Enhanced Precision in Chinese Medical Text Mining Using the ALBERT+Bi-LSTM+CRF Model
Abstract
:1. Introduction
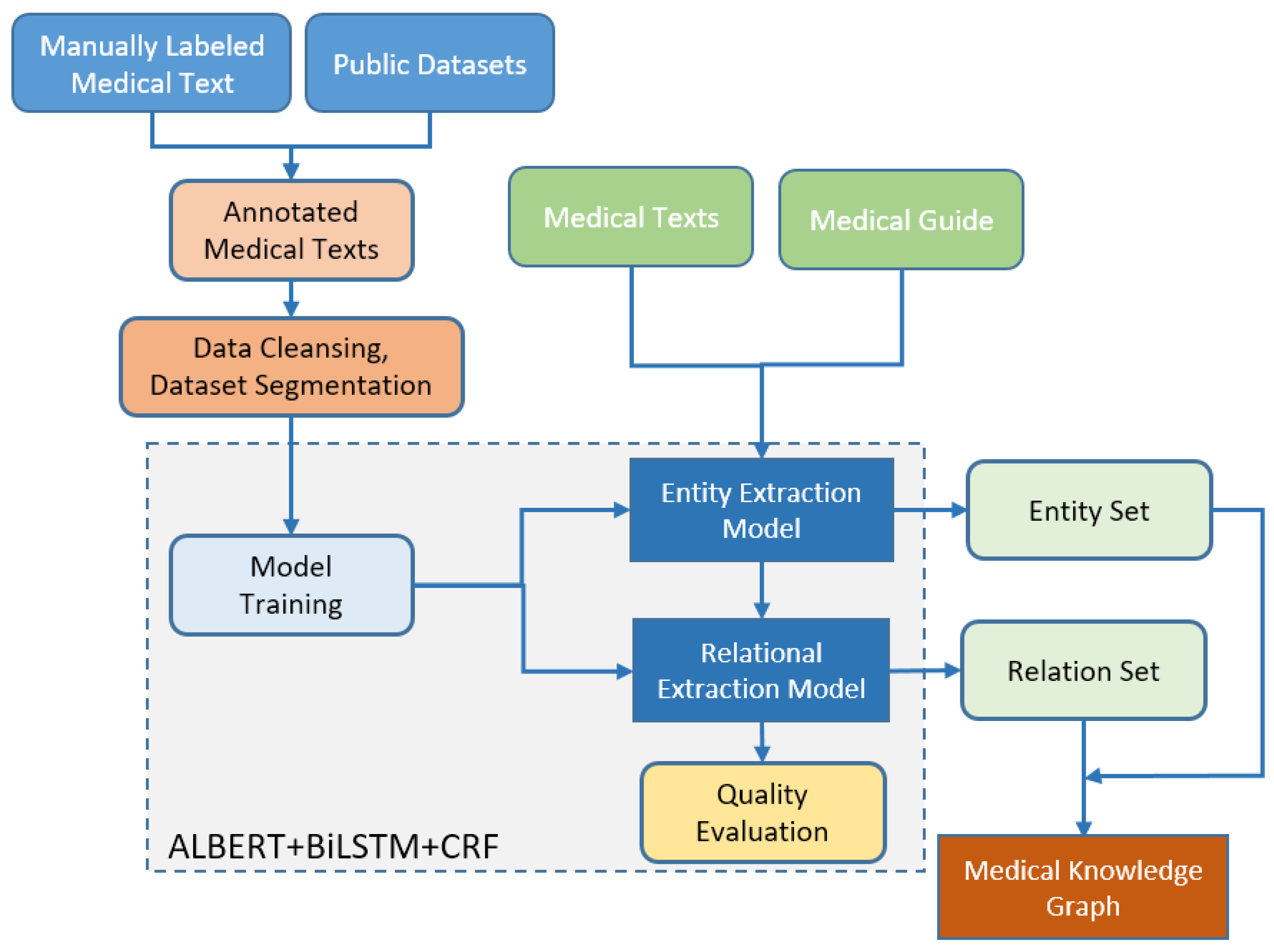
2. Construction of a Knowledge Graph
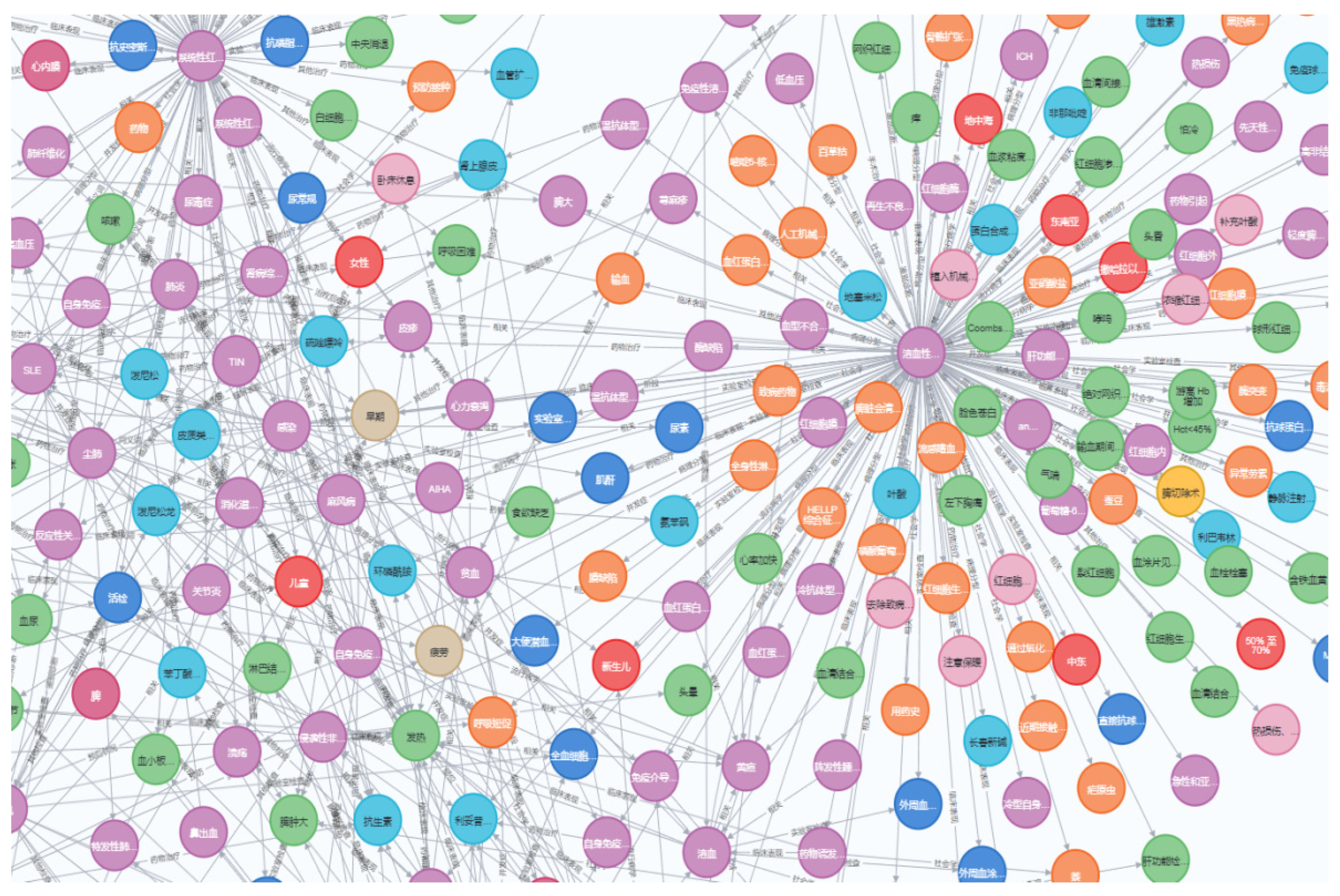
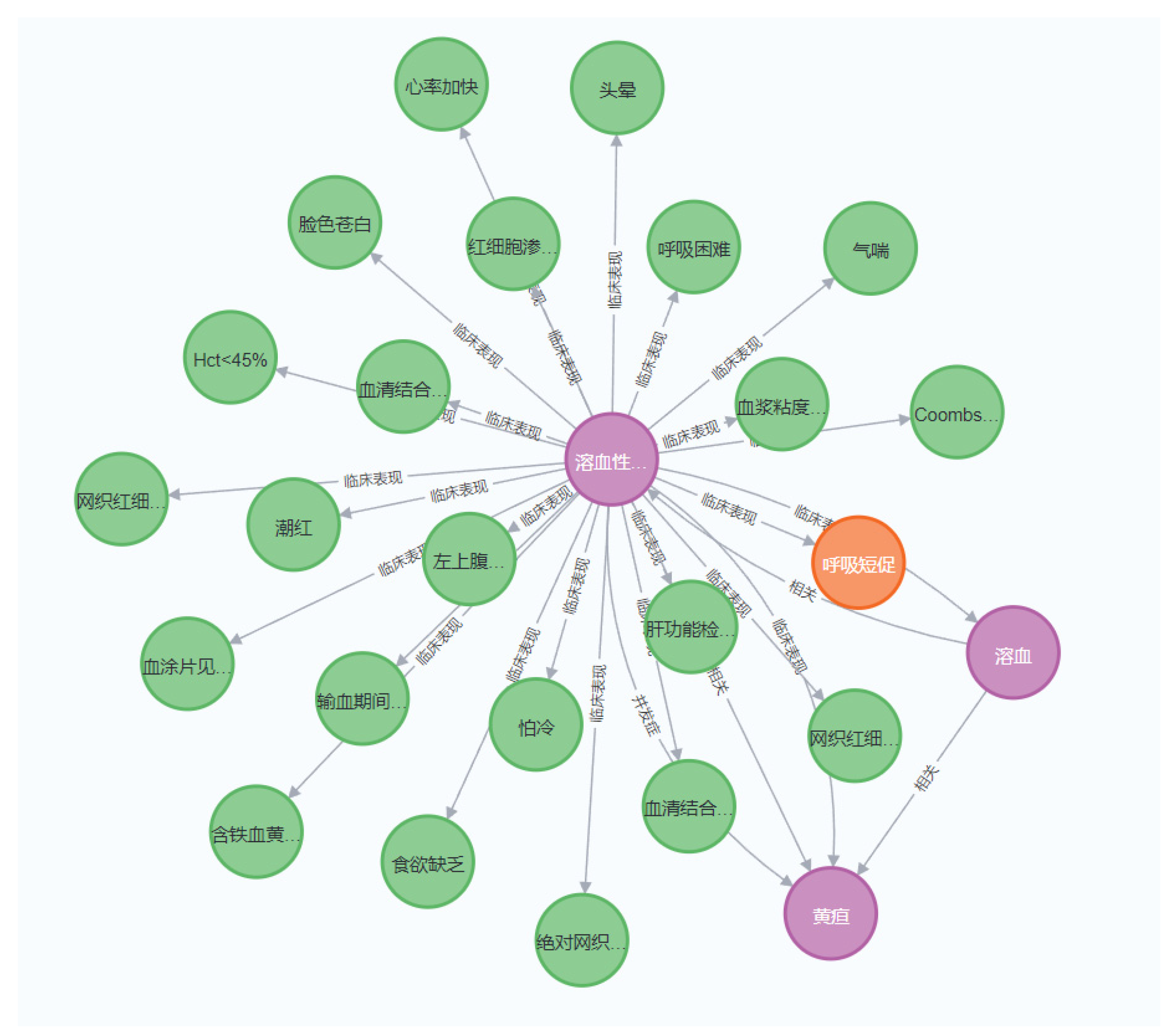
2.1. The Core Concepts of a Knowledge Graph
2.2. Architectural Design of a Knowledge Graph
3. Model Training
3.1. Choice of the ALBERT Algorithm
3.2. The Integration of Bi-LSTM with CRF
4. Experimental Process
4.1. Dataset
4.1.1. The Dataset for Entity Extraction
4.1.2. The Dataset for Relation Extraction
4.1.3. Annotation of Real Medical Texts
4.2. Data Preprocessing and Label Definition
4.2.1. The Definition of Entity Labels
4.2.2. The Definition of Relation Labels
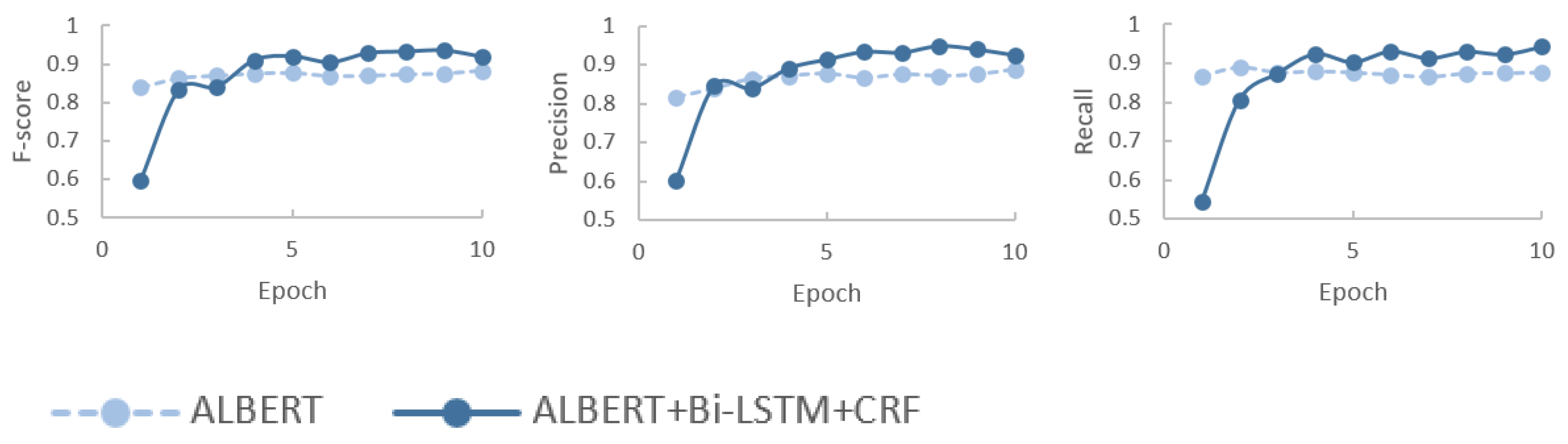
4.3. Results and Analysis
4.3.1. Results
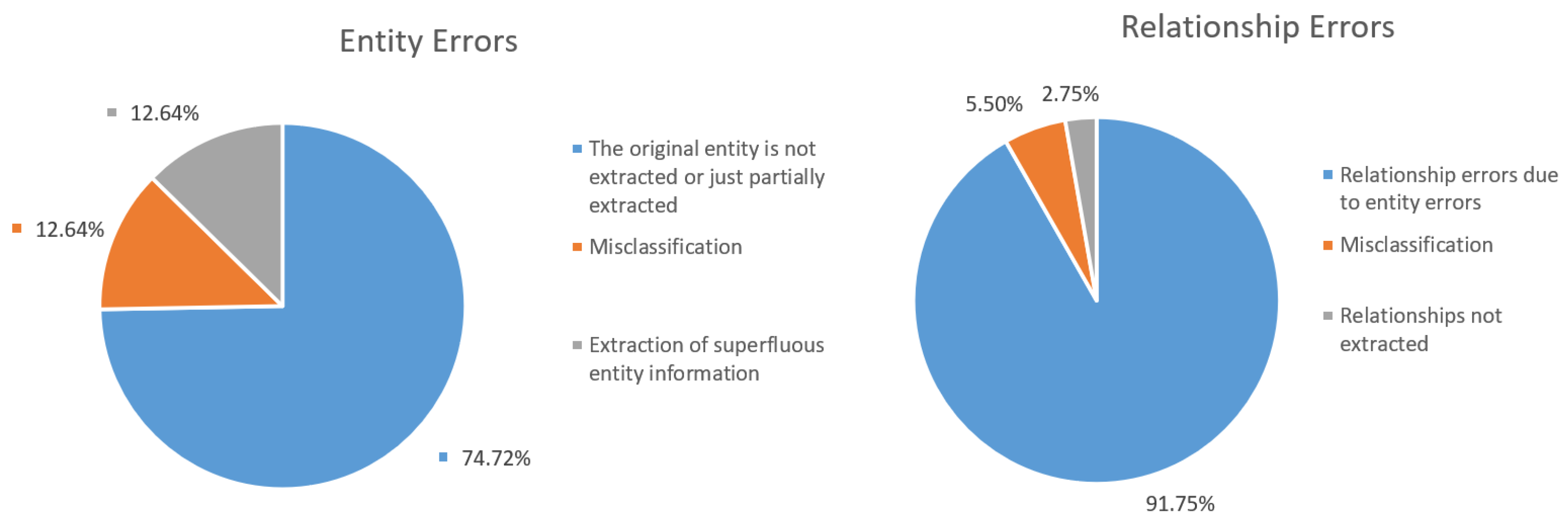
4.3.2. Error Analysis of Cases
5. Conclusions and Future Prospects
5.1. Discussion
5.2. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cai, Z.X.; Cai, Y.F. Clinical application and technology of smart medical. J. Med. Inform. 2021, 42, 48–53. (In Chinese) [Google Scholar]
- Li, C.L.; Zhao, C.; Si, Q.; Yan, M.L.; Li, Y.T.; Zhang, X. Development status and the future of smart medical treatment. Life Sci. Instrum. 2021, 19, 4–13. (In Chinese) [Google Scholar]
- Zhang, H.; Zong, Y.; Chang, B.B.; Sui, Z.F.; Zan, H.Y.; Zhang, K.L. Medical entity annotation specification for medical text processing. In Proceedings of the Chinese National Conference on Computational Linguistics, Haikou, China, 30 October–1 November 2020. (In Chinese). [Google Scholar]
- Wang, H.C.; Zhao, T.J. Research and development of biomedical text mining. J. Chin. Inf. Process. 2008, 22, 89–98. (In Chinese) [Google Scholar]
- Sun, Z.; Wang, H.L. Overview of the advance of the research on named entity recognition. Data Anal. Knowl. Discov. 2010, 6, 42–47. (In Chinese) [Google Scholar]
- Berners-Lee, T.; Hendler, J.; Lassila, O. The Semantic Web. Scientific American Magazine. Available online: https://www.scientificamerican.com/article/the-semantic-web/ (accessed on 1 May 2001).
- Sheth, A.; Thirunarayan, K. Semantics Empowered Web 3.0: Managing Enterprise, Social, Sensor, and Cloud-Based Data and Service for Advanced Applications; Morgan and Claypool: San Rafael, CA, USA, 2013. [Google Scholar]
- Amit, S. Introducing the Knowledge Graph. Official Blog of Google. Available online: http://googleblog.blogspot.pt/2012/05/introducing-knowledge-graph-things-not.html (accessed on 2 January 2015).
- Zhao, Y.H.; Liu, L.; Wang, H.L.; Han, H.Y.; Pei, D.M. Survey of knowledge graph recommendation system research. J. Front. Comput. Sci. Technol. 2023, 17, 771–791. (In Chinese) [Google Scholar]
- Mi, Z.H.; Qian, A.B. Research status and trend of smart healthcare: A literature review. Chin. Gen. Pract. 2019, 22, 366–370. (In Chinese) [Google Scholar]
- Wang, Z.C.; Feng, J.Y. Application of a digital health system based on the Internet of Things in China. Chin. Med. Devices 2022, 37, 174–179. (In Chinese) [Google Scholar]
- Lan, G.; Hu, M.T.; Li, Y.; Zhang, Y.Z. Contrastive knowledge integrated graph neural networks for Chinese medical text classification. Eng. Appl. Artif. Intell. 2023, 122, 106057. [Google Scholar] [CrossRef]
- Dong, X.L.; Zheng, W.F. Emerging technologies for drug repurposing: Harnessing the potential of text and graph embedding approaches. Artif. Intell. Chem. 2024, 2, 100060. [Google Scholar] [CrossRef]
- Guo, L.; Li, X.L.; Yan, F.; Lu, Y.Q.; Shen, W.P. A method for constructing a machining knowledge graph using an improved transformer. Expert Syst. Appl. 2024, 237, 121448. [Google Scholar] [CrossRef]
- Lin, Y.C.; Lu, K.M.; Yu, S.; Cai, T.X.; Zitnik, M. Multimodal learning on graphs for disease relation extraction. J. Biomed. Inform. 2023, 143, 104415. [Google Scholar] [CrossRef]
- Sung, Y.W.; Park, D.S.; Kim, C.G. A study of BERT-based classification performance of text-based health counseling data. Comput. Model. Eng. Sci. 2022, 135, 795–808. [Google Scholar]
- Wang, Y.M.; Zhang, J.D.; Yang, Z.Y.; Wang, B.; Jin, J.Y.; Liu, Y.T. Improving extractive summarization with semantic enhancement through topic-injection based BERT model. Inf. Process. Manag. 2024, 61, 103677. [Google Scholar] [CrossRef]
- Shiney, J.; Raghuveera, T. COVID-based question criticality prediction with domain adaptive BERT embeddings. Eng. Appl. Artif. Intell. 2024, 132, 107913. [Google Scholar]
- Devlin, J.; Chang, M.W.; Lee, K.; Toutanova, K. BERT: Pre-Training of Deep Bidirectional Transformers for Language Understanding. Available online: http://arxiv.org/abs/1810.04805 (accessed on 11 October 2018).
- Wu, Z.M.; Liang, J.; Zhang, Z.A.; Lei, J.B. Exploration of text matching methods in Chinese disease Q&A systems: A method using ensemble based on BERT and boosted tree models. J. Biomed. Inform. 2021, 115, 103683. [Google Scholar]
- Yang, P.R.; Wang, H.J.; Huang, Y.Z.; Yang, S.; Zhang, Y.; Huang, L.; Zhang, Y.S.; Wang, G.X.; Yang, S.Z.; He, L.; et al. LMKG: A large-scale and multi-source medical knowledge graph for intelligent medicine applications. Knowl. Based Syst. 2024, 284, 111323. [Google Scholar] [CrossRef]
- Xu, Z.L.; Sheng, Y.P.; He, L.R.; Wang, Y.F. Review on knowledge graph techniques. J. Univ. Electron. Sci. Technol. China 2016, 45, 589–606. (In Chinese) [Google Scholar]
- Liu, Q.; Li, Y.; Duan, H.; Liu, Y.; Qin, Z.G. Knowledge graph construction techniques. J. Comput. Res. Dev. 2016, 53, 582–600. (In Chinese) [Google Scholar]
- Huang, M.; Xu, G.J.; Li, H.L. Construction of personalized learning service system based on deep learning and knowledge graph. Appl. Math. Nonlinear Sci. 2024, 9. [Google Scholar] [CrossRef]
- Lan, Z.Z.; Chen, M.D.; Goodman, S.; Gimpel, K.; Sharma, P.; Soricut, R. ALBERT: A lite BERT for self-supervised learning of language representations. In Proceedings of the International Conference on Learning Representations, Virtual, 26 April–1 May 2020. [Google Scholar]
- Tu, Y.; Chi, M. E-Business. Digital Empowerment for an Intelligent Future; Springer: Cham, Switzerland, 2023. [Google Scholar]
- China Medical Knowledge Graph Research Association. CCKS2020 dataset. In Proceedings of the 8th Chinese Conference on Natural Language Processing and Chinese Computing, Dunhuang, China, 9–14 October 2019; pp. 100–110. [Google Scholar]
- Guan, P.; Zan, H.; Zhou, X.; Xu, H.; Zhang, K. CMeIE: Construction and evaluation of Chinese medical information extraction dataset. In Natural Language Processing and Chinese Computing, 9th CCF International Conference, Zhengzhou, China, 14–18 October 2020; Springer International Publishing: Berlin/Heidelberg, Germany, 2020. [Google Scholar]
- Li, Z.M.; Yun, H.Y.; Wang, Y.Z. Medical named entity recognition based on BERT with multi-feature fusion. J. Qingdao Univ. (Nat. Sci. Ed.) 2021, 34, 23–29. (In Chinese) [Google Scholar]
- Gao, W.C.; Zheng, X.H.; Zhao, S.S. Named entity recognition method of Chinese EMR based on BERT-BiLSTM-CRF. J. Phys. Conf. Ser. 2021, 1848, 012083. [Google Scholar] [CrossRef]
- Hou, M.W.; Wei, R.; Lu, L.; Lan, X.; Cai, H.W. A survey of knowledge graph research and its application in the medical field. Comput. Res. Dev. 2018, 55, 2587–2599. (In Chinese) [Google Scholar]
- Li, M.D.; Zhang, P.; Li, G.L.; Jiang, W.; Li, K.; Cai, P.Q. Study on Chinese medical named entity recognition algorithm. J. Med. Inform. 2022, 43, 45–51. (In Chinese) [Google Scholar]
- Tan, L.; E, H.H.; Kuang, Z.M.; Song, M.N.; Liu, Y.; Chen, Z.Y.; Xie, X.X.; Li, J.D.; Fan, J.W.; Wang, Q.C.; et al. Construction technologies and research development of medical knowledge graph. Appl. Res. Comput. 2021, 7, 80–104. (In Chinese) [Google Scholar]
- Huang, H.X.; Wang, X.Y.; Gu, Z.W.; Liu, J.; Zang, Y.N.; Sun, X. Research on construction technology and development status of the medical knowledge graph. Comput. Eng. Appl. 2023, 59, 33–48. (In Chinese) [Google Scholar]


| Entity Type | Train Set | Validation Set | Test Set | |||
|---|---|---|---|---|---|---|
| Entries | Percentage/% | Entries | Percentage/% | Entries | Percentage/% | |
| Body | 6061 | 45.4 | 1965 | 43.1 | 1990 | 43.8 |
| Operation | 606 | 4.5 | 216 | 4.7 | 217 | 4.8 |
| Disease | 3032 | 22.7 | 1069 | 23.4 | 932 | 20.5 |
| Examination | 860 | 6.4 | 282 | 6.2 | 252 | 5.6 |
| Drug | 2006 | 15.0 | 790 | 17.3 | 863 | 19.0 |
| Item | 776 | 5.8 | 214 | 4.7 | 313 | 6.9 |
| Relation Type | Train Set | Validation Set | Test Set | |||
|---|---|---|---|---|---|---|
| Entries | Percentage/% | Entries | Percentage/% | Entries | Percentage/% | |
| Epidemiological | 3895 | 4.5 | 1002 | 4.6 | 1043 | 4.8 |
| Prognosis | 388 | 0.1 | 110 | 0.5 | 107 | 0.5 |
| Sociological | 6649 | 7.6 | 1720 | 7.9 | 1756 | 8.1 |
| Synonyms | 4848 | 5.6 | 1144 | 5.2 | 1084 | 5.0 |
| Prevention | 2479 | 2.9 | 540 | 2.5 | 567 | 2.6 |
| Post-treatment | 147 | 0.1 | 37 | 0.1 | 55 | 0.3 |
| Imaging | 2238 | 2.6 | 505 | 2.3 | 521 | 2.4 |
| Medication | 6690 | 7.7 | 1547 | 7.1 | 1488 | 6.9 |
| Symptom | 22,373 | 25.7 | 5678 | 26.0 | 5621 | 25.9 |
| Pathological staging | 4304 | 4.9 | 1182 | 5.4 | 1145 | 5.3 |
| Stage | 1216 | 1.4 | 318 | 1.5 | 309 | 1.4 |
| Complications | 5187 | 6.0 | 1324 | 6.1 | 1297 | 6.0 |
| Other inspections | 1576 | 1.8 | 394 | 1.8 | 408 | 1.9 |
| Operation | 1681 | 1.9 | 404 | 1.8 | 391 | 1.8 |
| Diagnosis | 2900 | 3.3 | 754 | 3.4 | 776 | 3.6 |
| Body part | 4376 | 5.0 | 1130 | 5.2 | 1106 | 5.3 |
| Other treatments | 2848 | 3.3 | 715 | 3.3 | 703 | 3.2 |
| Laboratory | 3210 | 3.7 | 851 | 3.9 | 844 | 3.9 |
| Correlation | 9971 | 11.5 | 2529 | 11.6 | 2487 | 11.5 |
| Model | F1-Score/% | Precision/% | Recall/% |
|---|---|---|---|
| Word2vec-BiLSTM-CRF | 75.1 | 73.2 | 72.8 |
| Bi-LSTM+CRF | 88.6 | 88.2 | 88.4 |
| BERT+Bi-LSTM+CRF | 90.1 | 94.9 | 92.3 |
| ALBERT | 90.5 | 87.2 | 89.6 |
| ALBERT+Bi-LSTM+CRF | 91.8 | 92.5 | 94.3 |
| Model | F/% | Precision/% | Recall/% |
|---|---|---|---|
| ALBERT+Bi-LSTM+CRF | 88.3 | 88.1 | 88.4 |
| Data Source | Entries | Number of Triplets |
|---|---|---|
| Public datasets | 14,335 | 64,757 |
| Real hospital data | 13,580 | 172,354 |
| Diagnosis and treatment guidelines | 357 | 1010 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fang, T.; Yang, Y.; Zhou, L. Enhanced Precision in Chinese Medical Text Mining Using the ALBERT+Bi-LSTM+CRF Model. Appl. Sci. 2024, 14, 7999. https://doi.org/10.3390/app14177999
Fang T, Yang Y, Zhou L. Enhanced Precision in Chinese Medical Text Mining Using the ALBERT+Bi-LSTM+CRF Model. Applied Sciences. 2024; 14(17):7999. https://doi.org/10.3390/app14177999
Chicago/Turabian StyleFang, Tianshu, Yuanyuan Yang, and Lixin Zhou. 2024. "Enhanced Precision in Chinese Medical Text Mining Using the ALBERT+Bi-LSTM+CRF Model" Applied Sciences 14, no. 17: 7999. https://doi.org/10.3390/app14177999





