On-Edge Deployment of Vision Transformers for Medical Diagnostics Using the Kvasir-Capsule Dataset
Abstract
:1. Introduction
- Conduct a comprehensive evaluation of several state-of-the-art transformer-based CNN architectures specifically built for on-hand deployment on the Kvasir-Capsule dataset, namely, EfficientViT, MobileViT, EfficientFormer, and RepViT;
- Compare model performance based on different quantization methods available within the TFLite framework, namely, F16 quantization and QAT.
2. Literature Review
2.1. Kvasir-Capsule
2.2. On-Edge Deployment for Medical Diagnostics
3. Proposed System Architecture
3.1. Dataset
3.2. Neural Network Architectures
3.2.1. EfficientFormerV2
3.2.2. EfficientViT
3.2.3. MobileViT
3.2.4. RepViT
3.3. Quantization for On-Edge Classification
3.4. Evaluation Metrics
4. Results and Discussion
4.1. Model Evaluation
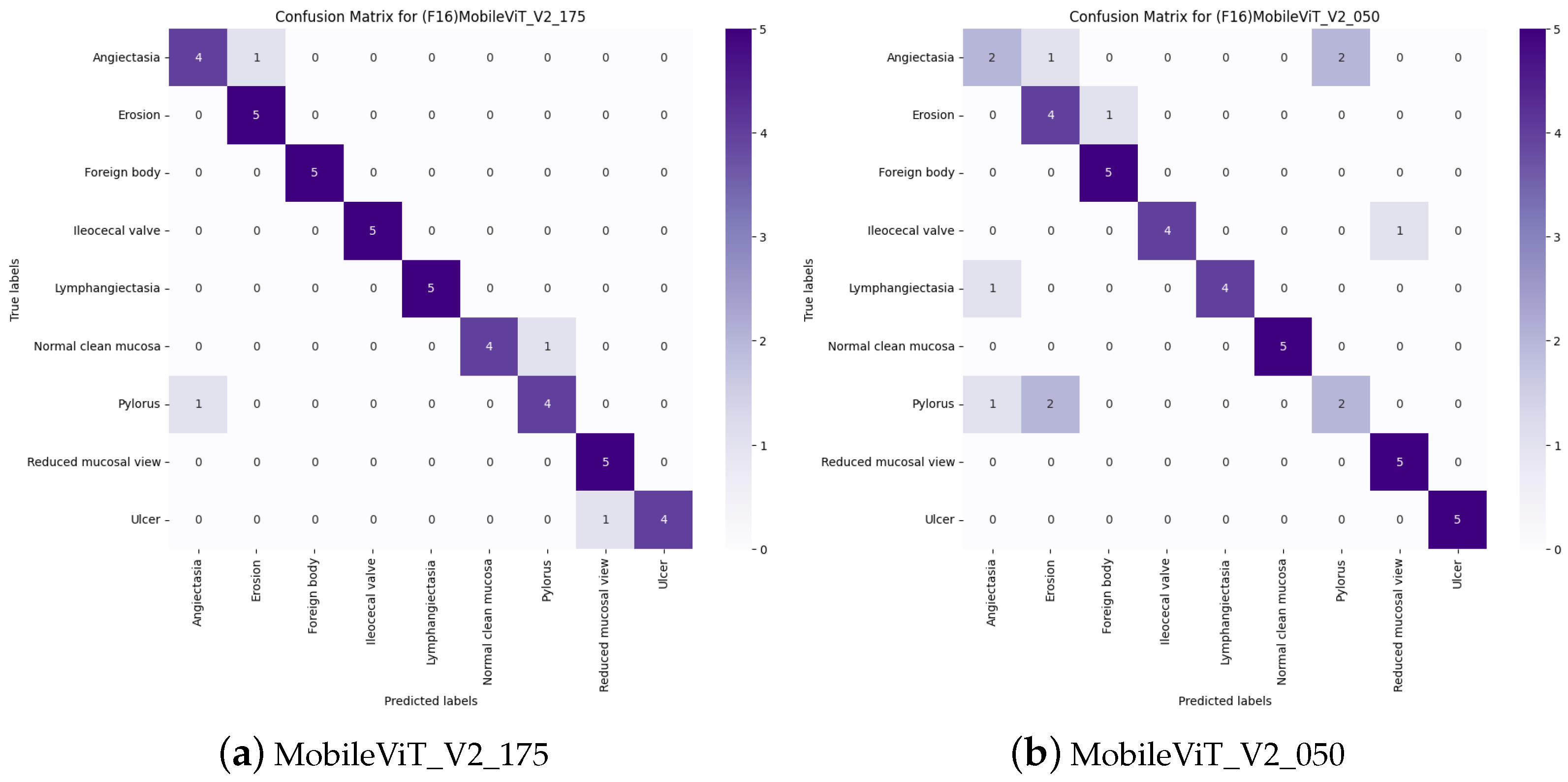
4.1.1. Model F1-Scores
4.1.2. Model Sizes
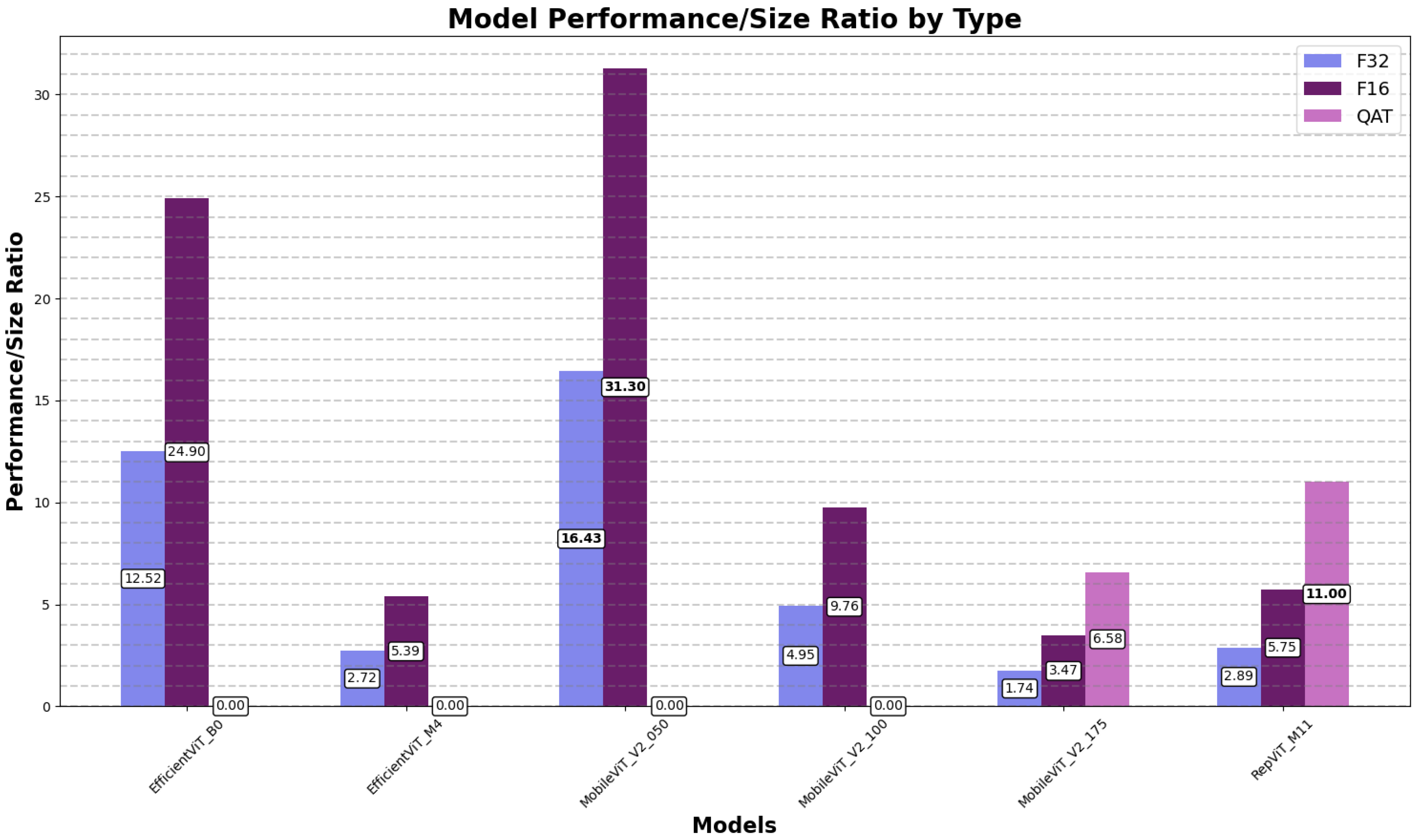
4.1.3. Model Performance-to-Size Ratios
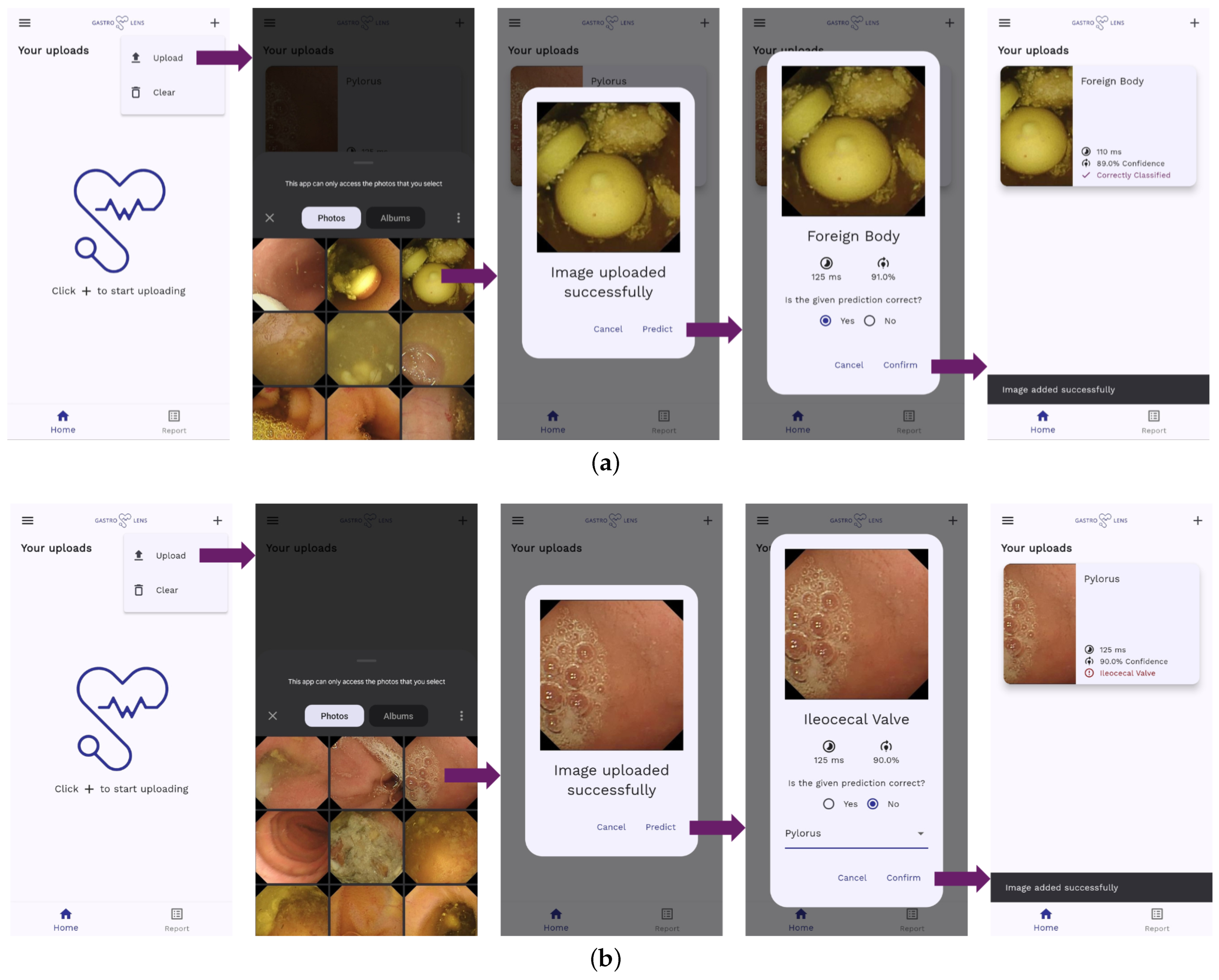
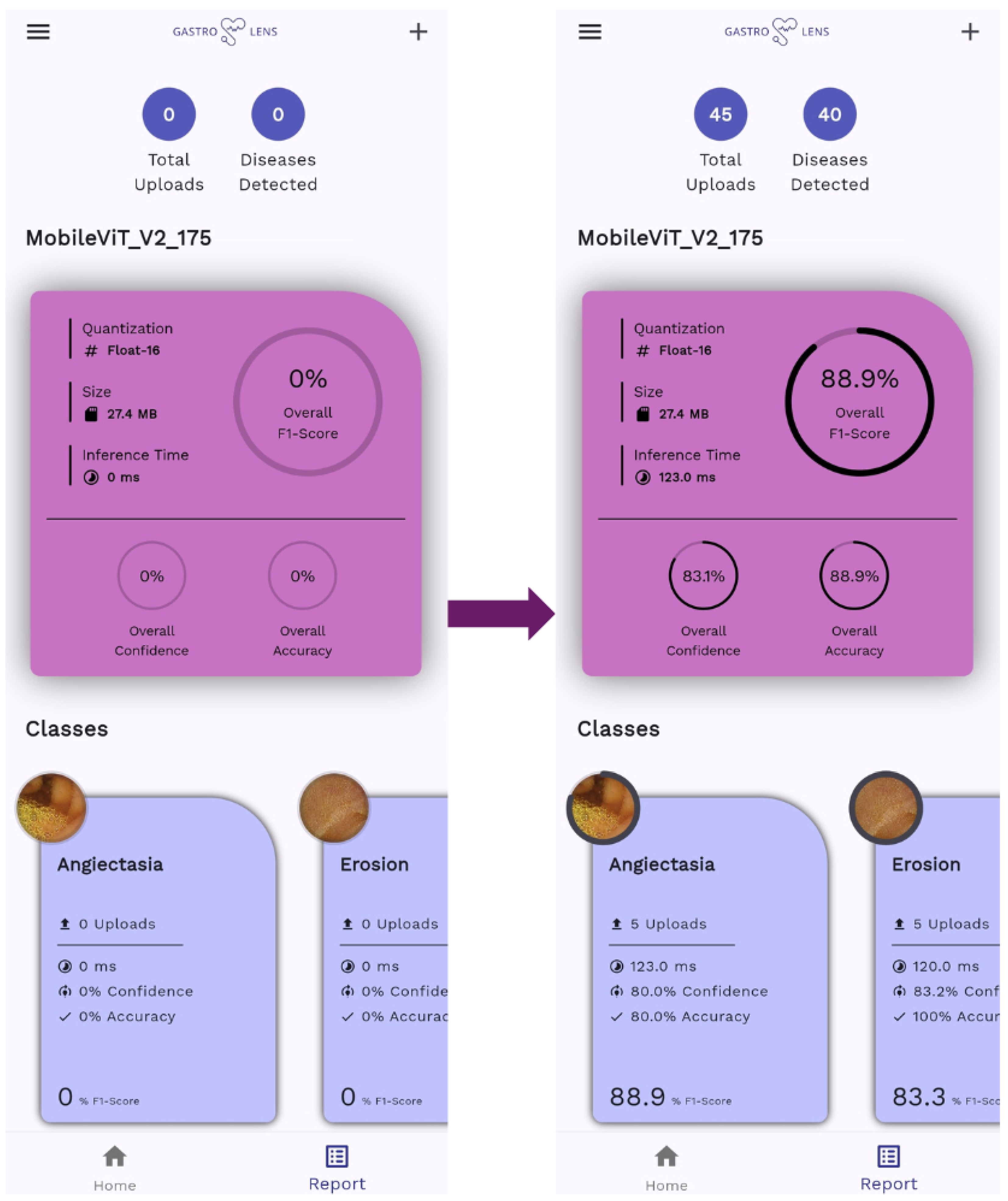
4.2. On-Edge Testing
4.3. Performance on Other Datasets
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A

References
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- TensorFlow Lite Guide. 2024. Available online: https://www.tensorflow.org/lite/guide (accessed on 10 July 2024).
- Min, J.K.; Kwak, M.S.; Cha, J.M. Overview of deep learning in gastrointestinal endoscopy. Gut Liver 2019, 13, 388. [Google Scholar] [CrossRef] [PubMed]
- Mall, P.K.; Singh, P.K.; Srivastav, S.; Narayan, V.; Paprzycki, M.; Jaworska, T.; Ganzha, M. A comprehensive review of deep neural networks for medical image processing: Recent developments and future opportunities. Healthc. Anal. 2023, 4, 100216. [Google Scholar] [CrossRef]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An Image is Worth 16 × 16 Words: Transformers for Image Recognition at Scale. arXiv 2021, arXiv:2010.11929. [Google Scholar]
- Liu, S.; Ha, D.S.; Shen, F.; Yi, Y. Efficient neural networks for edge devices. Comput. Electr. Eng. 2021, 92, 107121. [Google Scholar] [CrossRef]
- Han, S.; Mao, H.; Dally, W.J. Deep Compression: Compressing Deep Neural Networks with Pruning, Trained Quantization and Huffman Coding. arXiv 2016, arXiv:1510.00149. [Google Scholar]
- Park, E.; Yoo, S.; Vajda, P. Value-aware Quantization for Training and Inference of Neural Networks. arXiv 2018, arXiv:1804.07802. [Google Scholar]
- Smedsrud, P.H.; Thambawita, V.; Hicks, S.A.; Gjestang, H.; Nedrejord, O.O.; Næss, E.; Borgli, H.; Jha, D.; Berstad, T.J.D.; Eskeland, S.L.; et al. Kvasir-Capsule, a video capsule endoscopy dataset. Sci. Data 2021, 8, 142. [Google Scholar] [CrossRef]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, L.; Polosukhin, I. Attention Is All You Need. arXiv 2023, arXiv:1706.03762. [Google Scholar]
- Wang, X.; Zhang, L.L.; Wang, Y.; Yang, M. Towards efficient vision transformer inference: A first study of transformers on mobile devices. In Proceedings of the 23rd Annual International Workshop on Mobile Computing Systems and Applications, New York, NY, USA, 9–10 March 2022; pp. 1–7. [Google Scholar]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.C. MobileNetV2: Inverted Residuals and Linear Bottlenecks. arXiv 2019, arXiv:1801.04381. [Google Scholar]
- Xie, Q.; Luong, M.T.; Hovy, E.; Le, Q.V. Self-training with Noisy Student improves ImageNet classification. arXiv 2020, arXiv:1911.04252. [Google Scholar]
- Pogorelov, K.; Randel, K.R.; Griwodz, C.; Eskeland, S.L.; de Lange, T.; Johansen, D.; Spampinato, C.; Dang-Nguyen, D.T.; Lux, M.; Schmidt, P.T.; et al. KVASIR: A Multi-Class Image Dataset for Computer Aided Gastrointestinal Disease Detection. In Proceedings of the 8th ACM on Multimedia Systems Conference, New York, NY, USA, 20–23 June 2017; MMSys’17. pp. 164–169. [Google Scholar] [CrossRef]
- Borgli, H.; Thambawita, V.; Smedsrud, P.H.; Hicks, S.; Jha, D.; Eskeland, S.L.; Randel, K.R.; Pogorelov, K.; Lux, M.; Nguyen, D.T.D.; et al. HyperKvasir, a comprehensive multi-class image and video dataset for gastrointestinal endoscopy. Sci. Data 2020, 7, 283. [Google Scholar] [CrossRef] [PubMed]
- Varam, D.; Mitra, R.; Mkadmi, M.; Riyas, R.A.; Abuhani, D.A.; Dhou, S.; Alzaatreh, A. Wireless Capsule Endoscopy Image Classification: An Explainable AI Approach. IEEE Access 2023, 11, 105262–105280. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv 2015, arXiv:1409.1556. [Google Scholar]
- Oukdach, Y.; Kerkaou, Z.; El Ansari, M.; Koutti, L.; Fouad El Ouafdi, A.; De Lange, T. ViTCA-Net: A framework for disease detection in video capsule endoscopy images using a vision transformer and convolutional neural network with a specific attention mechanism. Multimed. Tools Appl. 2024, 83, 1–20. [Google Scholar] [CrossRef]
- Qu, A.; Wu, Q.; Wang, J.; Yu, L.; Li, J.; Liu, J. TNCB: Tri-Net With Cross-Balanced Pseudo Supervision for Class Imbalanced Medical Image Classification. IEEE J. Biomed. Health Inform. 2024, 28, 2187–2198. [Google Scholar] [CrossRef]
- Ghubaish, A.; Salman, T.; Zolanvari, M.; Unal, D.; Al-Ali, A.; Jain, R. Recent Advances in the Internet-of-Medical-Things (IoMT) Systems Security. IEEE Internet Things J. 2021, 8, 8707–8718. [Google Scholar] [CrossRef]
- Datta Gupta, K.; Sharma, D.K.; Ahmed, S.; Gupta, H.; Gupta, D.; Hsu, C.H. A novel lightweight deep learning-based histopathological image classification model for IoMT. Neural Process. Lett. 2023, 55, 205–228. [Google Scholar] [CrossRef] [PubMed]
- Orlov, N.V.; Chen, W.W.; Mark Eckley, D.; Macura, T.J.; Shamir, L.; Jaffe, E.S.; Goldberg, I.G. Automatic Classification of Lymphoma Images With Transform-Based Global Features. IEEE Trans. Inf. Technol. Biomed. 2010, 14, 1003–1013. [Google Scholar] [CrossRef]
- Shreyas, S.; Rao, J.K. Diagnostic Decision Support for Medical Imaging and COVID-19 Image Classification on ARM Mali GPU. In Proceedings of the 2021 IEEE Globecom Workshops (GC Wkshps), Madrid, Spain, 7–11 December 2021; pp. 1–6. [Google Scholar] [CrossRef]
- Cohen, J.P.; Morrison, P.; Dao, L. COVID-19 image data collection. arXiv 2020, arXiv:2003.11597. [Google Scholar]
- Aldamani, R.; Abuhani, D.A.; Shanableh, T. LungVision: X-ray Imagery Classification for On-Edge Diagnosis Applications. Algorithms 2024, 17, 280. [Google Scholar] [CrossRef]
- Feltrin, F. X-ray Lung Diseases Images (9 Classes)—kaggle.com. 2023. Available online: https://www.kaggle.com/datasets/fernando2rad/x-ray-lung-diseases-images-9-classes (accessed on 10 July 2024).
- Pei, S.C.; Lin, C.N. Image normalization for pattern recognition. Image Vis. Comput. 1995, 13, 711–723. [Google Scholar] [CrossRef]
- Li, Y.; Hu, J.; Wen, Y.; Evangelidis, G.; Salahi, K.; Wang, Y.; Tulyakov, S.; Ren, J. Rethinking Vision Transformers for MobileNet Size and Speed. In Proceedings of the IEEE International Conference on Computer Vision, Paris, France, 2–6 October 2023. [Google Scholar]
- Cai, H.; Li, J.; Hu, M.; Gan, C.; Han, S. EfficientViT: Multi-Scale Linear Attention for High-Resolution Dense Prediction. arXiv 2024, arXiv:2205.14756. [Google Scholar]
- Liu, X.; Peng, H.; Zheng, N.; Yang, Y.; Hu, H.; Yuan, Y. EfficientViT: Memory Efficient Vision Transformer with Cascaded Group Attention. arXiv 2023, arXiv:2305.07027. [Google Scholar]
- Mehta, S.; Rastegari, M. Separable Self-attention for Mobile Vision Transformers. arXiv 2022, arXiv:2206.02680. [Google Scholar]
- Wang, A.; Chen, H.; Lin, Z.; Han, J.; Ding, G. RepViT: Revisiting Mobile CNN From ViT Perspective. arXiv 2024, arXiv:2307.09283. [Google Scholar]
- Leondgarse. leondgarse/keras_cv_attention_models: Zenodo (zenodo). Zenodo. 2022. [Google Scholar] [CrossRef]
- Li, Y.; Yuan, G.; Wen, Y.; Hu, J.; Evangelidis, G.; Tulyakov, S.; Wang, Y.; Ren, J. Efficientformer: Vision transformers at mobilenet speed. Adv. Neural Inf. Process. Syst. 2022, 35, 12934–12949. [Google Scholar]
- Mehta, S.; Rastegari, M. MobileViT: Light-weight, General-purpose, and Mobile-friendly Vision Transformer. arXiv 2022, arXiv:2110.02178. [Google Scholar]
- Model Optimization in TensorFlow Lite. 2024. Available online: https://www.tensorflow.org/lite/performance/model_optimization (accessed on 10 July 2024).
- Gholami, A.; Kim, S.; Dong, Z.; Yao, Z.; Mahoney, M.W.; Keutzer, K. A Survey of Quantization Methods for Efficient Neural Network Inference. arXiv 2021, arXiv:2103.13630. [Google Scholar]
- Villalobos, P.; Sevilla, J.; Besiroglu, T.; Heim, L.; Ho, A.; Hobbhahn, M. Machine Learning Model Sizes and the Parameter Gap. arXiv 2022, arXiv:2207.02852. [Google Scholar]






| Paper | Images Used | Best Model | Performance | Model Size (MB) |
|---|---|---|---|---|
| Varam et al. [16] | Top 9 classes: 4500 | ViT | F1-score: 0.97 | Not reported |
| Oukdach et al. [18] | 12 classes: 6990 | ViTCA-Net | F1-score: 0.97 | Not reported |
| Qu et al. [19] | Top 10 classes: 47,002 | TNCB | F1-score: 0.95 | Not reported |
| Paper | Dataset | Model | Quantization | Performance | Model Size | Inference Time |
|---|---|---|---|---|---|---|
| Gupta et al. [21] | Malignant Lymphoma | ReducedFireNet | F32 | F1-score: 0.958 | 42.8 KB | Not reported |
| Shreya et al. [23] | COVID-19 chest X-rays | DDSM++ | F16 | Accuracy: 0.936 | Not reported | 280 ms |
| Aldamani et al. [25] | X-ray lung diseases | EfficientNetB2V2 | QAT | F1-score: 0.975 | 9.9 MB | 470 ms |
| Class Name | Original Dataset | Training Dataset |
|---|---|---|
| Normal Clean Mucosa | 34,338 | 500 |
| Ileocecal Valve | 4189 | 500 |
| Reduced Mucosal View | 2906 | 500 |
| Pylorus | 1529 | 500 |
| Angiectasia | 866 | 500 |
| Ulcer | 854 | 500 |
| Foreign Body | 776 | 500 |
| Lymphangiectasia | 592 | 500 |
| Erosion | 506 | 500 |
| Blood—Fresh | 446 | 0 |
| Erythema | 159 | 0 |
| Polyp | 55 | 0 |
| Blood—Hematin | 12 | 0 |
| Ampulla of Vater | 10 | 0 |
| Total | 47,238 | 4500 |
| Model | Params | FLOPs | Input | Top1 Acc | T4 Inference |
|---|---|---|---|---|---|
| EfficientFormerV2S2 | 12.70 M | 1.27 G | 224 × 224 | 0.820 | 573.90 qps |
| EfficientViT_B0 | 3.41 M | 0.12 G | 224 × 224 | 0.716 | 1581.76 qps |
| EfficientViT_M4 | 8.80 M | 299 M | 224 × 224 | 0.743 | 672.89 qps |
| MobileViT_V2_050 | 1.37 M | 0.47 G | 256 × 256 | 0.702 | 718.34 qps |
| MobileViT_V2_100 | 4.90 M | 1.83 G | 256 × 256 | 0.781 | 591.22 qps |
| MobileViT_V2_175 | 14.3 M | 5.52 G | 256 × 256 | 0.808 | 412.76 qps |
| RepViT_M11 | 8.29 M | 1.35 G | 224 × 224 | 0.812 | 846.68 qps |
| Technique | Size Reduction | Accuracy |
|---|---|---|
| Post-training float-16 quantization | Up to 50% | Insignificant accuracy loss |
| Quantization-aware training | Up to 75% | Smallest accuracy loss |
| Quantization Technique | |||
|---|---|---|---|
| Model | F32 | F16 | QAT |
| EfficientFormerV2S2 | 82.59 | 82.60 | 25.11 |
| EfficientViT_B0 | 90.02 | 90.50 | 15.65 |
| EfficientViT_M4 | 91.91 | 91.69 | 89.44 |
| MobileViT_V2_050 | 90.34 | 90.33 | 70.02 |
| MobileViT_V2_100 | 93.93 | 93.93 | 85.64 |
| MobileViT_V2_175 | 95.34 | 95.34 | 94.42 |
| RepViT_M11 | 91.72 | 91.72 | 93.11 |
| Quantization Technique | |||
|---|---|---|---|
| Model | F32 | F16 | QAT |
| EfficientFormerV2S2 | 48.04 | 24.22 | 13.71 |
| EfficientViT_B0 | 7.19 | 3.63 | 1.98 |
| EfficientViT_M4 | 33.81 | 17.01 | 9.09 |
| MobileViT_V2_050 | 5.50 | 2.89 | 1.70 |
| MobileViT_V2_100 | 18.98 | 9.63 | 5.22 |
| MobileViT_V2_175 | 54.66 | 27.47 | 14.36 |
| RepViT_M11 | 31.72 | 15.96 | 8.46 |
| Model | F16 (% of F32) | QAT (% of F32) |
|---|---|---|
| EfficientFormerV2S2 | 50.41% | 28.55% |
| EfficientViT_B0 | 50.54% | 27.49% |
| EfficientViT_M4 | 50.33% | 26.89% |
| MobileViT_V2_050 | 52.51% | 30.88% |
| MobileViT_V2_100 | 50.71% | 27.49% |
| MobileViT_V2_175 | 50.26% | 26.27% |
| RepViT_M11 | 50.33% | 26.68% |
| Quantization Technique | |||
|---|---|---|---|
| Model | F32 | F16 | QAT |
| EfficientFormerV2S2 | 1.72 | 3.41 | 1.83 |
| EfficientViT_B0 | 12.52 | 24.90 | 7.91 |
| EfficientViT_M4 | 2.72 | 5.39 | 9.84 |
| MobileViT_V2_050 | 16.43 | 31.30 | 41.25 |
| MobileViT_V2_100 | 4.95 | 9.76 | 16.42 |
| MobileViT_V2_175 | 1.74 | 3.47 | 6.58 |
| RepViT_M11 | 2.89 | 5.75 | 11.00 |
| Paper | Model | F1-Score | Params | Quantization |
|---|---|---|---|---|
| [16], 2023 | ViT | 0.9700 | ≈86 M | None |
| [18], 2024 | ViTCA-Net | 0.9700 | ≈86 M | None |
| [19], 2024 | TNCB | 0.9500 | 12.2 M | None |
| Proposed solution | MobileViT_V2_175 | 0.9534 | 14.3 M | F16 |
| Model | Quantization | F1-Score | Size (MB) | Performance-to-Size Ratio |
|---|---|---|---|---|
| MobileViT_V2_175 | F16 | 0.9534 | 27.47 | 3.47 |
| MobileViT_V2_050 | F16 | 0.9033 | 2.89 | 31.30 |
| Model | Quantization | F1-Score | Inference Time (ms) |
|---|---|---|---|
| MobileViT_V2_175 | F16 | 0.9107 | 677.2 |
| MobileViT_V2_050 | F16 | 0.7946 | 80.8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Varam, D.; Khalil, L.; Shanableh, T. On-Edge Deployment of Vision Transformers for Medical Diagnostics Using the Kvasir-Capsule Dataset. Appl. Sci. 2024, 14, 8115. https://doi.org/10.3390/app14188115
Varam D, Khalil L, Shanableh T. On-Edge Deployment of Vision Transformers for Medical Diagnostics Using the Kvasir-Capsule Dataset. Applied Sciences. 2024; 14(18):8115. https://doi.org/10.3390/app14188115
Chicago/Turabian StyleVaram, Dara, Lujain Khalil, and Tamer Shanableh. 2024. "On-Edge Deployment of Vision Transformers for Medical Diagnostics Using the Kvasir-Capsule Dataset" Applied Sciences 14, no. 18: 8115. https://doi.org/10.3390/app14188115







