Vermicomposting Enhances Microbial Detoxification of Sewage Sludge, Enabling Potential Application of the Treated Product in Agroecosystems
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sewage Sludge, Vermicomposting Set-Up and Earthworm Cast Sampling
2.2. Amplification, Sequencing and Analysis of 16S and ITS rRNA
2.3. Bioinformatic and Statistical Analysis
3. Results
3.1. Changes in the Composition of Bacterial and Fungal Communities
3.2. Changes in the α- and β-Diversity of Bacterial and Fungal Communities
3.3. Shared Bacterial and Fungal ASVs among Sewage Sludge, Fresh Earthworm Casts and Vermicompost
3.4. Functional Diversity of the Bacterial Communities
3.5. Fungal Trophic Guilds
4. Discussion
4.1. Composition of Bacterial and Fungal Communities during Vermicomposting of Sewage Sludge
4.2. Changes in the α- and β-Diversity during Vermicomposting of Sewage Sludge
4.3. Bacterial Functional Diversity and Fungal Trophic Guilds
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Souza, A.C.Z.; Santos, J.E.; Marin-Morales, M.A.; Mazzeo, D.E.C. Ecotoxicological aspects and environmental implications of the use of water and sewage treatment sludges. Int. J. Environ. Sci. Technol. 2024, 21, 3527–3552. [Google Scholar] [CrossRef]
- Domínguez, J.; Aira, M.; Crandall, K.A.; Pérez-Losada, M. Earthworms drastically change fungal and bacterial communities during vermicomposting of sewage sludge. Sci. Rep. 2021, 11, 15556. [Google Scholar] [CrossRef] [PubMed]
- Košnář, Z.; Mercl, F.; Pierdonà, L.; Chane, A.D.; Míchal, P.; Tlustoš, P. Concentration of the main persistent organic pollutants in sewage sludge in relation to wastewater treatment plant parameters and sludge stabilisation. Environ. Pollut. 2023, 333, 122060. [Google Scholar] [CrossRef] [PubMed]
- Wluka, A.-K.; Huang, Y.; Coenen, L.; Dsikowitzky, L.; Schwarzbauer, J. Structural Diversity of Organic Contaminants in Sewage Sludge: A Comparison of Sewage Fingerprints from Germany and China. Discov. Water 2021, 1, 4. [Google Scholar] [CrossRef]
- Domínguez, J.; Gómez-Brandón, M. Vermicomposting: Composting with earthworms to recycle organic wastes. In Management of Organic Waste; Kumar, S., Ed.; InTech: London, UK, 2012; ISBN 978-953-307-925-7. [Google Scholar]
- Swati, A.; Hait, S. A Comprehensive Review of the Fate of Pathogens during Vermicomposting of Organic Wastes. J. Environ. Qual. 2018, 47, 16–29. [Google Scholar] [CrossRef]
- Rodríguez-Canché, L.G.; Cardoso Vigueros, L.; Maldonado-Montiel, T.; Martínez-Sanmiguel, M. Pathogen Reduction in Septic Tank Sludge through Vermicomposting Using Eisenia Fetida. Bioresour. Technol. 2010, 101, 3548–3553. [Google Scholar] [CrossRef] [PubMed]
- Hénault-Ethier, L.; Martin, V.J.J.; Gélinas, Y. Persistence of Escherichia Coli in Batch and Continuous Vermicomposting Systems. Waste Manag. 2016, 56, 88–99. [Google Scholar] [CrossRef]
- Lv, B.; Xing, M.; Yang, J. Exploring the effects of earthworms on bacterial profiles during vermicomposting process of sewage sludge and cattle dung with high-throughput sequencing. Environ. Sci. Pollut. Res. 2018, 25, 12528–12537. [Google Scholar] [CrossRef]
- Chen, J.; Xia, H.; Huang, K.; Li, J.; Xie, J. Earthworms restructure the distribution of extracellular antibiotics resistance genes of sludge by modifying the structure of extracellular polymeric substances during vermicomposting. J. Hazard. Mater. 2023, 452, 131315. [Google Scholar] [CrossRef]
- Zhu, L.; Wu, Z.; Chen, J.; Huang, K. Removal of Antibiotic Resistance Genes in Sewage Sludge Vermicomposting. In Occurrence and Behavior of Emerging Contaminants in Organic Wastes and Their Control Strategies; Elsevier: Amsterdam, The Netherlands, 2024; pp. 169–177. ISBN 978-0-443-13585-9. [Google Scholar]
- El-Hassanin, A.S.; Samak, M.R.; Ahmed, S.M.; Afifi, M.M.I.; Abd El-Satar, A.M. Bioaccumulation of heavy metals during composting and vermicomposting processes of sewage sludge. Egypt J. Chem. 2022, 65, 1155–1162. [Google Scholar] [CrossRef]
- Dume, B.; Hanč, A.; Švehla, P.; Michal, P.; Pospíšil, V.; Grasserová, A.; Cajthaml, T.; Chane, A.D.; Nigussie, A. Influence of earthworms on the behaviour of organic micropollutants in sewage sludge. J. Clean. Prod. 2023, 416, 137869. [Google Scholar] [CrossRef]
- Lim, S.L.; Wu, T.Y.; Lim, P.N.; Shak, K.P.Y. The use of vermicompost in organic farming: Overview, effects on soil and economics. J. Sci. Food Agric. 2015, 95, 1143–1156. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Prakash, C.H.B.; Brar, N.S.; Kumar, B. Potential of Vermicompost for Sustainable Crop Production and Soil Health Improvement in Different Cropping Systems. Int. J. Curr. Microbiol. Appl. Sci. 2018, 7, 1042–1055. [Google Scholar] [CrossRef]
- Scaglia, B.; Nunes, R.R.; Rezende, M.O.O.; Tambone, F.; Adani, F. Investigating organic molecules responsible of auxin-like activity of humic acid fraction extracted from vermicompost. Sci. Total Environ. 2016, 562, 289–295. [Google Scholar] [CrossRef] [PubMed]
- Nsiah-Gyambibi, R.; Essandoh, H.M.K.; Asiedu, N.Y.; Fei-Baffoe, B. Valorization of fecal sludge stabilization via vermicomposting in microcosm enriched substrates using organic soils for vermicompost production. Heliyon 2021, 7, e06422. [Google Scholar] [CrossRef]
- Vuković, A.; Velki, M.; Ečimović, S.; Vuković, R.; Štolfa Čamagajevac, I.; Lončarić, Z. Vermicomposting—Facts, Benefits and Knowledge Gaps. Agronomy 2021, 11, 1952. [Google Scholar] [CrossRef]
- Lei, X.; Cui, G.; Sun, H.; Hou, S.; Deng, H.; Li, B.; Yang, Z.; Xu, Q.; Huo, X.; Cai, J. How do earthworms affect the pathway of sludge bio-stabilization via vermicomposting? Sci. Total Environ. 2024, 916, 170411. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, V.; Squartini, A.; Masi, A.; Sarkar, A.; Singh, R.P. Metabarcoding Analysis of the Bacterial Succession during Vermicomposting of Municipal Solid Waste Employing the Earthworm Eisenia fetida. Sci. Total Environ. 2021, 766, 144389. [Google Scholar] [CrossRef]
- Xing, M.; Zhao, R.; Yang, G.; Li, Z.; Sun, Y.; Xue, Z. Elimination of Antibiotic-Resistant Bacteria and Resistance Genes by Earthworms during Vermifiltration Treatment of Excess Sludge. Environ. Sci. Pollut. Res. 2024, 31, 7853–7871. [Google Scholar] [CrossRef]
- Zhang, H.; Li, J.; Zhang, Y.; Huang, K. Quality of Vermicompost and Microbial Community Diversity Affected by the Contrasting Temperature during Vermicomposting of Dewatered Sludge. Int. J. Environ. Res. Public Health 2020, 17, 1748. [Google Scholar] [CrossRef]
- Hu, X.; Zhang, T.; Tian, G.; Zhang, L.; Bian, B. Pilot-scale vermicomposting of sewage sludge mixed with mature vermicompost using earthworm reactor of frame composite structure. Sci. Total Environ. 2021, 767, 144217. [Google Scholar] [CrossRef]
- Zhang, Y.; Yuan, H.; Peng, S.; Wang, Z.; Cai, S.; Chen, Z.; Yang, B.; Yang, P.; Wang, D.; Guo, J.; et al. Vermicomposting Preferably Alters Fungal Communities in Wasted Activated Sludge and Promotes the Production of Plant Growth-Promoting Biostimulants in the Vermicompost. Chem. Eng. J. 2024, 495, 153232. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Holmes, S.P. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J. 2017, 11, 2639–2643. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. Updating the 97% Identity Threshold for 16S Ribosomal RNA OTUs. Bioinformatics 2018, 34, 2371–2375. [Google Scholar] [CrossRef]
- Bartlett, A.; Padfield, D.; Lear, L.; Bendall, R.; Vos, M. A Comprehensive List of Bacterial Pathogens Infecting Humans. Microbiology 2022, 168, 001269. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; McGowan, L.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the Tidyverse. J. Open Source Softw. 2019, 4, 1686. [Google Scholar] [CrossRef]
- Neuwirth, E. R Color Brewer: Color Brewer Palettes 2022. Available online: https://cran.r-project.org/web/packages/RColorBrewer/RColorBrewer.pdf (accessed on 18 March 2024).
- Pedersen, T.L. Patchwork: The Composer of Plots 2024. Available online: https://cran.r-project.org/web/packages/patchwork/patchwork.pdf (accessed on 18 March 2024).
- Oksanen, J.; Simpson, G.L.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’Hara, R.B.; Solymos, P.; Stevens, M.H.H.; Szoecs, E.; et al. Vegan: Community Ecology Package 2022. Available online: https://www.researchgate.net/publication/360782912_vegan_community_ecology_package_version_26-2_April_2022 (accessed on 18 March 2024).
- Galili, T. Dendextend: An R package for visualizing, adjusting and comparing trees of hierarchical clustering. Bioinformatics 2015, 31, 3718–3720. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z. Complex heatmap visualization. iMeta 2022, 1, e43. [Google Scholar] [CrossRef] [PubMed]
- R Core Team R: A Language and Environment for Statistical Computing 2023. Available online: https://www.R-project.org/ (accessed on 18 March 2024).
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Faith, D.P. Conservation evaluation and phylogenetic diversity. Biol. Conserv. 1992, 61, 1–10. [Google Scholar] [CrossRef]
- Lozupone, C.A.; Hamady, M.; Kelley, S.T.; Knight, R. Quantitative and qualitative β diversity measures lead to different insights into factors that structure microbial communities. Appl. Environ. Microbiol. 2007, 73, 1576–1585. [Google Scholar] [CrossRef]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2: An improved and extensible approach for metagenome inference. BioRxiv 2019, 672295. [Google Scholar] [CrossRef]
- Rosado, D.; Lores, M.; Ramos-Tapia, I.; Crandall, K.A.; Pérez-Losada, M.; Domínguez, J. Integrated Fertilization with Bagasse Vermicompost Changes the Microbiome of Mencía Must and Wine. Fermentation 2022, 8, 357. [Google Scholar] [CrossRef]
- Rosado, D.; Ramos-Tapia, I.; Crandall, K.A.; Pérez-Losada, M.; Domínguez, J. Grapevine treatment with bagasse vermicompost changes the microbiome of Albariño must and wine and improves wine quality. OENO One 2022, 56, 219–230. [Google Scholar] [CrossRef]
- Douglas, G.M.; Kim, S.; Langille, M.G.I.; Shapiro, B.J. Efficient computation of contributional diversity metrics from microbiome data with FuncDiv. Bioinformatics 2023, 39, btac809. [Google Scholar] [CrossRef]
- Tenenbaum, D. KEGGREST: Client-Side REST Access to the Kyoto Encyclopedia of Genes and Genomes (KEGG). R Package Version 1.44.0. Available online: https://bioconductor.org/packages/release/bioc/html/KEGGREST.html (accessed on 3 June 2024).
- Nguyen, N.H.; Song, Z.; Bates, S.T.; Branco, S.; Tedersoo, L.; Menke, J.; Schilling, J.S.; Kennedy, P.G. FUNGuild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 2016, 20, 241–248. [Google Scholar] [CrossRef]
- Kolbe, A.R.; Aira, M.; Gómez-Brandón, M.; Pérez-Losada, M.; Domínguez, J. Bacterial succession and functional diversity during vermicomposting of the white grape marc Vitis vinifera v. Albariño. Sci. Rep. 2019, 9, 7472. [Google Scholar] [CrossRef] [PubMed]
- Gómez Brandón, M.; Aira, M.; Kolbe, A.R.; de Andrade, N.; Pérez-Losada, M.; Domínguez, J. Rapid Bacterial Community Changes during Vermicomposting of Grape Marc Derived from Red Winemaking. Microorganisms 2019, 7, 473. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Brandón, M.; Aira, M.; Santana, N.; Pérez-Losada, M.; Domínguez, J. Temporal Dynamics of Bacterial Communities in a Pilot-Scale Vermireactor Fed with Distilled Grape Marc. Microorganisms 2020, 8, 642. [Google Scholar] [CrossRef] [PubMed]
- Domínguez, J.; Aira, M.; Kolbe, A.R.; Gómez-Brandón, M.; Pérez-Losada, M. Changes in the composition and function of bacterial communities during vermicomposting may explain beneficial properties of vermicompost. Sci. Rep. 2019, 9, 9657. [Google Scholar] [CrossRef]
- Rosado, D.; Pérez-Losada, M.; Aira, M.; Domínguez, J. Bacterial Succession during Vermicomposting of Silver Wattle (Acacia dealbata Link). Microorganisms 2021, 10, 65. [Google Scholar] [CrossRef]
- Yang, J.; Huang, K.; Peng, L.; Li, J.; Liu, A. Fate of Functional Bacterial and Eukaryotic Community Regulated by Earthworms during Vermicomposting of Dewatered Sludge, Studies Based on the 16S rDNA and 18S rDNA Sequencing of Active Cells. Int. J. Environ. Res. Public Health 2021, 18, 9713. [Google Scholar] [CrossRef]
- Lores, M.; Gómez-Brandón, M.; Pérez-Díaz, D.; Domínguez, J. Using FAME profiles for the characterization of animal wastes and vermicomposts. Soil. Biol. Biochem. 2006, 38, 2993–2996. [Google Scholar] [CrossRef]
- Commission Decision (EU) 2022/1244 of 13 July 2022 Establishing the EU Ecolabel Criteria for Growing Media and Soil Improvers (Notified under Document C(2022) 4758) (Text with EEA Relevance). 2022; Volume 190. Available online: https://www.isprambiente.gov.it/it/attivita/certificazioni/files/ecolabel/criteri/ammendanti/eur-lex-32022d1244-en-eur-lex.pdf (accessed on 29 August 2024).
- Directive (EU) 2015/2366 of the European Parliament and of the Council of 25 November 2015 on Payment Services in the Internal Market, Amending Directives 2002/65/EC, 2009/110/EC and 2013/36/EU and Regulation (EU) No 1093/2010, and Repealing Directive 2007/64/EC (Text with EEA Relevance). 2024. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/PDF/?uri=CELEX:32015L2366 (accessed on 29 August 2024).
- Cui, G.; Bhat, S.A.; Li, W.; Wei, Y.; Kui, H.; Fu, X.; Gui, H.; Wei, C.; Li, F. Gut digestion of earthworms significantly attenuates cell-free and -associated antibiotic resistance genes in excess activated sludge by affecting bacterial profiles. Sci. Total Environ. 2019, 691, 644–653. [Google Scholar] [CrossRef]
- Huang, K.; Xia, H.; Zhang, Y.; Li, J.; Cui, G.; Li, F.; Bai, W.; Jiang, Y.; Wu, N. Elimination of antibiotic resistance genes and human pathogenic bacteria by earthworms during vermicomposting of dewatered sludge by metagenomic analysis. Bioresour. Technol. 2020, 297, 122451. [Google Scholar] [CrossRef]
- Duan, Z.; Zhu, Y.; Xia, H.; Huang, K.; Peng, L. A Novel Strategy for Eliminating Antibiotic Resistance Genes during Fertilization of Dewatered Sludge by Earthworms: Vermicomposting Practice Using Chinese Herbal Residues Derived from Lianhua Qingwen as a Bulking Material. J. Environ. Manag. 2024, 349, 119444. [Google Scholar] [CrossRef]
- Huang, K.; Xia, H.; Wu, Y.; Chen, J.; Cui, G.; Li, F.; Chen, Y.; Wu, N. Effects of earthworms on the fate of tetracycline and fluoroquinolone resistance genes of sewage sludge during vermicomposting. Bioresour. Technol. 2018, 259, 32–39. [Google Scholar] [CrossRef] [PubMed]
- Xia, H.; Chen, J.; Chen, X.; Huang, K.; Wu, Y. Effects of tetracycline residuals on humification, microbial profile and antibiotic resistance genes during vermicomposting of dewatered sludge. Environ. Pollut. 2019, 252, 1068–1077. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Mao, H.; Li, X. Functional characteristics and influence factors of microbial community in sewage sludge composting with inorganic bulking agent. Bioresour. Technol. 2018, 249, 527–535. [Google Scholar] [CrossRef] [PubMed]
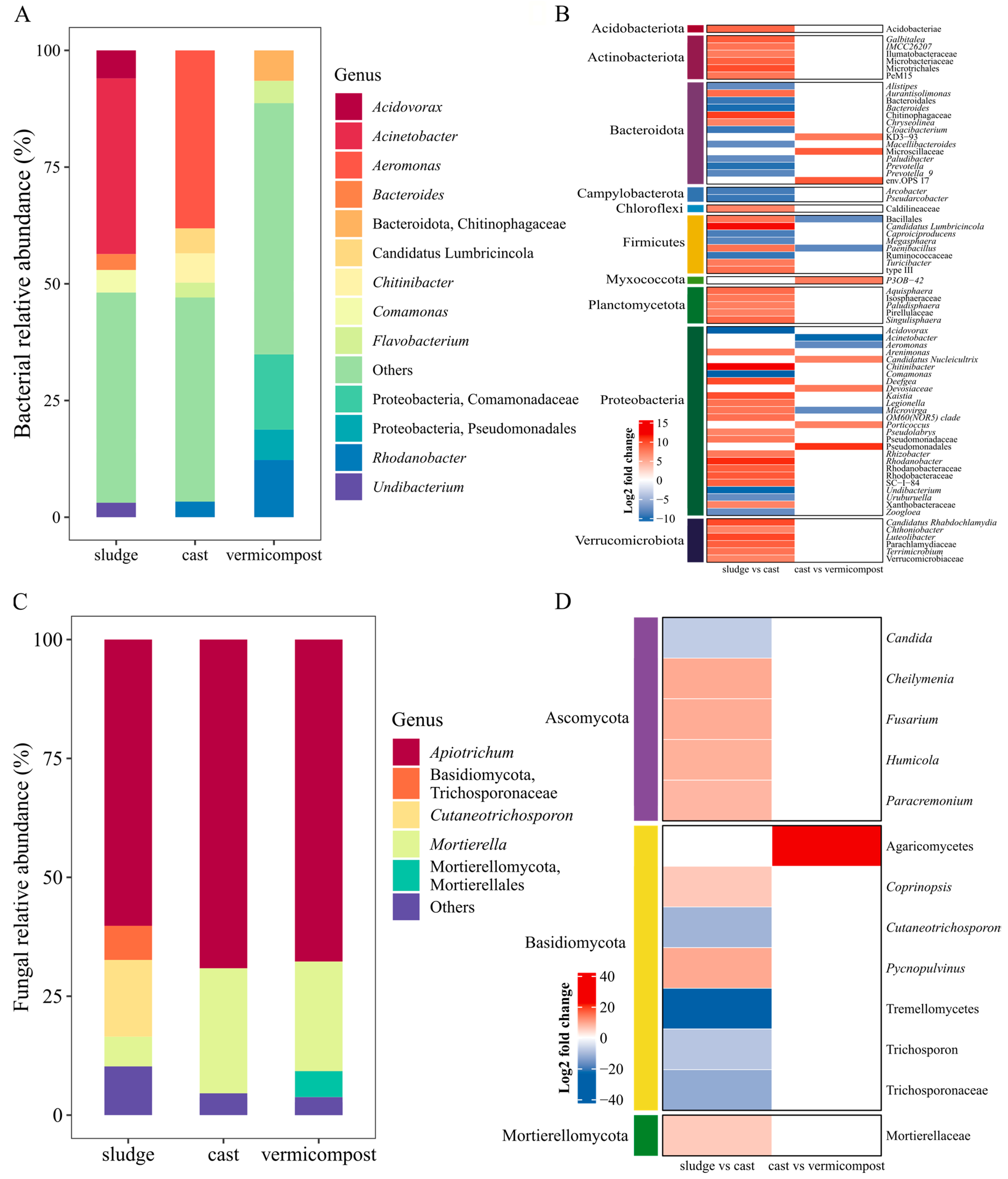
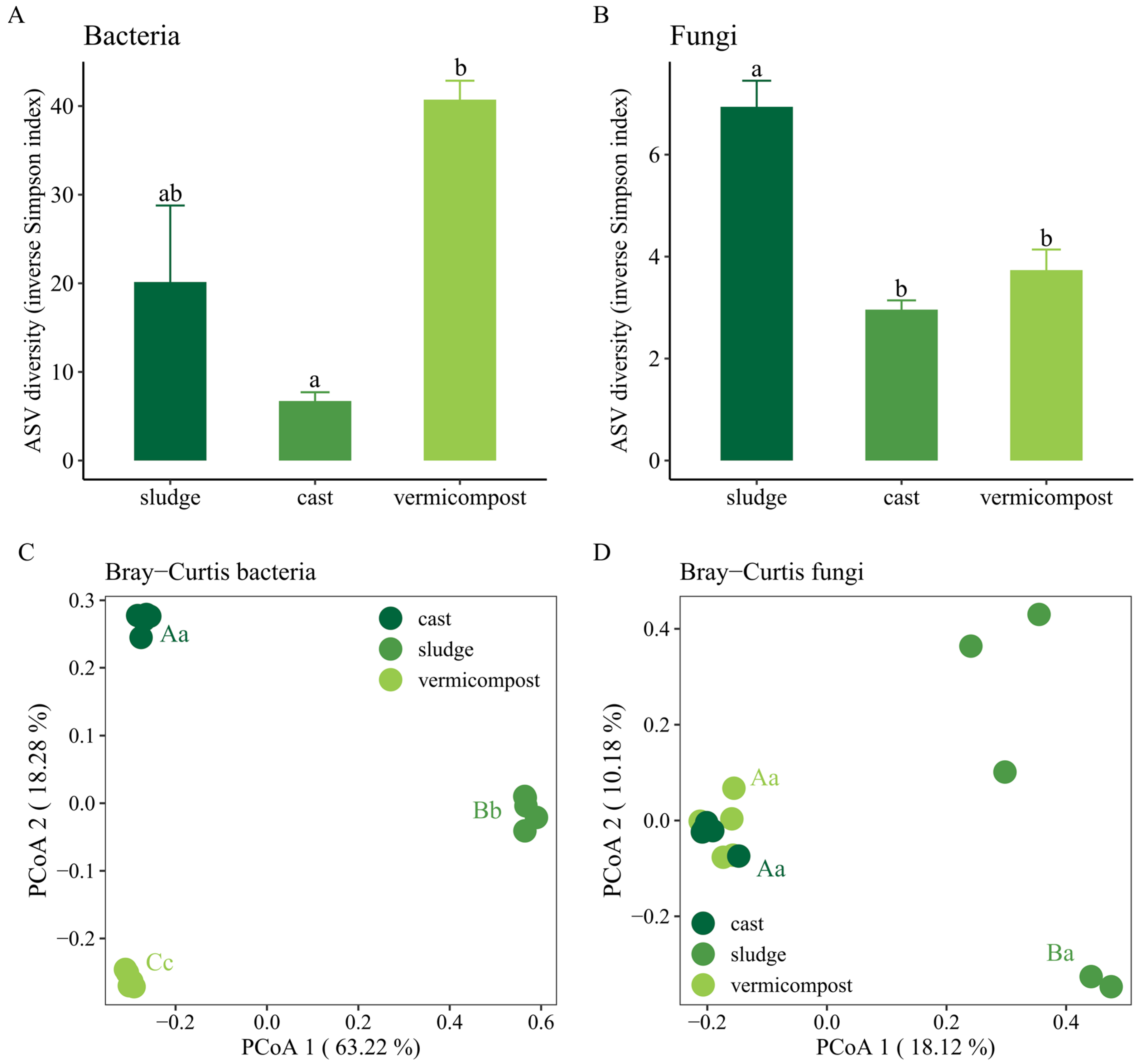
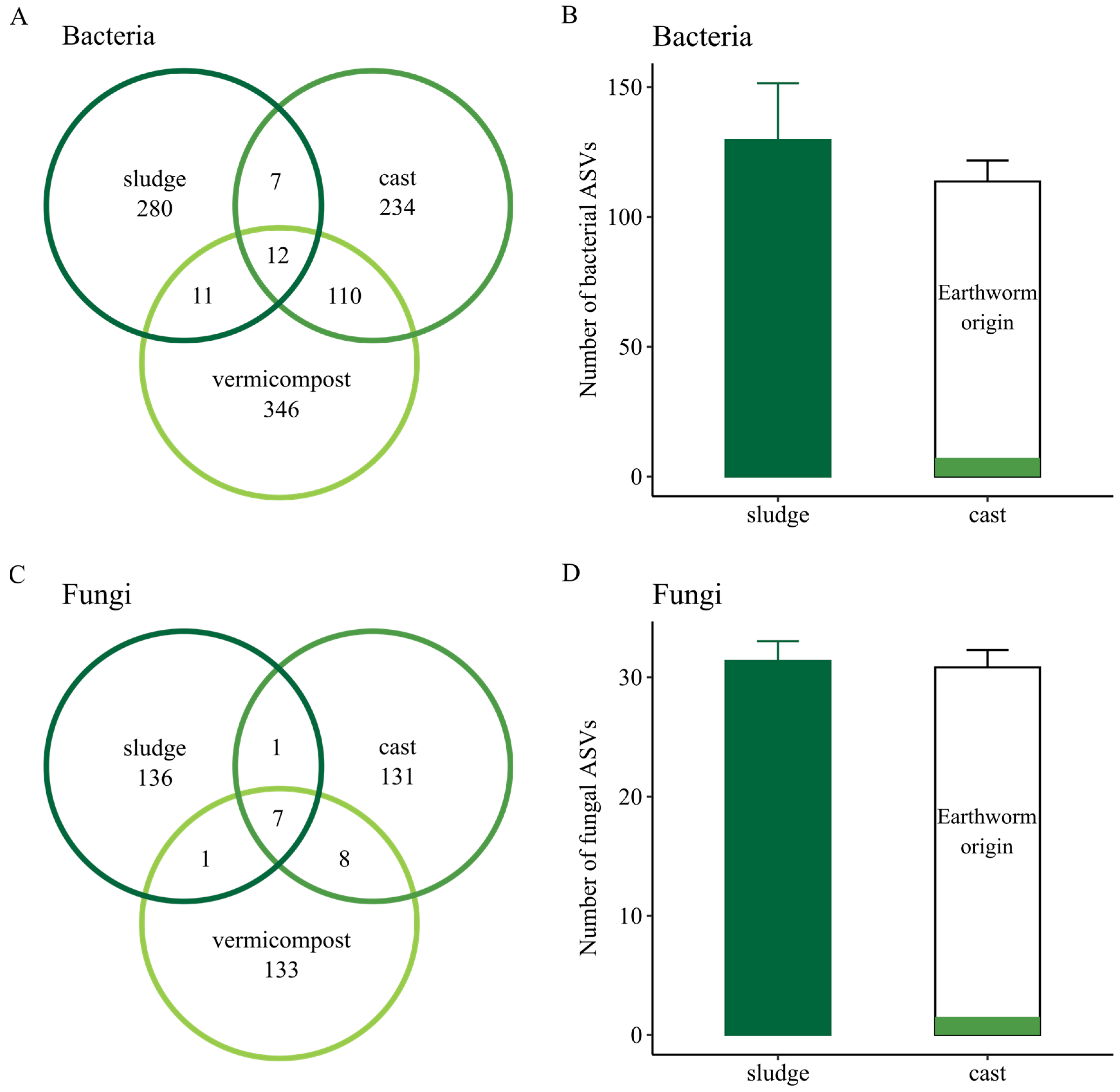
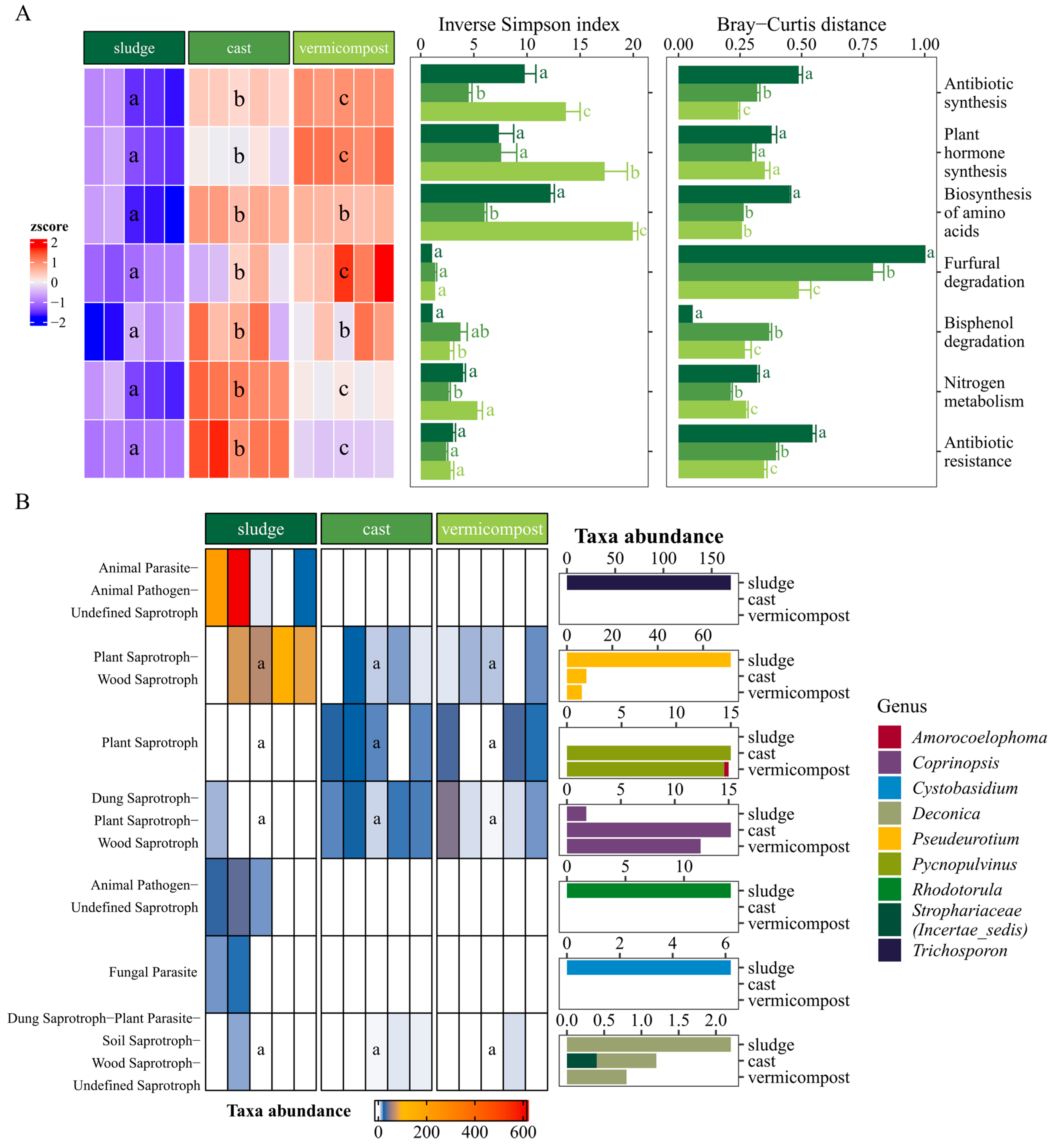
| Kingdom | Phylum | Sewage Sludge | Cast | Vermicompost |
|---|---|---|---|---|
| Bacteria | Proteobacteria | 60.13 ± 9.78 | 66.86 ± 1.37 | 57.91 ± 0.64 |
| Bacteria | Bacteroidota | 18.78 ± 4.22 | 7.07 ± 1.05 | 25.35 ± 0.64 |
| Bacteria | Firmicutes | 13.18 ± 3.19 | 11.66 ± 0.99 | 2.66 ± 0.18 |
| Bacteria | Campylobacterota | 3.96 ± 1.28 | - | - |
| Bacteria | Verrucomicrobiota | - | 5.08 ± 0.21 | 4.50 ± 0.17 |
| Bacteria | Actinobacteriota | - | 4.94 ± 0.7 | 1.91 ± 0.15 |
| Bacteria | Planctomycetota | - | 2.42 ± 0.16 | 3.22 ± 0.16 |
| Fungi | Basidiomycota | 87.80 ± 3.31 | 69.78 ± 2.44 | 68.53 ± 4.14 |
| Fungi | Mortierellomycota | 6.81 ± 4.21 | 27.68 ± 2.33 | 29.68 ± 4.11 |
| Fungi | Ascomycota | 5.32 ± 1.69 | 2.54 ± 0.28 | 1.80 ± 0.15 |
| Fungi | Mucoromycota | 0.06 ± 0.04 | - | - |
| ASVs | Species | Sludge | Cast | Vermicompost |
|---|---|---|---|---|
| ASV43 | Arcobacter cryaerophilus | 1.34 ± 0.72 | - | 0.01 ± 0.03 |
| ASV57 | Bacteroides vulgatus | 0.94 ± 0.46 | - | - |
| ASV67 | Bacteroides uniformis | 0.81 ± 0.4 | - | - |
| ASV194 | Collinsella aerofaciens | 0.2 ± 0.19 | - | 0.02 ± 0.02 |
| ASV208 | Sebaldella termitidis | 0.2 ± 0.06 | - | - |
| ASV246 | Bacteroides massiliensis | 0.16 ± 0.12 | - | - |
| ASV282 | Dialister invisus | 0.12 ± 0.08 | - | - |
| ASV480 | Bacteroides caccae | 0.04 ± 0.09 | - | - |
| ASV547 | Laribacter hongkongensis | 0.02 ± 0.06 | - | - |
| ASV643 | Bilophila wadsworthia | 0.02 ± 0.03 | - | - |
| ASV130 | Turicibacter sanguinis | - | 0.21 ± 0.14 | 0.14 ± 0.05 |
| ASV290 | Cellulosimicrobium cellulans | - | 0.1 ± 0.13 | - |
| ASV390 | Turicibacter sanguinis | - | 0.06 ± 0.14 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gómez-Roel, A.; Aira, M.; Domínguez, J. Vermicomposting Enhances Microbial Detoxification of Sewage Sludge, Enabling Potential Application of the Treated Product in Agroecosystems. Appl. Sci. 2024, 14, 7894. https://doi.org/10.3390/app14177894
Gómez-Roel A, Aira M, Domínguez J. Vermicomposting Enhances Microbial Detoxification of Sewage Sludge, Enabling Potential Application of the Treated Product in Agroecosystems. Applied Sciences. 2024; 14(17):7894. https://doi.org/10.3390/app14177894
Chicago/Turabian StyleGómez-Roel, Ana, Manuel Aira, and Jorge Domínguez. 2024. "Vermicomposting Enhances Microbial Detoxification of Sewage Sludge, Enabling Potential Application of the Treated Product in Agroecosystems" Applied Sciences 14, no. 17: 7894. https://doi.org/10.3390/app14177894






