37], while the Illumina technology was far more utilized, especially the MiSeq platform (in 19 studies). In the third-generation sequencing, Illumina metagenomic shotgun sequencing (NextSeq500, Illumina Inc., San Diego, California, USA) was used in a single study [24], while the fourth-generation Nanopore (MinION Mk1C, Oxford Nanopore Technologies, Oxford, UK) was used in two studies, performed by Delegou et al. [44] and by Rabbachin et al. [15]. These studies revealed longer sequence reads and did not acquire prior PCR amplification; therefore, the outcome sequences were not biased by this intermediate step.
3.4.3. Following the Biodeterioration and Obtaining Insight into the Microbial Community Function
In the case of prior knowledge on the specific biodeteriogen and its genome, specific DNA probes were developed and real-time PCR was conducted, resulting in quantitative data on a selected species. By real-time PCR, Ochroconis lascauxensis, an invasive black fungus, was studied to score the extent of its colonization in Lascaux Cave [8].
Although the 16S rRNA gene does not provide direct evidence of a community’s functional capabilities, there are tools that do allow such a prediction [6]. Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt 1.1.4) can be used to predict the functional composition of a metagenome using the data of the marker gene and a database of reference genomes [58], while the Functional Annotation of Prokaryotic Taxa (FAPROTAX 1.2.10) can provide information on microbial functions [6]. As far as we know, sequencing of environmental RNA, which would reveal the actual functional genes being transcribed, has not been applied to biodeteriorated CHL yet.
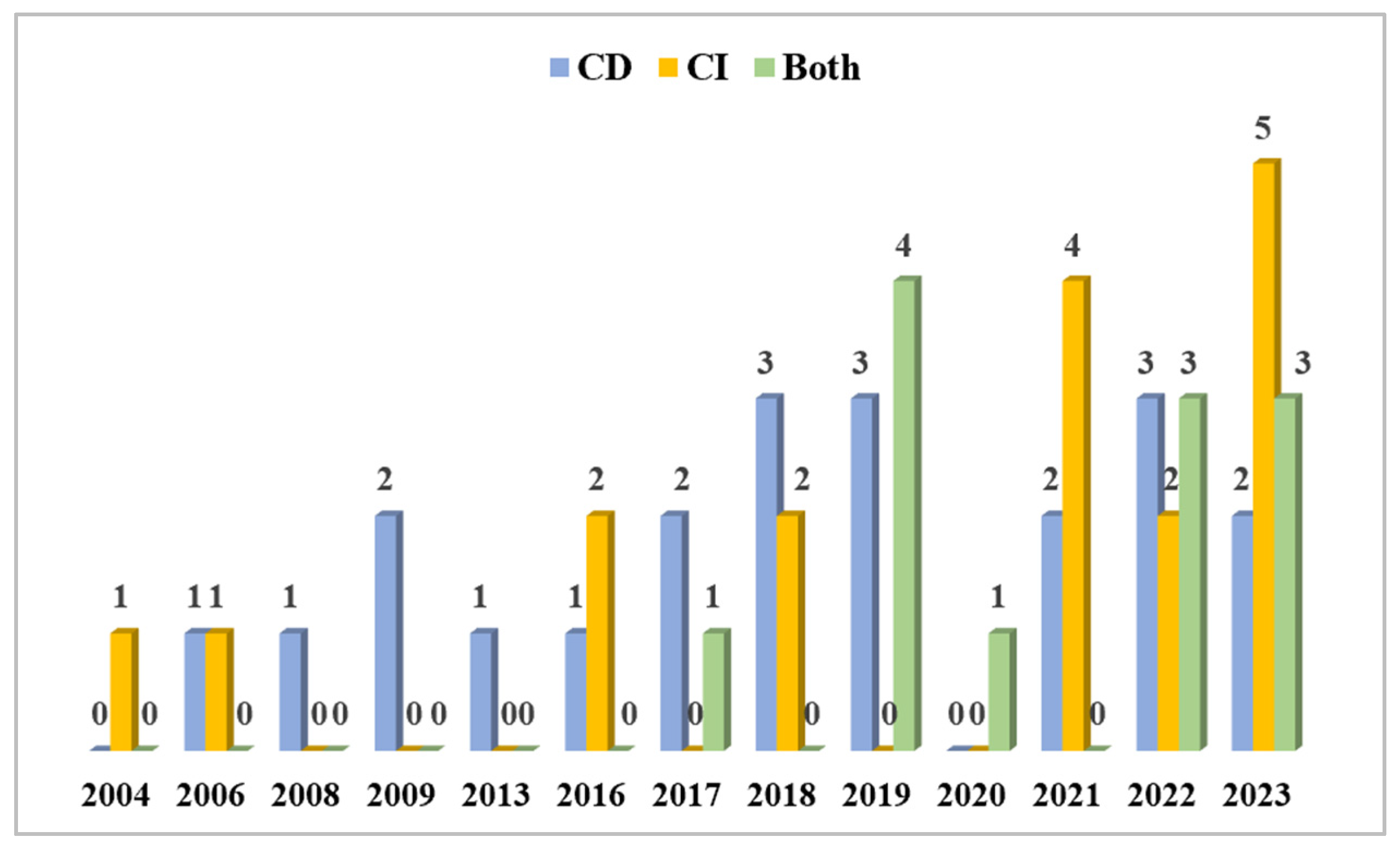
3.4.4. The Use of Methodology Studying Biodeteriorated CHL Microbiomes
Our strategy of bibliographic search (see Section 2) detected 50 articles from 2004 to 2023 (Figure 9). Figure 9 corresponds to Table 1, with a slight modification, as follows: the entries are adjusted for cases where both CD and CI approaches were used and compared to the number of studies using one of these approaches only. A total of 17 articles were found using CI techniques. Starting in 2004, the CI methods appeared in all of the reported years, apart from the years 2008, 2009, 2013, 2017, 2019, and 2020, and increased over the last three years, as 11 out of 17 CI articles were published after the year 2021. However, the majority of studies, namely 21, were conducted with CD methods, which appeared rather constantly through the years, with the exception of the years 2004 and 2020, when there were no CD-based studies published. There were also 12 articles that employed both CI and CD approaches within a single study. The first one was published in 2017, and, in 2019, a peak of four articles used this combined approach. By 2020, the only article found in our bibliographic search belonged to this category. Over the past two years (2022 and 2023), six articles, three from each year, were identified. This trend may indicate that more researchers are recognizing the necessity of using both CI and CD approaches to gain comprehensive insights into the microbiota present in cultural heritage limestone.
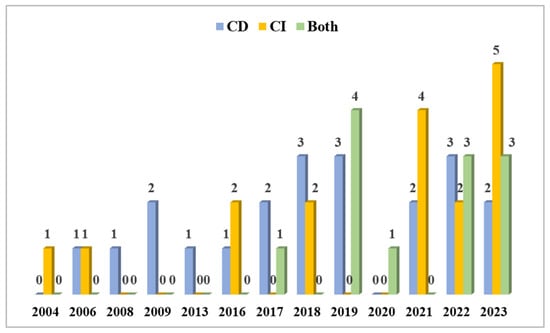
Figure 9.
The number of molecular biology studies on CHL conducted using CD approaches, CI approaches, and both CD and CI approaches together through the years in ascending order.
4. Discussion
4.1. Microorganisms as the Agents of Biodeterioration
This work summarizes the literature results concerning microorganism biodiversity in a limestone substrate of cultural heritage significance and presents a crucial foundation for further explorations for further characterizing the diversity of the bacteria, cyanobacteria, fungi, algae, archaea, and lichens involved in the deterioration of limestone monuments or buildings. Several authors [26,59,60] consider lichens as one of the most severe stone biodeterioratiors and saxicolous crustose lichens as the most difficult organisms to remove from limestone. However, some authors [15,61,62,63] have also emphasized the bioprotective role that lichens possess when present on cultural heritage assets. Lichens can, for example, soothe the abrasive effect of airborne sand particles, reduce temperature fluctuations, especially where freeze–thaw cycles occur, and even limit the dissolution of stone material when in contact with water [15,61,62]. The different microbial communities coexisting in the biofilm have different metabolic functions (they can include phototrophs, chemolithotrophs, and chemoorganotrophs) and they cooperate to achieve substrate utilization and the recycling of nutrients for their common survival [64]. On the surface of rocks, microorganisms always exist as a community and collaborate to perform various tasks. Therefore, to comprehend the impact of the microbial community on rock, studies should begin with a functional analysis of the entire population [6]. The alterations caused by microbial biodeterioration are, according to Mihajlovski et al. [18], classified into the following three different categories, namely: (i) biophysical, (ii) biochemical, and (iii) aesthetic changes, all of which can occur simultaneously or separately. Biophysical changes such as exfoliation and biopitting happen due to the penetration of fungal hyphae through the limestone surface [46], while biochemical alterations occur due to the production of organic acids, metabolites, exoenzymes, and metal-chelating compounds [65].
4.1.1. Biophysical and Biochemical Alterations
Stone substrates represent oligotrophic environments, since the nutrient availability is scarce, the ultraviolet exposure is high, and the moisture content is usually low [66], yet the formation of microbial communities occurs, starting with the occurrence of autotrophic microorganisms, such as cyanobacteria and algae, which can grow in the complete absence of organic matter [67]. The sulfur-oxidizing bacteria (SOB) and nitrifying bacteria involved in limestone weathering are autotrophic as well [57]. Fungi are also considered to be common limestone inhabitants, mainly due to the transportaition of their spores by air, atmospheric water, insects, and other biotic vectors [19] and because they possess the ability to excrete different types of acids, such as the following: citric, formic, fumaric, gluconic, lactic, malic, succinic, oxalic, and tartaric acids, which act as metal chelators or molecules involved in the dissolution of limestone [21,51]. Oxalic acid excreted from fungal hyphae has long been known for its ability to dissolve calcium carbonate and form secondary minerals such as calcium oxalates (whewellite and weddellite) on limestone cultural heritage assets, which causes the formation of a white patina on them [68,69]. Hyphae are not the only calcium oxalate that excrete fungal structure, as the crystalline can be excreted from fungal strands, cords and rhizomorphs, fruiting bodies, and lichen thalli as well [69]; however, they are the only fungal structures that are capable of enforcing mechanical pressure onto limestone by drilling into the substrate, due to the high mechanical resistance caused by the turgor pressure exerted by the protoplast on the inner side of the rigid fungal cell wall [70].
4.1.2. Aesthetic Changes Caused by Pigments
The most noticeable aesthetic changes are colored patinas, which can be of bacterial, cyanobacterial, fungal, algal, or even archaeal origin [4]. Black crusts are common for fungi-infested limestone, infested with namely Aspergillus versicolor, Penicillium brevicompactum [51], and the Ochroconis genera [8]. The black discoloration caused by fungi is often a consequence of strongly melanized cell walls. Melanin is a stress-protective pigment, which is beneficial for the survival of certain extremophilic fungi, among which belong the so-called “black fungi”. Black fungi genera represent a heterogeneous taxonomic group, including orders within the classes Dothideomycetes and Eurotiomycetes [48]. Orange-colored pigments, carotenoids, serve the same purpose as melanin—the protection of the cell [48,71]—but they are mainly excreted by other inhabitants of CHL, such as the algal Trentepohlia genera [15] and some bacterial species, namely the following: Staphylococcus aureus, Dietzia maris, Gordonia rubripertincta, Rhodococcus corynebacterioides [4], Rhodococcus cerastii, and Rhodococcus fascians [7]. Pink discoloration, which was previously assigned to a non-biological origin only, was later found to be excreted by bacterial genera Rubrobacter, Halobacillus [4], Micrococcus [54], and Arthrobacter [4,7], and by taxons belonging to archaea, such as genera Halobacterium, Halococcus, and Haloferax [4]. Green patinas on CHL are a consequence of chlorophyll production by algae and cyanobacteria [1,6,39] or a consequence of different metabolite production by fungal genera such as Aspergillus and Penicillium [51]. White coloration can occur due to a calcium-oxalate-producing genera that include mainly Streptomyces [54], Aspergillus, Penicillium, and Colletotrichum [21], or due to the growth of white-looking lichen with a fungus from the Leprocaulon genera [6]. Interestingly Zhu et al. [6] found that some of the white patches in the West Lake Cultural Landscape were transitional.
4.2. Microbial Diversity Uncovered by the Application of Molecular Approaches
Metagenomics analyses typically provide diversity indexes such as Simpson’s index, Shannon–Weiner Index, etc. They allow us to better circumscribe the diversity of species occurring in an environmental niche, but they fail to provide species level identifications [36,52,72]. Especially in bacteria, the above-species-level-identified taxa doubled in comparison to CD-based studies. Besides obtaining a greater depth into the microbial diversity, CD approaches allow species level identifications and results from culture-obtained metabolite profiles [26,36,73]. Pure cultures should best be obtained using the spread plate technique for obtaining the highest diversity. The isolation of the DNA from a pure culture can then be carried out, and later the isolated DNA can be amplified via PCR. A vast majority of articles using CI approaches used the Illumina MiSeq. One of the newer sequencing platforms, the so-called fourth-generation sequencing [28], is Nanopore sequencing, which was used only in two articles written by Delegou et al. and Rabbachin et al. [15,44]. It provides long reads and might, therefore, be the method of choice in future, as it also allows species-level identifications.
4.2.1. Discovered Microbial Biodiversity on CHL
As evidenced by this article, fungi and algae were mainly detected using CD methods, while archaea and bacteria were detected using CI methods. The identification of cyanobacteria was conducted with a relatively balanced employment of both kinds of techniques, while the identification of lichens was based on morphology and rarely employed molecular methods. Our results show that there were only two studies on lichen identification carried out using molecular approaches, one with CD [22] and one with CI methods [6]. Among the fungi in the cited articles (Table 1), which are also considered to play one of the major roles of limestone biodeterioration, Cladosporium cladosporioides is the most common fungal species found in biodeteriorated limestone, having been detected and identified using culture-dependent approaches. However, C. cladosporioides presents a complex of more than 200 species [62]. As this is obviously an important limestone colonizer, future research needs to focus on identifying the phylogenetic species involved. Algae were determined mainly by the use of CD methods. The most frequently found algae in the cited articles (Table 1) were Acutodesmus bajacalifornicus and Pseudostichococcus monallantoides. The bacterial taxa in the cited articles (Table 1) were identified mainly by using CI methods. Arthrobacter agilis was the most frequently identified species in limestone. Cyanobacterial biodiversity was identified using CD and CI methods (Figure 6A,B). As noticed by Ortega-Morales et al. [17] in the cyanobacterial community in cultural heritage stone biofilms in six different tropical and subtropical countries, statistically significant differences were calculated according to the type of stone (calciferous or siliceous), as well as according to the climatic regions and atmospheric pollution. This was the first meta-analytical study performed on stone biofilm data obtained using the same sampling, isolation, and identification techniques. Regarding the archaea that are usually considered difficult to grow on culture media, the only species able to grow on axenic culture was Haloferax mediterannei.
4.2.2. The Discovery of Novel Species on CHL
The exploration of microbial biodiversity on CHL resulted in the discovery of three new taxa, including one cyanobacteria and two fungi. During the identification of the phototrophic community of biodeteriorated cathedral walls, a hitherto unknown Nostoc/Komarekiella-like cyanobacteria was isolated, for which new genus and species, Parakomarekiella sesnandensis were proposed [53]. Similarly, the efforts of uncovering the fungal diversity on the walls of a Roman cryptoporticus gave rise to the discovery of a previously unknown strain related to Bionectriaceae (Hypocreales). Based on morphological and phylogenetic analyses, a novel genus and species, Circumfusicillium cavernae gen. et sp. nov. were proposed [56]. With a similar approach, a new genus, Saxispiralis gen. nov., and a new species, Saxispiralis lemnorum sp. nov., were discovered during the microbial study of a funerary art piece at the Lemos Pantheon [48].
5. Conclusions
There is a wide variety of methods that can be used to unravel microbial diversity with culture-dependent (CD) or/and culture-independent (CI) approaches. Out of the 50 articles that were included in our review, 12 articles contained both approaches, showing the ever-growing effort of discovering the overall microbiome of the biodeteriorated cultural heritage assets. The trend of utilizing only CD methods has persisted rather constantly through the years, while the usage of CI has gained its popularity in the last 4 years. The microbial biodiversity of CHL is mostly studied in bacteria and fungi, however, phototrophic cyanobacteria and algae are also studied well, and even previously poorly studied archaea are becoming a point of interest in some recent studies on CHL. The authors of this review article believe that it is crucial to keep exploring microbial biodiversity, preferably with a combined CD and CI approach, not only with the intention of uncovering the taxa and microbial communities responsible for the biodeterioration processes, but also with the curiosity that builds up the exiting knowledge of taxonomy, since the efforts of studying the microbial diversity on CHL have even resulted in the discovery and description of three new species in three novel genera, one cyanobacterial and two fungal, which would have otherwise remained unnoticed. The carrying out of studies on the topic of biodeterioration on CHL using molecular approaches is still rather limited to certain countries, even though limestone is widespread worldwide as a common building and artistic material. Therefore, we encourage conservators and biologists worldwide to keep exploring this field on their own cultural heritage assets; moreover, as for future directions, we encourage further studies on microbial biodeterioration to use CD and CI methods hand-in-hand and couple them with carefully monitored environmental and substrate characteristics. With the growing popularity of building predictive models that integrate biology datasets with data mining statistical algorithms to predict future outcomes of certain phenomena, we believe that predictive models could also be used in the field of microbial biodeterioration in the future to provide novel insights and answer questions such as how different combinations of microbial taxa commonly found on CHL alternate their substrate under different environmental factors.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/app14167429/s1. Table S1: Fungi detected on limestone cultural heritage assets using molecular methods in the literature; Table S2: Bacteria detected on limestone cultural heritage assets using molecular methods in the literature; Table S3: Cyanobacteria detected on limestone cultural heritage assets using molecular methods; Table S4: Algae detected on limestone cultural heritage assets using molecular methods; Table S5: Archaea detected on limestone cultural heritage using molecular methods; Table S6: Culture media for microorganisms used in biodeteriorated culture heritage limestone studies.
Author Contributions
H.S., P.Z. and M.F.M. contributed to the conceptualization, investigation, writing, and editing; software: Microsoft Word Version 2407, Microsoft Excel Version 2407, Microsoft PowerPoint Version 2407, NCBI Taxonomy Browser, Data Wrapper; funding acquisition, H.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by The EUTOPIA Student Individual Research Project, grant number 803-3/2023, by enabling H.S. the mobility from the home Biotechnical faculty of University of Ljubljana to the host university FCT-Portuguese Foundation for Science and Technology.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the references or in the Supplementary Material.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Miller, A.Z.; Macedo, M.F.; Dionísio, A.; Saiz-Jimenez, C. Primary Bioreceptivity of Limestones from the Mediterranean Basin to Phototrophic Microorganisms. Ph.D. Thesis, New University of Lisbon, Lisbon, Portugal, 2010. [Google Scholar]
- Oates, J.A.H. Lime and Limestone: Chemistry and Technology, Production and Uses, 2nd ed.; WILEY-VCH Verlag GmbH: Weinheim, Germany, 1998. [Google Scholar]
- ElBaghdady, K.Z.; Tolba, S.T.; Houssien, S.S. Biogenic Deterioration of Egyptian Limestone Monuments: Treatment and Conservation. J. Cult. Herit. 2019, 38, 118–125. [Google Scholar] [CrossRef]
- Tescari, M.; Visca, P.; Frangipani, E.; Bartoli, F.; Rainer, L.; Caneva, G. Celebrating Centuries: Pink-Pigmented Bacteria from Rosy Patinas in the House of Bicentenary (Herculaneum, Italy). J. Cult. Herit. 2018, 34, 43–52. [Google Scholar] [CrossRef]
- Elert, K.; Ruiz-Agudo, E.; Jroundi, F.; Gonzalez-Muñoz, M.T.; Fash, B.W.; Fash, W.L.; Valentin, N.; de Tagle, A.; Rodriguez-Navarro, C. Degradation of Ancient Maya Carved Tuff Stone at Copan and Its Bacterial Bioconservation. npj Mater. Degrad. 2021, 5, 44. [Google Scholar] [CrossRef]
- Zhu, C.; Wang, B.; Tang, M.; Wang, X.; Li, Q.; Hu, Y.; Zhang, B. Analysis of the Microbiomes on Two Cultural Heritage Sites. Geomicrobiol. J. 2023, 40, 203–212. [Google Scholar] [CrossRef]
- Eyssautier-Chuine, S.; Vaillant-Gaveau, N.; Charpentier, E.; Reffuveille, F. Comparison of Biofilm Development on Three Building and Restoration Stones Used in French Monuments. Int. Biodeterior. Biodegrad. 2021, 165, 105322. [Google Scholar] [CrossRef]
- Martin-Sanchez, P.M.; Bastian, F.; Alabouvette, C.; Saiz-Jimenez, C. Real-Time PCR Detection of Ochroconis Lascauxensis Involved in the Formation of Black Stains in the Lascaux Cave, France. Sci. Total Environ. 2013, 443, 478–484. [Google Scholar] [CrossRef] [PubMed]
- Bontemps, Z.; Prigent-Combaret, C.; Guillmot, A.; Hugoni, M.; Moënne-Loccoz, Y. Dark-Zone Alterations Expand throughout Paleolithic Lascaux Cave despite Spatial Heterogeneity of the Cave Microbiome. Environ. Microbiome 2023, 18, 31. [Google Scholar] [CrossRef]
- Alonso, L.; Creuzé-Des-Châtelliers, C.; Trabac, T.; Dubost, A.; Moënne-Loccoz, Y.; Pommier, T. Rock Substrate Rather than Black Stain Alterations Drives Microbial Community Structure in the Passage of Lascaux Cave 06 Biological Sciences 0605 Microbiology. Microbiome 2018, 6, 216. [Google Scholar] [CrossRef]
- Martin-Pozas, T.; Fernandez-Cortes, A.; Cuezva, S.; Cañaveras, J.C.; Benavente, D.; Duarte, E.; Saiz-Jimenez, C.; Sanchez-Moral, S. New Insights into the Structure, Microbial Diversity and Ecology of Yellow Biofilms in a Paleolithic Rock Art Cave (Pindal Cave, Asturias, Spain). Sci. Total Environ. 2023, 897, 165218. [Google Scholar] [CrossRef]
- Laiz, L.; Piñar, G.; Lubitz, W.; Saiz-Jimenez, C. Monitoring the Colonization of Monuments by Bacteria: Cultivation versus Molecular Methods. Environ. Microbiol. 2003, 5, 72–74. [Google Scholar] [CrossRef]
- Rappé, M.S.; Giovannoni, S.J. The Uncultured Microbial Majority. Annu. Rev. Microbiol. 2003, 57, 369–394. [Google Scholar] [CrossRef]
- Crispim, C.A.; Gaylarde, C.C. Cyanobacteria and Biodeterioration of Cultural Heritage: A Review. Microb. Ecol. 2005, 49, 1–9. [Google Scholar]
- Rabbachin, L.; Piñar, G.; Nir, I.; Kushmaro, A.; Eitenberger, E.; Waldherr, M.; Graf, A.; Sterflinger, K. Natural Biopatina on Historical Petroglyphs in the Austrian Alps: To Clean or Not to Clean? Int. Biodeterior. Biodegrad. 2023, 183, 105632. [Google Scholar] [CrossRef]
- Wu, Y.; Li, Q.; Tong, H.; He, Z.; Qu, J.; Zhang, B. Monitoring the Deterioration of Masonry Relics at a UNESCO World Heritage Site. KSCE J. Civ. Eng. 2021, 25, 3097–3106. [Google Scholar] [CrossRef]
- Ortega-Morales, B.O.; Narváez-Zapata, J.A.; Schmalenberger, A.; Sosa-López, A.; Tebbe, C.C. Biofilms Fouling Ancient Limestone Mayan Monuments in Uxmal, Mexico: A Cultivation-Independent Analysis. Biofilms 2004, 1, 79–90. [Google Scholar] [CrossRef]
- Mihajlovski, A.; Gabarre, A.; Seyer, D.; Bousta, F.; Di Martino, P. Bacterial Diversity on Rock Surface of the Ruined Part of a French Historic Monument: The Chaalis Abbey. Int. Biodeterior. Biodegrad. 2017, 120, 161–169. [Google Scholar] [CrossRef]
- Savković, Ž.; Unković, N.; Stupar, M.; Franković, M.; Jovanović, M.; Erić, S.; Šarić, K.; Stanković, S.; Dimkić, I.; Vukojević, J.; et al. Diversity and Biodeteriorative Potential of Fungal Dwellers on Ancient Stone Stela. Int. Biodeterior. Biodegrad. 2016, 115, 212–223. [Google Scholar] [CrossRef]
- Li, Q.; Zhang, B.; Wang, L.; Ge, Q. Distribution and Diversity of Bacteria and Fungi Colonizing Ancient Buddhist Statues Analyzed by High-Throughput Sequencing. Int. Biodeterior. Biodegrad. 2017, 117, 245–254. [Google Scholar] [CrossRef]
- Li, T.; Hu, Y.; Zhang, B.; Yang, X. Role of Fungi in the Formation of Patinas on Feilaifeng Limestone, China. Microb. Ecol. 2018, 76, 352–361. [Google Scholar] [CrossRef] [PubMed]
- Kirchhoff, N.; Hoppert, M.; Hallmann, C. Algal and Fungal Diversity on Various Dimension Stone Substrata in the Saale/Unstrut Region. Environ. Earth Sci. 2018, 77, 609. [Google Scholar] [CrossRef]
- Gholipour-Shahraki, M.; Mohammadi, P. The Study of Growth of Calogaya Sp. PLM8 on Cyrus the Great’s Tomb, UNESCO World Heritage Site in Iran. Int. J. Environ. Res. 2017, 11, 501–513. [Google Scholar] [CrossRef]
- Nir, I.; Barak, H.; Kramarsky-Winter, E.; Kushmaro, A.; de los Ríos, A. Microscopic and Biomolecular Complementary Approaches to Characterize Bioweathering Processes at Petroglyph Sites from the Negev Desert, Israel. Environ. Microbiol. 2022, 24, 967–980. [Google Scholar] [CrossRef]
- Pinheiro, A.C.; Mesquita, N.; Trovão, J.; Soares, F.; Tiago, I.; Coelho, C.; de Carvalho, H.P.; Gil, F.; Catarino, L.; Piñar, G.; et al. Limestone Biodeterioration: A Review on the Portuguese Cultural Heritage Scenario. J. Cult. Herit. 2019, 36, 275–285. [Google Scholar] [CrossRef]
- Ding, Y.; Salvador, C.S.C.; Caldeira, A.T.; Angelini, E.; Schiavon, N. Biodegradation and Microbial Contamination of Limestone Surfaces: An Experimental Study from Batalha Monastery, Portugal. Corros. Mater. Degrad. 2021, 2, 31–45. [Google Scholar] [CrossRef]
- Pangallo, D.; Chovanová, K.; Šimonovičová, A.; Ferianc, P. Investigation of Microbial Community Isolated from Indoor Artworks and Air Environment: Identification, Biodegradative Abilities, and DNA Typing. Can. J. Microbiol. 2009, 55, 277–287. [Google Scholar] [CrossRef]
- Slatko, B.E.; Gardner, A.F.; Ausubel, F.M. Overview of Next-Generation Sequencing Technologies. Curr. Protoc. Mol. Biol. 2018, 122, e59. [Google Scholar] [CrossRef]
- McNamara, C.J.; Perry, T.D., IV; Bearce, K.A.; Hernandez-Duque, G.; Mitchell, R. Epilithic and Endolithic Bacterial Communities in Limestone from a Maya Archaeological Site. Microb. Ecol. 2006, 51, 51–64. [Google Scholar] [CrossRef]
- Chimienti, G.; Piredda, R.; Pepe, G.; van der Werf, I.D.; Sabbatini, L.; Crecchio, C.; Ricciuti, P.; D’Erchia, A.M.; Manzari, C.; Pesole, G. Profile of Microbial Communities on Carbonate Stones of the Medieval Church of San Leonardo Di Siponto (Italy) by Illumina-Based Deep Sequencing. Appl. Microbiol. Biotechnol. 2016, 100, 8537–8548. [Google Scholar] [CrossRef]
- Miller, A.Z.; Laiz, L.; Gonzalez, J.M.; Dionísio, A.; Macedo, M.F.; Saiz-Jimenez, C. Reproducing Stone Monument Photosynthetic-Based Colonization under Laboratory Conditions. Sci. Total Environ. 2008, 405, 278–285. [Google Scholar] [CrossRef]
- Miller, A.Z.; Laiz, L.; Dionísio, A.; Macedo, M.F.; Saiz-Jimenez, C. Growth of Phototrophic Biofilms from Limestone Monuments under Laboratory Conditions. Int. Biodeterior. Biodegrad. 2009, 63, 860–867. [Google Scholar] [CrossRef]
- Crispim, C.A.; Gaylarde, P.M.; Gaylarde, C.C.; Neilan, B.A. Deteriogenic Cyanobacteria on Historic Buildings in Brazil Detected by Culture and Molecular Techniques. Int. Biodeterior. Biodegrad. 2006, 57, 239–243. [Google Scholar] [CrossRef]
- Rizk, S.M.; Magdy, M.; De Leo, F.; Werner, O.; Rashed, M.A.-S.; Ros, R.M.; Urzì, C. Culturable and Unculturable Potential Heterotrophic Microbiological Threats to the Oldest Pyramids of the Memphis Necropolis, Egypt. Front. Microbiol. 2023, 14, 1167083. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Ruff, S.E.; Oskolkov, N.; Tierney, B.T.; Ryon, K.; Danko, D.; Mason, C.E.; Elhaik, E. The Microbial Biodiversity at the Archeological Site of Tel Megiddo (Israel). Front. Microbiol. 2023, 14, 1253371. [Google Scholar] [CrossRef] [PubMed]
- Skipper, P.J.A.; Skipper, L.K.; Dixon, R.A. A Metagenomic Analysis of the Bacterial Microbiome of Limestone, and the Role of Associated Biofilms in the Biodeterioration of Heritage Stone Surfaces. Sci. Rep. 2022, 12, 4877. [Google Scholar] [CrossRef]
- Lepinay, C.; Mihajlovski, A.; Touron, S.; Seyer, D.; Bousta, F.; Di Martino, P. Bacterial Diversity Associated with Saline Efflorescences Damaging the Walls of a French Decorated Prehistoric Cave Registered as a World Cultural Heritage Site. Int. Biodeterior. Biodegrad. 2018, 130, 55–64. [Google Scholar] [CrossRef]
- Li, T.; Cai, Y.; Ma, Q. Microbial Diversity on the Surface of Historical Monuments in Lingyan Temple, Jinan, China. Microb. Ecol. 2023, 85, 76–86. [Google Scholar] [CrossRef]
- Li, Q.; Zhang, B.; He, Z.; Yang, X. Distribution and Diversity of Bacteria and Fungi Colonization in Stone Monuments Analyzed by High-Throughput Sequencing. PLoS ONE 2016, 11, e0163287. [Google Scholar] [CrossRef] [PubMed]
- Gambino, M.; Lepri, G.; Štovícek, A.; Ghazayarn, L.; Villa, F.; Gillor, O.; Cappitelli, F. The Tombstones at the Monumental Cemetery of Milano Select for a Specialized Microbial Community. Int. Biodeterior. Biodegrad. 2021, 164, 105298. [Google Scholar] [CrossRef]
- Irit, N.; Hana, B.; Yifat, B.; Esti, K.-W.; Ariel, K. Insights into Bacterial Communities Associated with Petroglyph Sites from the Negev Desert, Israel. J. Arid. Environ. 2019, 166, 79–82. [Google Scholar] [CrossRef]
- Ponizovskaya, V.B.; Rebrikova, N.L.; Kachalkin, A.V.; Antropova, A.B.; Bilanenko, E.N.; Mokeeva, V.L. Micromycetes as Colonizers of Mineral Building Materials in Historic Monuments and Museums. Fungal Biol. 2019, 123, 290–306. [Google Scholar] [CrossRef]
- Trovão, J.; Gil, F.; Catarino, L.; Soares, F.; Tiago, I.; Portugal, A. Analysis of Fungal Deterioration Phenomena in the First Portuguese King Tomb Using a Multi-Analytical Approach. Int. Biodeterior. Biodegrad. 2020, 149, 104933. [Google Scholar] [CrossRef]
- Delegou, E.T.; Karapiperis, C.; Hilioti, Z.; Chasapi, A.; Valasiadis, D.; Alexandridou, A.; Rihani, V.; Kroustalaki, M.; Bris, T.; Ouzounis, C.A.; et al. Metagenomics of the Built Cultural Heritage: Microbiota Characterization of the Building Materials of the Holy Aedicule of the Holy Sepulchre in Jerusalem. Sci. Cult. 2022, 8, 59–83. [Google Scholar] [CrossRef]
- Mascaro, M.E.; Pellegrino, G.; Palermo, A.M. Analysis of Biodeteriogens on Architectural Heritage. An Approach of Applied Botany on a Gothic Building in Southern Italy. Sustainability 2022, 14, 34. [Google Scholar] [CrossRef]
- Paiva, D.S.; Fernandes, L.; Trovão, J.; Mesquita, N.; Tiago, I.; Portugal, A. Uncovering the Fungal Diversity Colonizing Limestone Walls of a Forgotten Monument in the Central Region of Portugal by High-Throughput Sequencing and Culture-Based Methods. Appl. Sci. 2022, 12, 10650. [Google Scholar] [CrossRef]
- Paiva, D.S.; Fernandes, L.; Pereira, E.; Trovão, J.; Mesquita, N.; Tiago, I.; Portugal, A. Exploring Differences in Culturable Fungal Diversity Using Standard Freezing Incubation—A Case Study in the Limestones of Lemos Pantheon (Portugal). J. Fungi 2023, 9, 501. [Google Scholar] [CrossRef] [PubMed]
- Paiva, D.S.; Trovão, J.; Fernandes, L.; Mesquita, N.; Tiago, I.; Portugal, A. Expanding the Microcolonial Black Fungi Aeminiaceae Family: Saxispiralis Lemnorum Gen. et Sp. Nov. (Mycosphaerellales), Isolated from Deteriorated Limestone in the Lemos Pantheon, Portugal. J. Fungi 2023, 9, 916. [Google Scholar] [CrossRef] [PubMed]
- Dias, L.; Rosado, T.; Candeias, A.; Mirão, J.; Caldeira, A.T. A Change in Composition, a Change in Colour: The Case of Limestone Sculptures from the Portuguese National Museum of Ancient Art. J. Cult. Herit. 2020, 42, 255–262. [Google Scholar] [CrossRef]
- Soares, F.; Portugal, A.; Trovão, J.; Coelho, C.; Mesquita, N.; Pinheiro, A.C.; Gil, F.; Catarino, L.; Cardoso, S.M.; Tiago, I. Structural Diversity of Photoautotrophic Populations within the UNESCO Site ‘Old Cathedral of Coimbra’ (Portugal), Using a Combined Approach. Int. Biodeterior. Biodegrad. 2019, 140, 9–20. [Google Scholar] [CrossRef]
- Trovão, J.; Portugal, A.; Soares, F.; Paiva, D.S.; Mesquita, N.; Coelho, C.; Pinheiro, A.C.; Catarino, L.; Gil, F.; Tiago, I. Fungal Diversity and Distribution across Distinct Biodeterioration Phenomena in Limestone Walls of the Old Cathedral of Coimbra, UNESCO World Heritage Site. Int. Biodeterior. Biodegrad. 2019, 142, 91–102. [Google Scholar] [CrossRef]
- Coelho, C.; Mesquita, N.; Costa, I.; Soares, F.; Trovão, J.; Freitas, H.; Portugal, A.; Tiago, I. Bacterial and Archaeal Structural Diversity in Several Biodeterioration Patterns on the Limestone Walls of the Old Cathedral of Coimbra. Microorganisms 2021, 9, 709. [Google Scholar] [CrossRef]
- Soares, F.; Ramos, V.; Trovão, J.; Cardoso, S.M.; Tiago, I.; Portugal, A. Parakomarekiella Sesnandensis Gen. et Sp. Nov. (Nostocales, Cyanobacteria) Isolated from the Old Cathedral of Coimbra, Portugal (UNESCO World Heritage Site). Eur. J. Phycol. 2021, 56, 301–315. [Google Scholar] [CrossRef]
- Ahmed, E.A.-E.; Mohamed, R.M. Bacterial Deterioration in the Limestone Minaret of Prince Muhammad and Suggested Treatment Methods, Akhmim, Egypt. Geomaterials 2022, 12, 37–58. [Google Scholar] [CrossRef]
- Soares, F.; Trovão, J.; Portugal, A. Phototrophic and Fungal Communities Inhabiting the Roman Cryptoporticus of the National Museum Machado de Castro (UNESCO Site, Coimbra, Portugal). World J. Microbiol. Biotechnol. 2022, 38, 157. [Google Scholar] [CrossRef]
- Trovão, J.; Soares, F.; Paiva, D.S.; Tiago, I.; Portugal, A. Circumfusicillium Cavernae Gen. et Sp. Nov. (Bionectriaceae, Hypocreales) Isolated from a Hypogean Roman Cryptoporticus. J. Fungi 2022, 8, 837. [Google Scholar] [CrossRef] [PubMed]
- Balland-Bolou-Bi, C.; Saheb, M.; Alphonse, V.; Livet, A.; Reboah, P.; Abbad-Andaloussi, S.; Verney-Carron, A. Effect of Cultivable Bacteria and Fungi on the Limestone Weathering Used in Historical Buildings. Diversity 2023, 15, 587. [Google Scholar] [CrossRef]
- Langille, M.G.I.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Vega Thurber, R.L.; Knight, R.; et al. Predictive Functional Profiling of Microbial Communities Using 16S RRNA Marker Gene Sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef]
- Scheerer, S.; Ortega-Morales, O.; Gaylarde, C. Chapter 5 Microbial Deterioration of Stone Monuments—An Updated Overview. Adv. Appl. Microbiol. 2009, 66, 97–139. [Google Scholar]
- Warscheid, T.; Braams, J. Biodeterioration of Stone: A Review. Int. Biodeterior. Biodegrad. 2000, 46, 343–368. [Google Scholar] [CrossRef]
- Gadd, G.M.; Dyer, T.D. Bioprotection of the Built Environment and Cultural Heritage. Microb. Biotechnol. 2017, 10, 1152–1156. [Google Scholar] [CrossRef]
- Cozzolino, A.; Adamo, P.; Bonanomi, G.; Motti, R. The Role of Lichens, Mosses, and Vascular Plants in the Biodeterioration of Historic Buildings: A Review. Plants 2022, 11, 3429. [Google Scholar] [CrossRef]
- Salvadori, O.; Municchia, A.C. The Role of Fungi and Lichens in the Biodeterioration of Stone Monuments. Open Conf. Proc. J. 2016, 7, 39–54. [Google Scholar] [CrossRef]
- Liu, X.; Qian, Y.; Wu, F.; Wang, Y.; Wang, W.; Gu, J.D. Biofilms on Stone Monuments: Biodeterioration or Bioprotection? Trends Microbiol. 2022, 30, 816–819. [Google Scholar] [CrossRef] [PubMed]
- Trovão, J.; Tiago, I.; Catarino, L.; Gil, F.; Portugal, A. In Vitro Analyses of Fungi and Dolomitic Limestone Interactions: Bioreceptivity and Biodeterioration Assessment. Int. Biodeterior. Biodegrad. 2020, 155, 105107. [Google Scholar] [CrossRef]
- Jin, C.; Yu, R.; Shui, Z. Fungi: A Neglected Candidate for the Application of Self-Healing Concrete. Front. Built Environ. 2018, 4, 62. [Google Scholar] [CrossRef]
- Tiano, P. Biodeterioration of Monumental Rocks: Decay Mechanisms and Control Methods. Sci. Technol. Cult. Herit. 1998, 7, 19–38. [Google Scholar]
- Monte, M.D.; Sabbioni, C. Chemical and Biological Weathering of an Historical Building: Reggio Emilia Cathedral; Elsevier: Amsterdam, The Netherlands, 1986; Volume 50. [Google Scholar]
- Gadd, G.M.; Bahri-Esfahani, J.; Li, Q.; Rhee, Y.J.; Wei, Z.; Fomina, M.; Liang, X. Oxalate Production by Fungi: Significance in Geomycology, Biodeterioration and Bioremediation. Fungal Biol. Rev. 2014, 28, 36–55. [Google Scholar] [CrossRef]
- Bindschedler, S.; Cailleau, G.; Verrecchia, E. Role of Fungi in the Biomineralization of Calcite. Minerals 2016, 6, 41. [Google Scholar] [CrossRef]
- Gorbushina, A.A.; Kotlova, E.R.; Sherstneva, O.A. Cellular Responses of Microcolonial Rock Fungi to Long-Term Desiccation and Subsequent Rehydration. Stud. Mycol. 2008, 61, 91–97. [Google Scholar] [CrossRef]
- Pyzik, A.; Ciuchcinski, K.; Dziurzynski, M.; Dziewit, L. The Bad and the Good—Microorganisms in Cultural Heritage Environments—An Update on Biodeterioration and Biotreatment Approaches. Materials 2021, 14, 177. [Google Scholar] [CrossRef]
- Trovão, J.; Portugal, A. Current Knowledge on the Fungal Degradation Abilities Profiled through Biodeteriorative Plate Essays. Appl. Sci. 2021, 11, 4196. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).