Abstract
Multiple Sequence Alignment (MSA) plays a pivotal role in bioinformatics, facilitating various critical biological analyses, including the prediction of unknown protein structures and functions. While numerous methods are available for MSA, bioinspired algorithms stand out for their efficiency. Despite the growing research interest in addressing the MSA challenge, only a handful of comprehensive reviews have been undertaken in this domain. To bridge this gap, this study conducts a thorough analysis of bioinspired-based methods for MSA through a systematic literature review (SLR). By focusing on publications from 2010 to 2024, we aim to offer the most current insights into this field. Through rigorous eligibility criteria and quality standards, we identified 45 relevant papers for review. Our analysis predominantly concentrates on bioinspired-based techniques within the context of MSA. Notably, our findings highlight Genetic Algorithm and Memetic Optimization as the most commonly utilized algorithms for MSA. Furthermore, benchmark datasets such as BAliBASE and SABmark are frequently employed in evaluating MSA solutions. Structural-based methods emerge as the preferred approach for assessing MSA solutions, as revealed by our systematic literature review. Additionally, this study explores current trends, challenges, and unresolved issues in the realm of bioinspired algorithms for MSA, offering practitioners and researchers valuable insights and comprehensive understanding of the field.
1. Introduction
MSA is a vital and complex task within the field of bioinformatics that plays a crucial role in uncovering the evolutionary connections between biological sequences [1,2]. While traditional algorithms, such as dynamic programming-based methods [3,4] can align a limited number of sequences, they struggle with the immense amounts of sequence data generated by modern sequencing technologies. As a result, researchers have turned to innovative, bioinspired algorithms [5] as a potential solution to the challenges of computational efficiency and scalability faced by conventional methods [6].
Bioinspired algorithms, such as Genetic Algorithms (GA), Simulated Annealing (SA), Particle Swarm Optimization (PSO), and Artificial Bee Colony Optimization (ABC), have been widely used in Multiple Sequence Alignment (MSA) research due to their remarkable ability to unearth optimal or near-optimal solutions [7,8]. These algorithms mimic natural processes or use population-based strategies, which help them find optimal solutions and avoid becoming stuck in local optima. Due to their adaptability and flexibility, these algorithms are well-suited for addressing the challenges and complexities of the MSA problem [9].
In the current era, the investigation of bioinspired algorithms for performing Multiple Sequence Alignment continues to be a thriving field of research [10,11,12]. This domain constantly evolves with a diverse range of methodologies, including traditional and innovative approaches, such as multi-objective and hybrid techniques [13]. With this in mind, we aim to conduct a thorough systematic literature review (SLR) that concentrates on bioinspired algorithms utilized in MSA. This review provides a methodical and updated view of the state-of-the-art techniques implemented to tackle MSA challenges. We have focused on studies conducted from 2010 to 2024 to ensure the latest advancements are included. Specifically, our study centers on the bioinspired algorithms applied to Multiple Sequence Alignment through a comprehensive analysis of existing research.
Our goal is to examine the strengths, limitations, and potential areas of improvement in the field of bioinspired algorithms for the MSA problem. By compiling knowledge from various research areas, we aim to deepen our understanding of how these algorithms can enhance the effectiveness and efficiency of MSA techniques. Furthermore, we address the challenges and unresolved issues that persist in this field, paving the way for future research to refine and advance the application of bioinspired techniques in the context of MSA. The key contributions of this paper can be summarized as follows:
- Conducting a novel SLR that identifies and summarizes bioinspired-based techniques popularly applied for solving the Multiple Sequence Alignment problem.
- Recognize and succinctly outline the benchmark datasets employed in evaluating Multiple Sequence Alignment within the framework of bioinspired algorithms.
- Identify and succinctly summarize the performance evaluation measures utilized for Multiple Sequence Alignment in the context of bioinspired algorithms.
- Explore and analyze the research challenges, open issues, and future directions for Multiple Sequence Alignment within the context of bioinspired algorithms.
The structure of this work is as follows: we briefly explain the Multiple Sequence Alignment problem in Section 2. Section 3 presents related recent publications that provide reviews of bioinspired approaches for MSA, highlighting our work’s differences and contributions. In Section 4, we provide the methods for conducting the proposed SLR. We present our SLR results in Section 5, emphasizing the figures associated with the publications identified in the SLR. Section 6 provides a view of the limitations of the proposed SLR. In Section 7, we finally give the conclusion and discussion of the paper.
2. Multiple Sequence Alignment
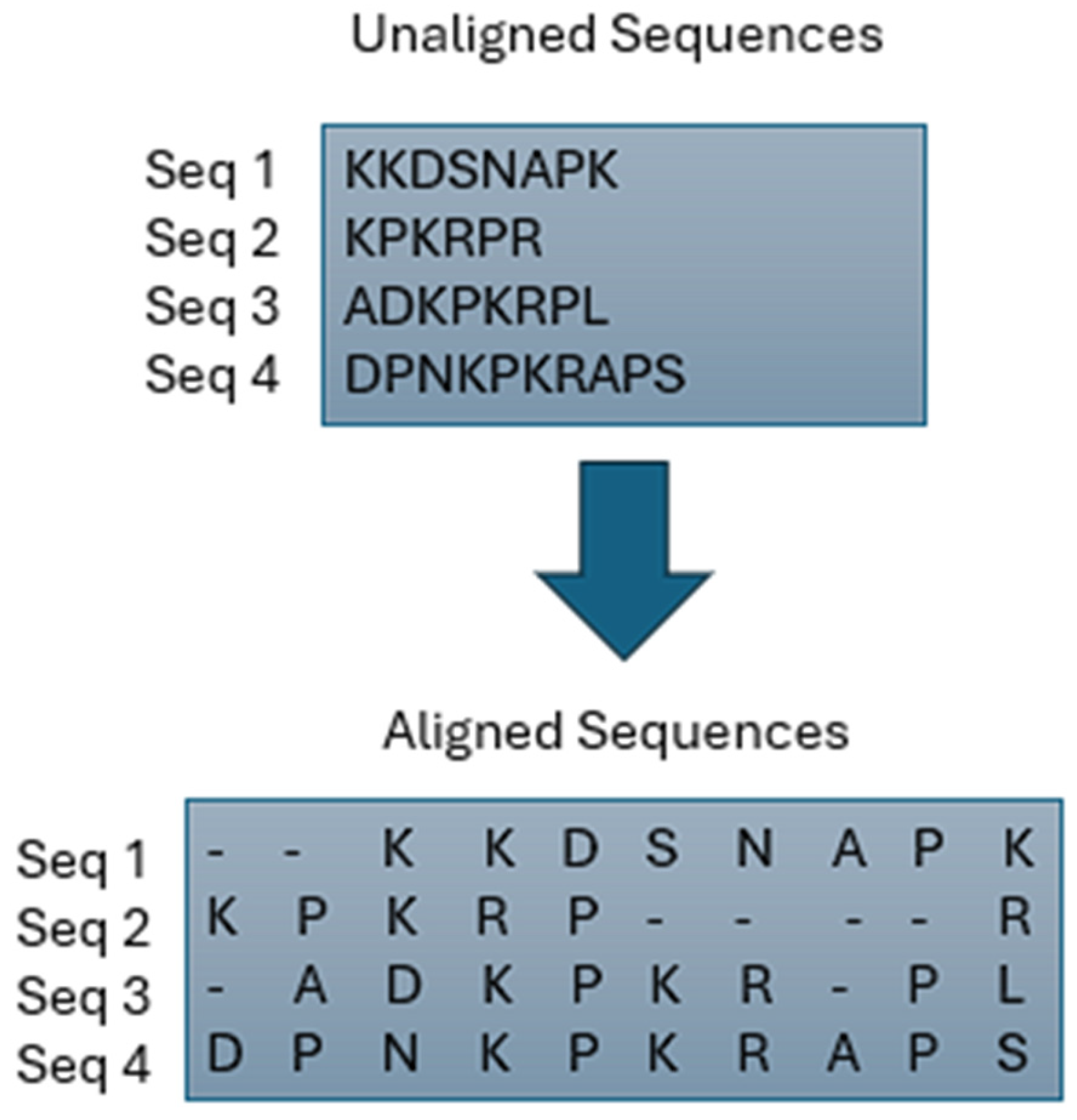
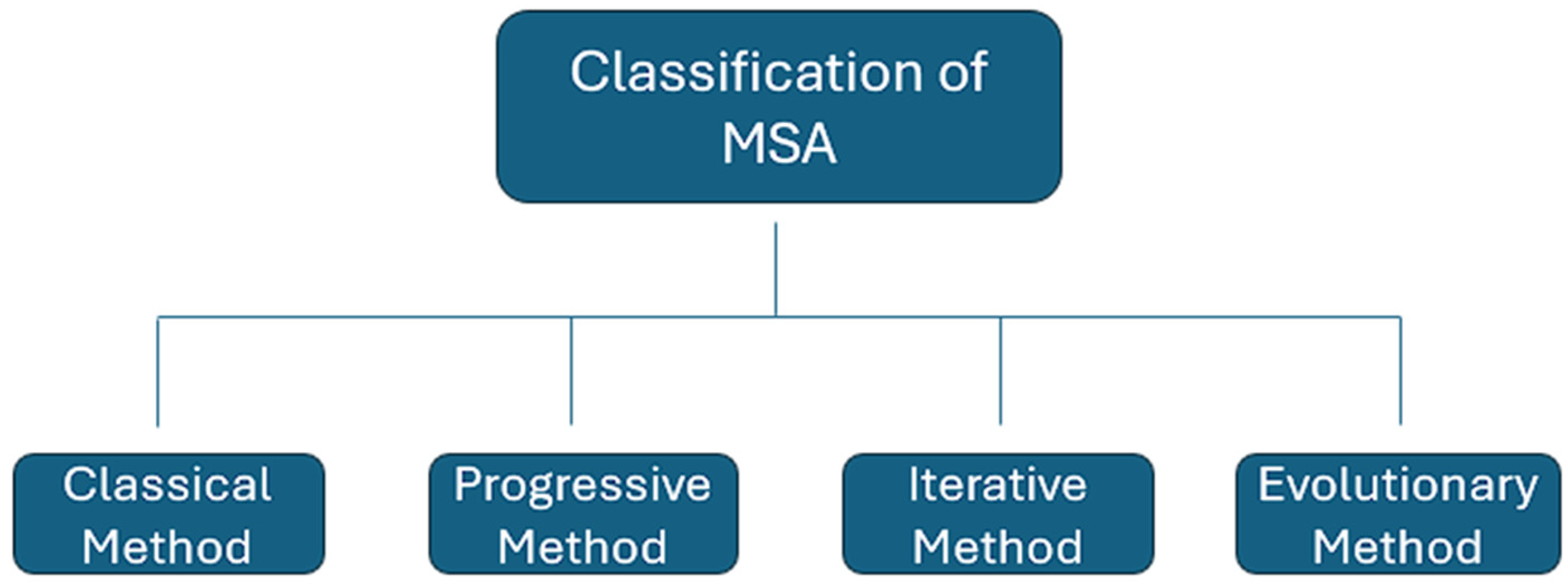
In bioinformatics, MSA is a vital process essential for evolutionary research [14,15] and aids in predicting the structure and function of unidentified proteins [16,17]. The MSA problem primarily involves aligning multiple biological sequences and optimizing a specific statistic, such as the count of aligned bases [18]. As the length and number of sequences in the issue space increase, implementing MSA with traditional methods becomes impractical [15]. Consequently, various methods have been proposed to address MSA challenges over the years. These approaches can be categorized as evolutionary, progressive, iterative, or classical. Each of these categories is briefly explained in this section. Refer to Figure 1 for a basic example of MSA and Figure 2 for the classification of MSA.
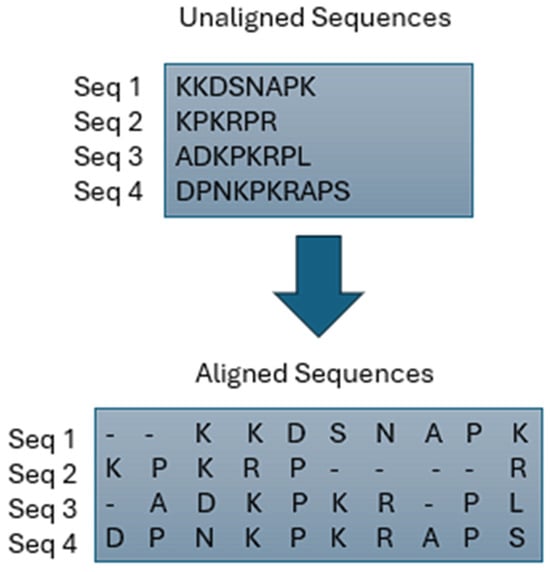
Figure 1.
Example of Multiple Sequence Alignment.
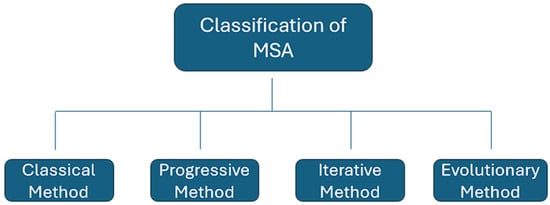
Figure 2.
Classification of the Multiple Sequence Alignment methods.
2.1. Classical Method
Dynamic programming effectively resolves Multiple Sequence Alignment (MSA) to obtain optimal alignments [19]. Moreover, dynamic programming employs a scoring function encompassing numerous domains. Originally developed to address challenges in two-sequence alignment [4], dynamic programming faced heightened difficulties with the increased length and number of sequences. This escalation in complexity became a significant issue associated with dynamic programming.
As a result, the MSA problem becomes NP-hard [4], posing a significant computational challenge. The primary limitation lies in the difficulty of efficiently utilizing computers to address the complexity associated with MSA. Typically, the classical technique requires increasing the alignment of amino acid or protein sequences with less complexity within a specified timeframe. The decision to transition to an alternative approach represents a critical juncture for researchers.
2.2. Progressive Method
The solution to the Multiple Sequence Alignment (MSA) problem aims to reduce complexity in terms of time and space [8]. The progressive technique initiates alignment by aligning identical sequences, adjusting its increment against divergent sequences within the alignment. The widely accepted representation of the progressive approach is ClustalW [20]. Initially, weights are assigned to each pair of sequences in a restricted alignment, with modest weights for identical sequences and significant weights for different sequences. Following the calculation of scores for two protein residues, a substitution matrix based on similarity is considered. The third stage introduces two specific gaps: the residual gap and the locally reduced specific gap penalty [21]. In the final stage, locally reduced specific gaps are encouraged to receive opening gaps at that spot. The ClustalW representation allows for the seamless integration of these four stages [20]. While the progressive technique offers superior alignment for MSA in terms of accuracy and time, it has certain drawbacks, such as dependence on the scoring scheme and initial alignments (i.e., matching comparable sequences at the beginning) [22]. Failure to meet specific requirements may lead to the recording of local optima results.
2.3. Iterative Method
The iterative method presents another approach for addressing Multiple Sequence Alignment (MSA) through algorithms, yielding an alignment [23]. It is advisable for the alignment to undergo multiple iterations until further improvement is no longer possible. The iterative approach for enhancing alignment can be either deterministic or stochastic, depending on the chosen strategy [24]. The deterministic iterative technique facilitates the extraction of one sequence at a time, which is then realigned to other sequences [25]. The iterative method aims to enhance results with each iteration to the maximum feasible extent without relying on the original alignments [24]. Instead, it initiates with new alignments tailored to specific challenges [23]. The primary objective of MSA is to ensure the quality of sequence alignment. Some approaches combine progressive and iterative techniques [26].
As a result, sequence alignment is halted when no further progress can be achieved. The Simulated Annealing method enhances alignment in MSA [27,28]. The Gibbs sampling technique has successfully identified barriers in local multiple alignments without gaps; however, dealing with gapped alignments posed challenges in their endurance and reproduction [5]. The best score of the goal function determines the fitness of an alignment. Based on this fitness measure, alignments may either persist or be discarded over multiple iterations. Stochastic modifications in the method can enhance and reproduce alignments by introducing crossovers and mutations. Despite the iterative alignment approach’s aim to provide high-quality alignments, it offers no guarantees regarding identifying exceptional alignments. Simulated Annealing (SA) [27] is a stochastic method for synthesizing multiple samples. The primary challenge with the iterative approach lies in capturing results at local optima.
2.4. Evolutionary Method
The strategy employed is population-based [21], initializing the population randomly in the first stage. In the subsequent step, which involves modifying the initial population, fundamental operators are utilized for successive generations, aiming to reach the global optimum [29]. Given the random initiation in the evolutionary technique for Multiple Sequence Alignment (MSA), the Evolutionary Algorithm for Sequences (EAS) has taken additional steps to enhance similarities in sequence alignment [9]. Evolutionary computation provides intriguing approaches for multisequence alignment [21], resulting in higher alignment accuracy during the MSA runtime process. The SAGA technique has seen improved utilization through evolutionary computation [30]. Incorporating 22 distinct crossover and mutation operators in SAGA demonstrated increased alignment fitness within the population. Addressing sequence alignment challenges, SAGA relies on a Weighted Sum-of-Pairs (WSP) model, wherein alignment sequences are scored and compared. Pairwise alignments are integrated into the overall alignment to produce classical scores.
3. Related Work
The application of bioinspired techniques in Multiple Sequence Alignment (MSA) has garnered considerable attention in the field of bioinformatics, driven by the proliferation of recent scholarly works [31,32,33]. Bioinspired algorithms possess robust search capabilities, rendering them well-suited for addressing optimization challenges like MSA [5,34,35]. The initial phase entails preparing the sequences for alignment, which encompasses tasks such as sequence preprocessing, selection, and weighting. Subsequently, an objective function is formulated to assess the alignment’s quality, typically gauging the similarity or dissimilarity between aligned symbols within the sequences. This objective function may be constructed based on diverse criteria, such as maximizing correspondence or minimizing gaps. Bioinspired algorithms are then employed to optimize the objective function and ascertain the optimal alignment. Leveraging search and optimization methodologies, these algorithms systematically explore the solution space to enhance the quality of the alignment [7,36].
Moreover, recent years have witnessed diverse literature appraisals emerging due to the subject’s pertinence. For instance, Ali and Hasanien [24] undertook a metaheuristic survey regarding applications in bioinformatics, elucidating the characteristics of various methods encompassing evolutionary algorithms, Simulated Annealing, and Particle Swarm Optimization. The authors exemplify how these methodologies address computational biology challenges such as MSA, structure prediction, and gene selection. However, it is noteworthy that the authors exclusively concentrate on Genetic Algorithms to address the MSA quandary.
Chatzou et al. [6] provided an overview of Multiple Sequence Alignment techniques and significant advancements within the past decade. However, their focus is primarily on illustrating heuristic-based tools such as MAFFT [37,38] and Clustal [39], among others. Zambrano-Vega et al. [8] contributed two separate comparative studies addressing the application of multi-objective bioinspired algorithms for MSA challenges. The first study delves into the formulation of MSA with three objectives, while the second study offers a more comprehensive examination of generic multi-objective methods for MSA. Nonetheless, instead of adopting a systematic review approach, both articles aim to deliver a thorough experimental assessment of reference techniques in sequence alignment. Chowdhury and Garai [40] reviewed various strategies to address MSA challenges, specifically focusing on Genetic Algorithms as the sole methodologies investigated.
A recent review by Chao [36] delves into various quality estimation techniques utilized in MSA tools and the fundamental concepts prevalent in sequence alignment research. Furthermore, an assessment of benchmarks for MSA was outlined in [26], concentrating solely on evaluation methods employed for resolving sequence alignments. Paruchuri [31] surveyed nature-inspired algorithms aimed at tackling Multiple Sequence Alignment problems; this article provided an overview of the state-of-the-art nature-inspired algorithms and implemented several algorithms to address MSA challenges, presenting the analyses and results of their experiments. Another review work introduced by Almanza [41] focused on parallel computing approaches applied to MSA.
The principal objective of our present investigation is to introduce a comprehensive systematic literature review aimed at compiling and synthesizing articles pertaining to bioinspired algorithms for Multiple Sequence Alignment. Consequently, our study distinguishes itself from recent reviews that tackle a multitude of biological topics by focusing specifically on this subject matter, as far as our knowledge extends. The Supplementary Materials provide a summary of the pertinent existing reviews in comparison to our study.
4. Methods
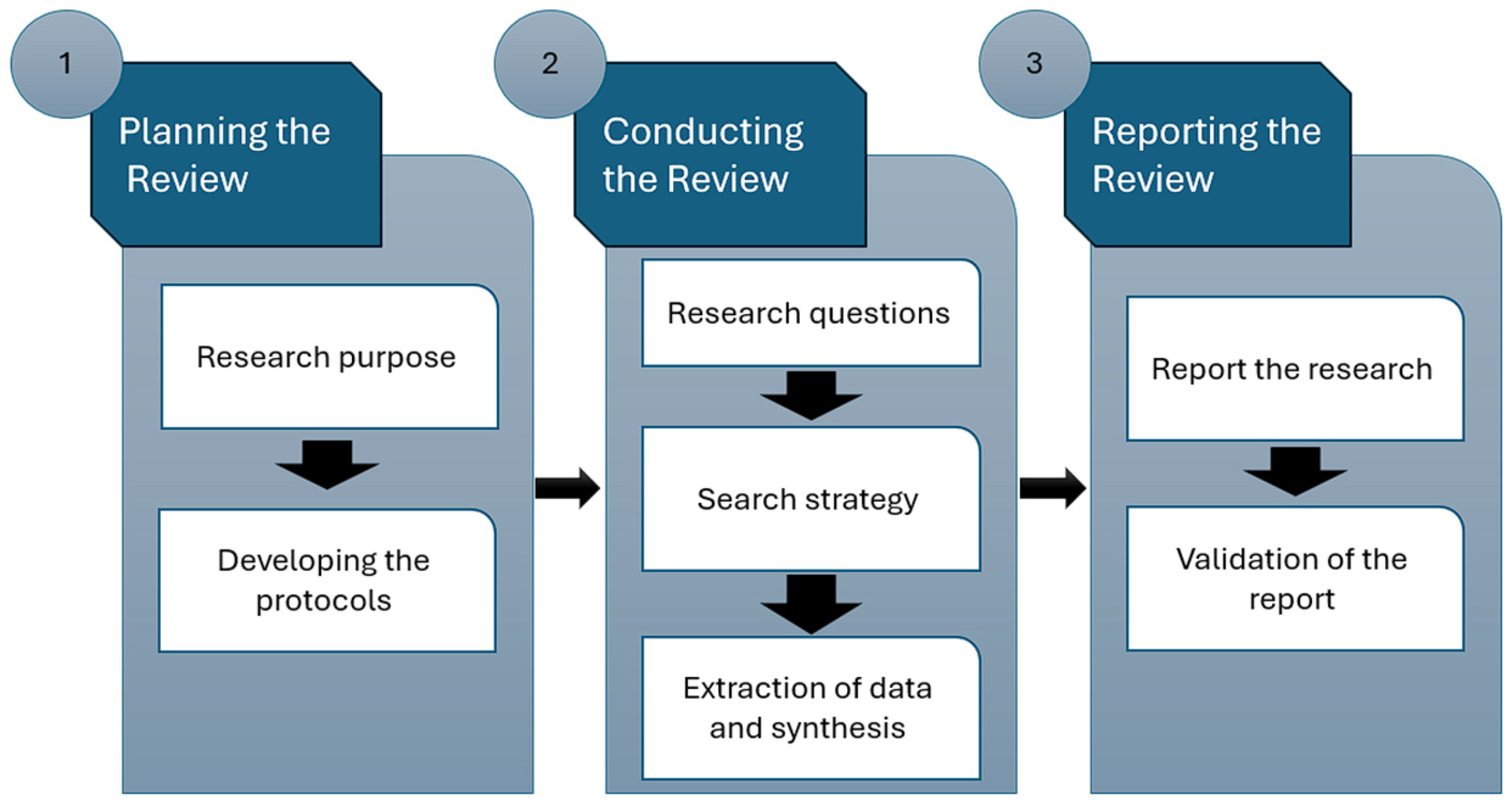
This study employed the systematic literature review (SLR) methodology, a rigorous approach for gathering and assessing all studies on specific research topics [42]. Utilizing SLR helps minimize biases by systematically identifying and gathering materials that address particular concerns [15,18], ensuring a review with high-quality evidence and allowing for scrutiny of reviewers’ decisions and conclusions [42]. The framework outlined in Figure 3 formed the basis for the proposed SLR, encompassing three main phases: review planning, execution, and documentation. Figure 2 depicts the primary stages of the proposed SLR.
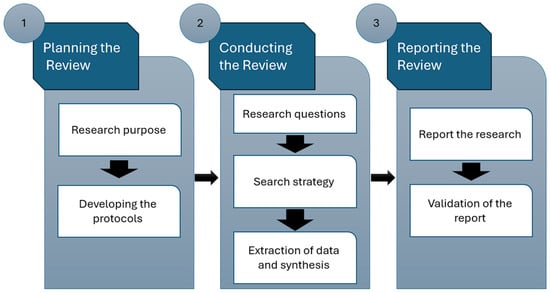
Figure 3.
Phases of the envisioned SLR.
4.1. Review Planning
The planning phase delineates the conception and preparation processes of the SLR, encompassing the identification of the study’s objectives and the development of the review protocol. An automated search was conducted across prominent bibliographic databases to identify additional relevant documents [15,18]. These digital repositories were selected due to their widespread usage and extensive literature pertinent to the research concerns of the study. To ensure a comprehensive collection of current and relevant articles for the SLR, the timeframe considered spans from 2010 to 2024.
4.2. Conducting the Review
Following the review planning, the subsequent step involves the actual conduct of the review. This phase entails executing the primary review method, which includes identifying the principal subjects to be reviewed and explored by defining the research questions (RQs) of the SLR. Furthermore, this stage encompasses formulating a search strategy and the data extraction and synthesis procedures, all elaborated upon in subsequent subsections.
4.3. Research Questions
In this systematic literature review (SLR), identifying research questions is the initial step in determining the issues to be explored and investigated. The selection of primary studies for inclusion in the review heavily relies on these research questions. Consequently, the development of research questions is typically the primary focal point of the SLR. The principal research questions (RQ) utilized in this study are presented in Table 1.

Table 1.
Research questions.
The initial query aims to ascertain the most frequently utilized bioinspired methods for Multiple Sequence Alignment (MSA). Research Question 2 (RQ2) delves into the benchmark datasets employed to assess the MSA problem. RQ3 seeks to examine the prevalent benchmark techniques utilized for MSA. Lastly, RQ4 addresses the research challenges, trends, and future directions within this topic area.
4.4. Search Strategy
Five distinct bibliographic databases were employed in the search process to identify relevant studies, see Table 2 below. These databases include Scopus, IEEE Xplore, SpringerLink, ACM, and Bioinformatic. The review’s timeframe was limited to the years 2010 to 2024 to ensure a comprehensive and up-to-date analysis. The following search strings were utilized:

Table 2.
The search terms utilized.
- String-A: “multiple sequence alignment” AND “bioinspired”
- String-B: “multiple sequence alignment” AND “genetic algorithm”
- String-C: “multiple sequence alignment” AND “particle swarm optimization”
- String-D: “multiple sequence alignment” AND “simulated annealing”
- String-E: “multiple sequence alignment” AND “bacterial foraging”
- String-F: “multiple sequence alignment” AND “artificial bee colony”
4.5. Study Selection Criteria
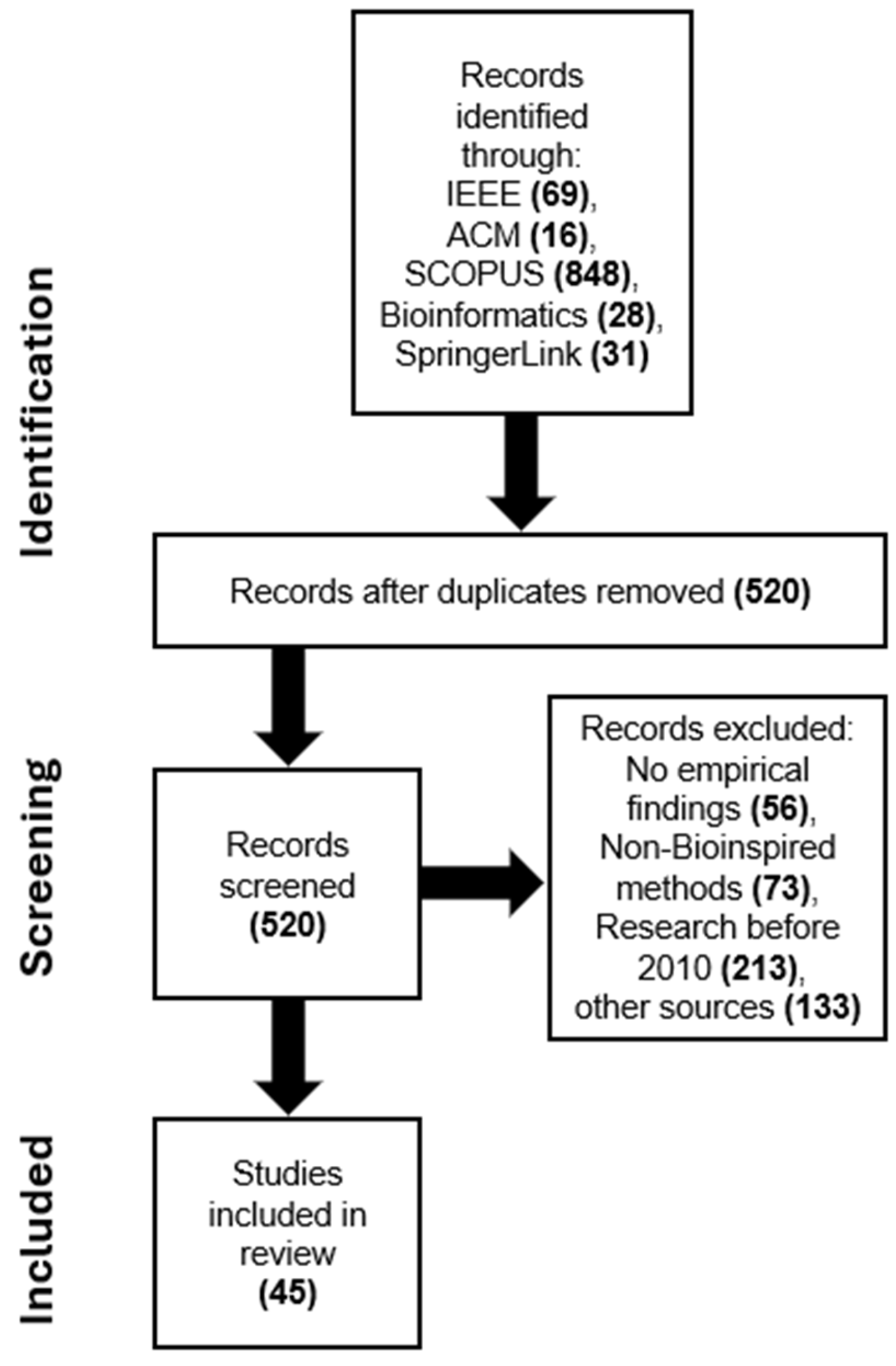
Following the application of the search criteria across the specified digital libraries, a total of 992 papers were initially retrieved. Upon further examination, numerous duplicate articles were identified and subsequently removed, resulting in 520 unique articles remaining for consideration. Through subsequent selection processes and eliminating duplicates, inclusion and exclusion criteria were established to identify the most relevant publications. Additionally, to ensure the quality of the selected papers, these studies were screened according to predefined quality assessment standards. The validity of the findings was further ensured through the cross-checking technique to verify whether the selected papers met these criteria. After applying the abovementioned criteria and the quality evaluation stage, 45 studies addressing the research topics were identified and utilized for the proposed SLR. The Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) is utilized to conduct an SLR as shown in Figure 4.
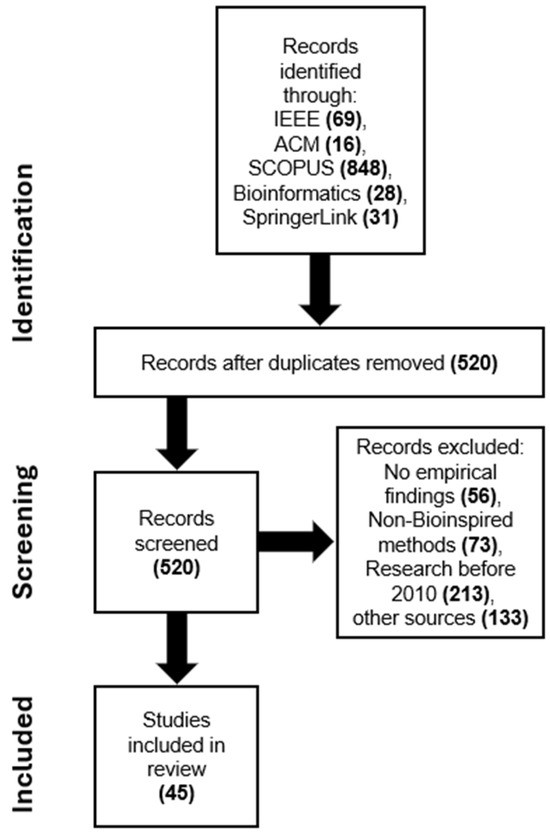
Figure 4.
The flowchart of the PRISMA approach used in the SLR study.
The Quality Standard Checklist (QSC) developed by Keele [43] was employed to ensure the quality of the selected publications. In accordance with this checklist, articles that answered “yes” to a minimum of seven questions were selected [44]. Both the quality assessment and data extraction processes were conducted in conjunction to ensure that the findings significantly contribute to the review [45]. The inclusion/exclusion criteria are provided in Table 3, and the Quality Standard Checklist is provided in the Supplementary Materials.

Table 3.
Inclusion/exclusion criteria.
4.6. Data Extraction and Synthesis
As part of the data extraction procedure for the systematic literature review (SLR), forms were created to collect data from the chosen publications [46]. By considering the information in the data extraction form, answers to the designated research questions for the SLR can be obtained. Table 4 presents the data that was extracted during the data extraction stage.

Table 4.
Data extraction form.
In Table 4, the first column delineates the search strategy employed to gather data for the data extraction process, encompassing both automatic and human-based extraction methods. The second and third columns illustrate the type of information extracted and the rationale behind each extraction. This data includes details related to the types of algorithms addressed, the benchmark methods utilized for validation, research challenges identified, and directions for future study. These extracted pieces of information are examined to facilitate grouping related studies based on their respective attributes.
5. Search Results and Metanalysis
In Table 4, the first column details the search techniques used to acquire the required data through manual and automatic extraction methods. The second column specifies the category of the extracted information, while the third column elucidates its intended purpose. This section describes the selected studies based on the extracted data.
5.1. Description of the Identified Articles
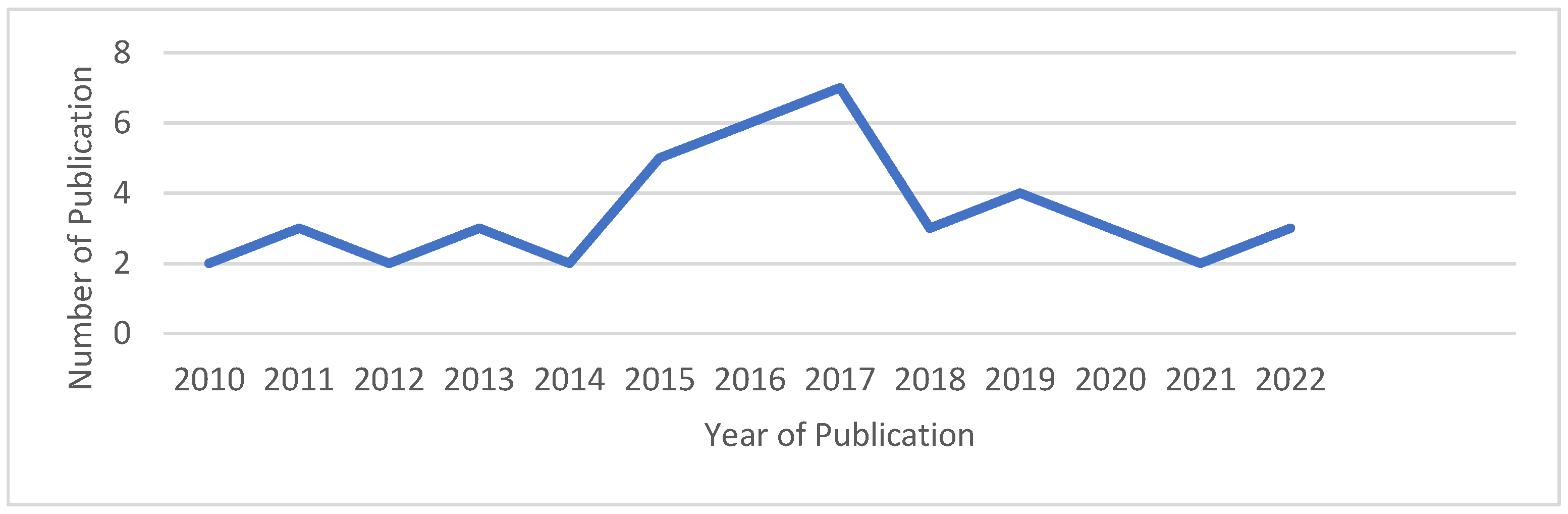
A chronological presentation of the published works selected for the proposed SLR is depicted in Figure 5. This figure illustrates the number of publications related to bioinspired algorithms used for Multiple Sequence Alignment (MSA) from 2010 to 2024. The figure indicates a rising trend in this field of study in recent years, particularly from 2014 to 2017, when a notable increase in the number of published articles occurred. Most of the publications considered for the study were added after 2014. Specifically, the highest number of published articles was observed in the years 2016 and 2017, with six papers published in 2016 and seven papers appearing in 2017. Figure 6 displays the overall numbers of relevant articles utilized in the review.
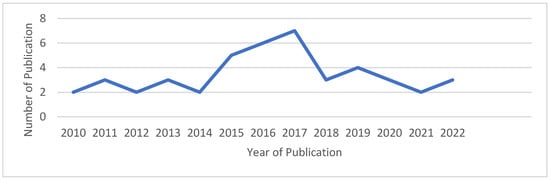
Figure 5.
Distribution of the publication and the corresponding years.
Figure 6.
Number of papers with the corresponding years of publication.
5.2. Synthesis Results
The SLR results of the data synthesis addressing the research questions derived from the chosen papers are presented in this section. As a result, the research questions created for the SLR will be addressed in this section.
5.2.1. RQ1: What Are the Common Bioinspired Algorithms Used for MSA?
This section attempts to answer research question 1 by exploring the popular bioinspired algorithms used for the MSA. Based on the reviewed articles, many bioinspired approaches have been proposed to solve the MSA problem. These approaches include Genetic Algorithm (GA), Particle Swarm Optimization (PSO), Artificial Bee Colony (ABC), Memetic Optimization (MO), and Biography-Based Optimization (BBO). The summary of the bioinspired algorithms for MSA identified in the selected studies is summarized in Table 5 and given below.

Table 5.
Summary of the bioinspired algorithms for MSA.
Genetic Algorithm (GA)
Natural selection, mutation, and crossover processes that individuals experience in evolution theory serve as the foundation for this method [3,70,71]. Each individual in the GA when solving the MSA problem is a potential alignment, and the optimization process is provided by the rearrangement of the sequence based on recombination and mutation operators [29]. Generally, the first operator changes an existing gap position, but the second generates new ones by joining two individuals. Several papers that used GA-based methods were identified based on the selected studies in this SLR. Kumar [47] designed a GA centered on the mutation and crossover process. The starting population is created randomly, and each individual’s quality is assessed using the sum-of-pairs objective function. Operators are put into action after a tournament selection process. The author used various mutation operators, which are empirically executed and dynamically selected, as well as vertical and recombination crossover. This approach was evaluated using the BAliBASE benchmark, with a maximum of 27 sequences assessed.
In contrast to other popular GA techniques for MSA, such as SAGA [30] and MSA-GA [72], this methodology produced good results in most situations. This approach was modified in [21] to use the COFFEE objective function [73] for DNA alignments. The Needleman–Wunsch algorithm is used to construct the GA’s initial population, and fitness values are computed for each individual. A modified version of the consistency-based COFFEE goal function replaces the Weighted Sum-of-Pairs technique. The approach was evaluated using the BAliBASE benchmark by applying the REFAB Q metric in DNA sequence alignment.
To obtain decent results, Rani and Ramyachitra [74] suggested an additional GA that concentrated on crossover operators. The first population is created randomly, and then, a multi-objective method that computes the sum-of-pairs and total column scores for each individual is used, then an elimination criterion is implemented. The authors used each test case for the BAliBASE datasets to evaluate the methodology. Chentouri et al. [48] developed a multi-objective GA for multiple RNA sequence alignment based on Pareto Optimality. The original population is randomly initialized, the non-dominant individuals are cloned into an archive population, and each individual’s fitness is calculated. The authors assessed this approach using the BAliBASE benchmark and SPSS and SCI metrics. The authors noted that the results are highly sensitive to the gap and mutation factors and recommended varying these parameters in subsequent research.
In a different study, the RNA alignment evaluation approach using a multi-objective GA with dynamic weight was proposed by Chentouri et al. [48]. The starting population is created randomly, and each individual is assessed for fitness using a multi-objective method that includes base pam score weighted completely matched column and entropy. The approach has been assessed using the BAliBASE benchmark. The model uses SPSS and SCI as the evaluation metrics. Based on NSGA-II, Kaya et al. [49] presented a multi-objective GA for MSA. Three objective functions were used to calculate the fitness of the initial population, which is recreated at random: support maximization, affine-gap penalty, and similarity. The authors evaluated the approach using the BAliBASE benchmark.
For MSA, Catteree et al. [33] suggested combining a hybrid GA with the Chemical Reaction Optimization (CRO) algorithm. Initially, a starting population is selected at random, the cut point is either an entire column match or a crossover performed with a single-point approach. Following the execution of mutation operators in the GA, the Chemical Reaction Optimization algorithm receives the output. DNA samples from the SWISS-PROT datasets were used to assess the technique. Amorim et al. [50] parallelized the GA’s primary stage in the MSA-GA tool through multithread programming. The individuals are initially formed using the Needleman–Wunsch algorithm. The authors parallelized the score matrix using the wavefront framework. The authors assessed the technique using the BAliBASE dataset. Based on the NSGA-II, ZambranoVega et al. [8] developed a parallel MSA employing a multi-objective GA. Pre-alignments computed by programs like ClustalW [75], MAFFT [37], MUSCLE [76], and T-Coffee are used to create the initial population [73]. Individuals from the original population are processed using crossover and mutation operators to form an auxiliary population. Each individual’s fitness is then determined using the STRIKE function, percentage of total columns, and percentage of non-gaps. The approach was created using the BAliBASE benchmark. It is noted, meanwhile, that processing medium- and large-scale datasets presents challenges to the approach.
Particle Swarm Optimization (PSO)
Another bioinspired algorithm used for solving the MSA problem is Particle Swarm Optimization (PSO), which is an approach to solving optimization issues based on a population of particles [77,78,79,80,81]. The optimization procedure is carried out with each particle’s position and velocity considered. Changes are therefore made to these values in accordance with each particle’s optimal position as well as the best available global knowledge [82]. Based on the selected studies in this SLR, several PSO-based methods were identified. For example, a PSO algorithm was created by Tran and Wallinga [65] to generate structurally and evolutionary optimal alignments. Then, each person’s fitness is determined using the STRIKE function, percentage of total columns, and percentage of non-gaps. This paper evaluated the approach using the BAliBASE dataset. Yang [83] presented a PSO technique for the MSA problem based on fish swarm intelligence. The plan was to retain the balance between local and global search capabilities while aiming for a faster convergence. The authors of this paper utilized the BAliBASE benchmark to assess the methodology. Lalwani [20] suggested a two-level PSO algorithm to execute MSA, where each level optimizes a distinct objective function. This tactic works well to stop a locally optimal solution from converging too soon. The authors evaluated the approach using the BAliBASE benchmark. The SP and TC were used as the metrics. Based on earlier research, the authors in [66] introduced a novel two-level PSO technique for RNA sequence-structure alignment. The PSO then executes independently for the two levels. Each alignment’s starting length is calculated at level one, and changes are made in the gap places at level two. The tests were conducted using the BAliBASE dataset.
We can also observe the application of hybrid PSO. Chaabane [10] presented a PSO model, the initial step of Simulated Annealing, to carry out MSA that meets the metropolitan criteria. Alignments are produced using the metropolis criterion and are utilized to populate the PSO algorithm’s initial population. The approach was evaluated using the BAliBASE dataset. The findings achieved were superior to those of other nature-inspired tools, such as GAPAM [51] and IBBOMSA [84], with an 8 percent rise in the SPS quality rating. A new PSO variation for MSA based on the free electron method and hybridized with HMM was proposed by Sun et al. [85]. The particles are started randomly, and the PSO is used in the HMM learning process. To spread out the search space, the mean best location is computed. This work assessed protein alignments using the BAliBASE benchmark, while DNA alignments were assessed using a simulated dataset. Zhan et al. [67] introduced a novel MSA approach that applies the partition function with a hybrid HMM and PSO technique. The PSO optimizes the HMM parameters, and the distance matrix for the Multiple Sequence Alignment is calculated using the findings. The three well-known protein benchmarks, BAliBASE, QXBench, and SABmark, were utilized to assess the method. This method was later enhanced by Zhan et al. [67]. The model was evaluated using other good techniques, such as the MSA generated by these methods, to reconstruct the phylogenetic trees.
Memetic Metaheuristic (MA)
The Memetic Metaheuristic is another bioinspired-based method for conducting MSA [13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87]. As an expansion of GA, this metaheuristic employs local search techniques to prevent premature convergence [87]. The Shuffled Frog-Leaping Technique (SFLA) is a well-known memetic algorithm for discrete optimization [100]. Multiple frogs jump in a wetland at SFLA. The frogs communicate with each other to determine which of the limited number of water lilies has the greatest food. When frogs are utilized to solve Multiple Sequence Alignment problems, gap locations are adjusted by the operators.
A hybridized multi-objective SFLA using Progressive Alignment was proposed by RubioLargo et al. [68]. Initially, a random initial population generator divides the people into subpopulations of water lilies, known as memeplexes. The suggested approach was assessed in this work using three benchmarks: BAliBASE, PREFAB, and SABmark. As a result, RubioLargo et al. [88] suggested utilizing OpenMP for distributed computing as a parallel solution to their method. The technique gives each memeplex its own thread, and the outcomes are processed concurrently. Here, the authors assessed the algorithm’s performance using the HOMSTRAD dataset. Subsequently, an expansion of the earlier work was published by Rubio-Largo et al. [89], the primary contribution of which is a compelling parallel version of the method for aligning data sets with a high sequence count.
Bacterial Foraging Optimization (BFOA)
Bacterial Foraging Optimization (BFO) is another essential bioinspired method identified in this SLR. A multi-objective BFO method was presented by Rani and Ramyachitra [74] for solving the MSA problem. The population’s individuals are first created, and then the chemotaxis phase starts. The BAliBASE benchmark was utilized in this work to assess the methodology. The authors compared the acquired results with other bioinspired-based approaches and commonly used MSA methods. A comparable technique for conducting MSA based on a hybrid BFO and GA was presented by Manikandan and Ramyachitra [99]. Consequently, each individual’s fitness is determined, and the top two are chosen for reproduction, which will occur according to a predetermined probability. The suggested approach was assessed using the BAliBASE, PREFAB, SABmark, and OXBench benchmarks.
Other Bioinspired Techniques
In addition to the methods mentioned above, other bioinspired algorithms are identified in this SLR. For example, the authors in [101] introduced Biogeography-Based Optimization for the MSA. The distribution of species across territory serves as the model for this strategy. In this way, through processes known as migration and emigration, species travel between the regions. Zemali and Boukra [98] presented a novel hybrid method for performing MSA using an optimization algorithm based on biogeography. The method creates the initial population using Progressive Alignment, which means that a distinct set of parameters is utilized for each territory. The authors have assessed the suggested approach using the BAliBASE benchmark. To tackle the MSA problem, [98] combined earlier research with Talbi and Draa’s Quantum Evolutionary approach [102]. Pairwise alignments and the hill climbing process construct the initial population. A Simulated Annealing technique is then used to initialize the quantum population, determining which individuals will be taken to the population. The method was evaluated using the Baliscore quality metric.
Another bioinspired-based method is the Simulated Annealing to carry out MSA. It involves adjusting the potential solution, which is governed by a temperature parameter, to investigate the solution space [91]. The methods mostly consist of controlled modifications to the alignment’s gap positions. Yao et al. [91] suggested a hybrid method combining Star Alignment and Simulated Annealing to solve the MSA problem. Ultimately, the Multiple Sequence Alignment is obtained by applying this approach. Another bioinspired algorithm applicable to MSA is the Flower Pollination algorithm (FPA) [92,93]. It is based on the mechanisms of self-pollination and cross-pollination that occur during flower pollination, among others [103]. Each flower represents a potential alignment when used to solve the MSA, and the pollination procedures essentially involve altering the gap positions either globally or locally. To achieve protein MSA, the authors in [92] suggested a novel FPA that involves two main processes: self-pollination and cross-pollination. BAliBASE was utilized in this paper to assess the approach. The outcomes demonstrated that this method might yield to better MSA. But the biological significance of the data was lacking. As a result, the authors suggested expanding on the earlier research by employing a multi-objective, hybrid EPA approach, as shown in [93]; here, the authors evaluated the approach using BAliBASE.
Similarly, a multi-objective hybrid ABC was proposed by Rubio-Largo et al. [15] to carry out MSA. First, each member of the original population is created randomly and referred to as an employed bee to create a new individual. A crossover operator is carried out on a randomly selected bee for each bee engaged during this procedure. The Kalign method [104] establishes a random alignment segment, and the realigned section is returned to its original position. This paper assessed the approach using the BAliBASE benchmark. The authors used PREFAB Q and TC quality indicators.
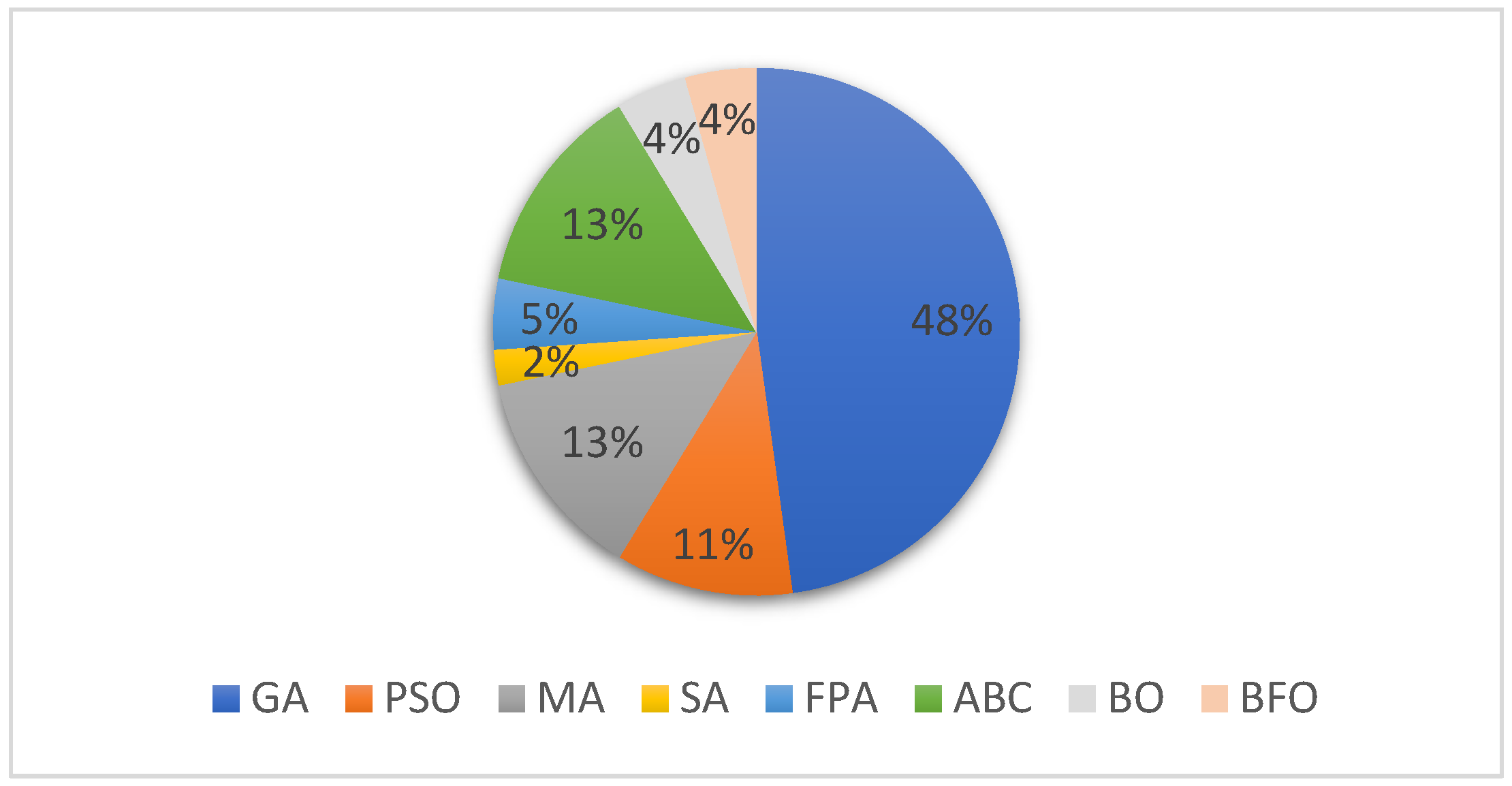
Figure 7 shows the distributions of papers for each category of the bioinspired-based methods for MSA. As can be seen from the figure, the percentage of each of the algorithms identified in the selected article for this SLR is indicated accordingly. The chart shows that the Genetic Algorithm is the most used bioinspired-based algorithm for solving the MSA problem with 48%, followed by the Memetic Algorithm and ABC representing 13% each, of the total papers identified for the MSA solutions. Next are the PSO and FPA with 11% and 5%, respectively, of the total articles identified in the review, followed by the BO and BFO which represent 4% each of the total number of bioinspired-based algorithms applied for solving the MSA problem. Finally, we can see that based on the reviewed literature, Simulated Annealing is the least utilized algorithm for MSA with 2% of totally reviewed papers.
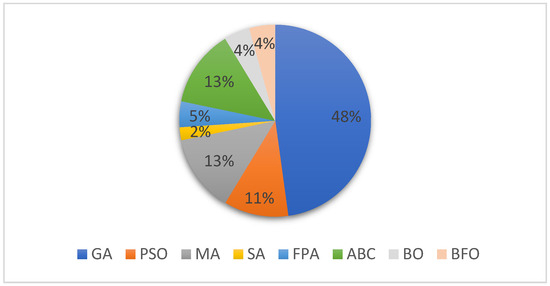
Figure 7.
Distribution of the bioinspired algorithms used in MSA.
5.2.2. RQ2: Benchmark Methods Used in MSA
Benchmarks for Multiple Sequence Alignment are used to evaluate the effectiveness of various MSA techniques and algorithms. There must be some benchmark sequences and “gold standard” reference alignments of those sequences to compare various alignment methods efficiently. It is necessary to determine a quantifiable accuracy concerning gold standard reference alignments to evaluate the performance of various automated alignment systems.
Several benchmarks are already available, with the primary objective being to evaluate the quality of the various Multiple Sequence Alignment tools. MSA solutions have been assessed using a variety of benchmarks based on the papers chosen for this SLR. Thus, the effectiveness of various MSA techniques and algorithms is evaluated using benchmarks. These benchmarks usually comprise a selection of well-chosen reference alignments for a wide range of protein or nucleotide sequences. Many benchmarks are available for free, all of which aim to evaluate the various Multiple Sequence Alignment tools. Several benchmarks have been utilized to assess MSA solutions based on the papers that were chosen for this SLR. This includes HOMSTRAD [64], BAliBASE 2.0 [59], OXBench [105], PREFAB [90], SABmark [94], and BAliBASE version 3 [71]. Table 6 shows the summary of the benchmarks identified in the selected studies and their explanation given as follows:

Table 6.
Widely used Multiple Sequence Alignment benchmarks.
- HOMSTRAD [64]: This database contains protein domains grouped according to structural and sequence similarities. Several authors have used HOMSTRAD as a benchmark database, even though it was not intended to be one. The database offers combined information on protein structure and sequence that has been taken from the PDB [95] as well as other databases, such as Pfam [96] and SCOP [97]; the most recent version of the database includes 9602 single-member families and 1032 domain families, with each family containing 2 to 41 sequences.
- BAliBASE [106,107]: This is the first extensive benchmark created especially for the alignment of multiple sequences. To guarantee the proper alignment of conserved residues, the alignment test cases are based on 3D structural superpositions that are manually improved. A wide range of issues encountered by multiple alignment methods is represented by the 217 alignments in the current version of BAliBASE, ranging from 4 to 142 sequences. These alignments are arranged into six reference sets, which include sequences with large NC-terminal extensions or internal insertions, transmembrane regions, repeated or inverted domains, and eukaryotic linear motifs.
- OXBench [105]: This offers several automatically constructed protein alignments based on aligning techniques for sequence and structure. Three data sets comprise the benchmark; between two and twenty-two sequences per alignment, the master collection now consists of 673 alignments of protein domains with known three-dimensional structures. Sequences with an unknown structure are added to the master set to create the extended dataset. Lastly, the full-length sequences for the domains in the master data set are included in the full-length data collection.
- PREFAB [90]: This dataset has 1932 multiple alignments and was built with a fully automated protocol. Two approaches of 3D structure superposition were used to identify and align pairs of sequences with known 3D structures. Next, for every pair of structures, multiple alignments were created using 50 homologous sequences found through sequence database searches; since the building is automatic, many tests can be incorporated. This benchmark’s drawback is that it only infers multiple alignment accuracy from aligning the first two sequences with known 3D structures.
- SABmark [94]: This includes reference sequence sets obtained from the SCOP protein structure categorization, separated into two groups: superfamilies and the twilight zone (Blast E-value 1) (residue identity 50 percent); each reference set’s sequence pairs are aligned using the consensus of two distinct 3D structure superposition programs. Once more, the benchmark offers pairings of sequences in only “gold standard” alignments. Even though the sequences are arranged into families, each containing a maximum of 25 sequences, a consistent multiple-alignment solution is not offered.
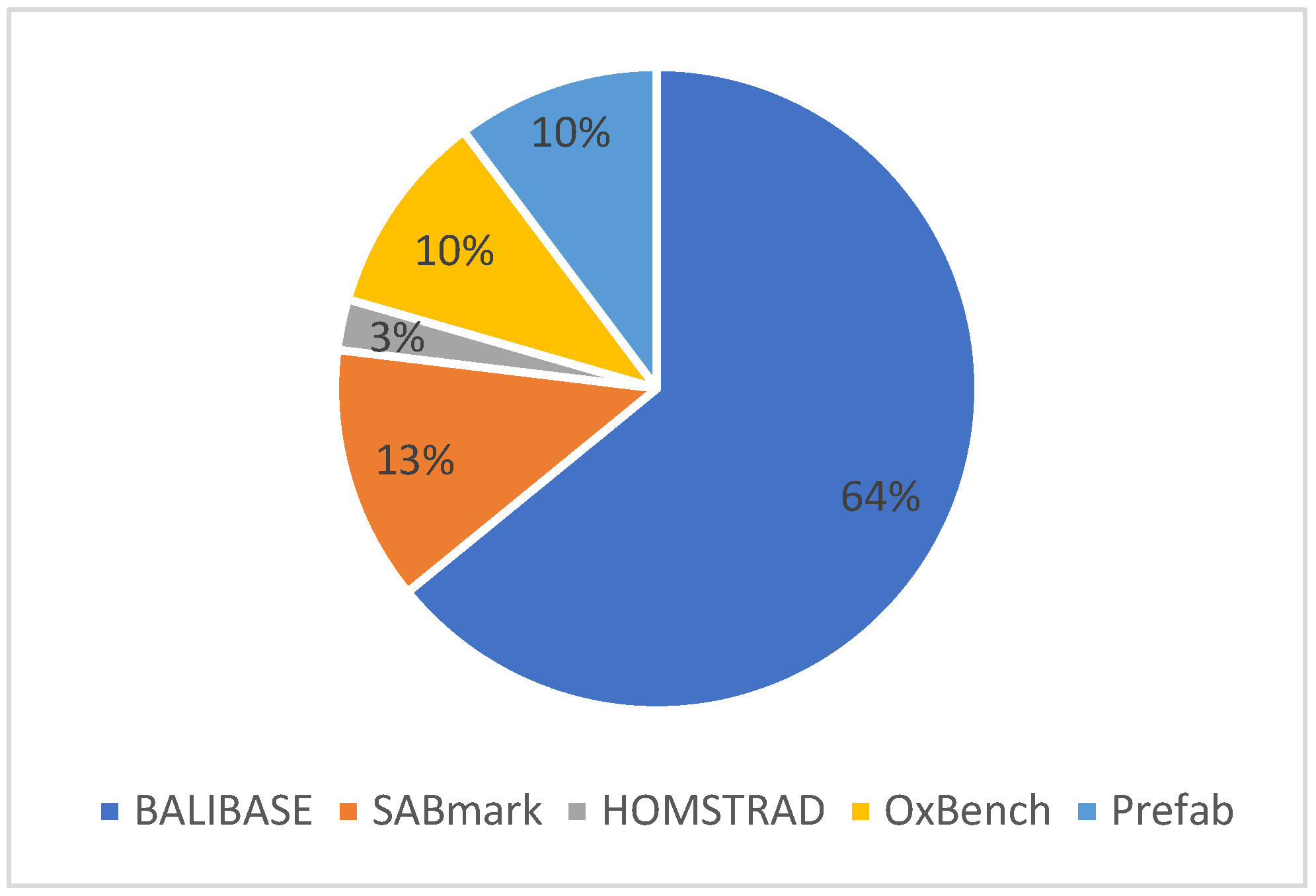
Figure 8 shows the performance evaluation measures used for the MSA problem. The chart shows that several benchmark datasets are used to validate the effectiveness of the Multiple Sequence Alignment solutions. Derived from the literature chosen for the proposed SLR investigation, prominent benchmark techniques encompass BAliBASE, SABmark, HOMSTRAD, OXBench, and Prefab. The data indicate that BAliBASE and SABmark are the prevailing benchmark datasets, utilized in 64% and 13% of cases, respectively, followed by OXBench and Prefab, each accounting for 10%. Conversely, HOMSTRAD emerges as the least utilized benchmark dataset for MSA, representing only 3% of cases.
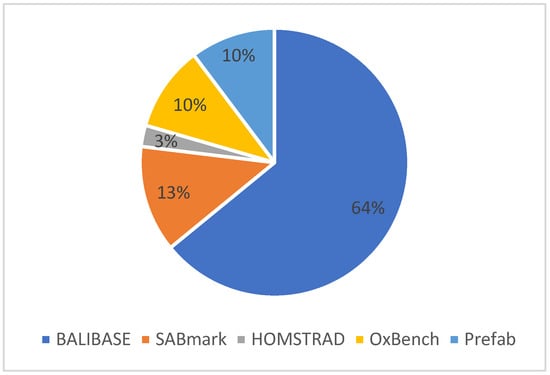
Figure 8.
Distribution of benchmark datasets.
5.2.3. RQ3: Performance Evaluation Measures for MSA
Due to the utilization of diverse algorithms by different Multiple Sequence Alignment tools, the quality of alignments may vary significantly. In response, various quality estimation techniques have been devised to evaluate and provide guidelines for the design and enhancement of sequence alignment software [55]. This section endeavors to address RQ3 by examining classical quality estimation methods [54] and providing a summary of Table 3 along with their descriptions [55]. Drawing from the literature, prevalent quality estimation techniques encompass structured-based, simulated, and consistency-based approaches. Subsequent subsections will delve into the specifics of these techniques:
Structural-Based Method
In particular, benchmarks were created using information on protein structure. The sort of benchmarks that are most commonly used are structural benchmarks [2]. These most frequently use the superposition of known protein structures as a separate alignment method, to which sequence analysis alignments can be compared using the previously mentioned sum-of-pairs and true column metrics. Structural standards are naturally quite relevant when looking for structural concordance across amino acid sequence alignments. One of the most often utilized structured benchmarks for protein alignment software is BAliBASE [56]. These benchmarks provide preset test sets and reference alignments that have been manually and automatically revised based on the three-dimensional structure of proteins. The concept behind these benchmarks is that amino acid residues corresponding to the same position in the three-dimensional structure should be aligned [55].
Many sequence alignment software developers optimize their programs on a limited number of data sets, which can lead to the “high in score but low in ability” phenomenon. This is true even though BAliBASE and other similar benchmarks are widely used and provide fixed test sets and reference alignments if these benchmarks are not updated regularly. Other specifically designed structural benchmarks are HOMSTRAD, PREFA, and SABmark, which are not generated by hand annotation of protein alignments like BAliBASE. Reference sets are also available for RNA structures [41,106].
Simulation-Based Method
Another technique uses produced data sets to score Multiple Sequence Alignment software. One of the main goals of MSA is to find residues that have developed from a common ancestor. One benchmarking method is to create artificial sequence families by simulating evolution along a known tree. This simulation-based method tracks the “real” homology relationships between specific residue sites while describing nucleotide substitution, deletion, and insertion rates using a probabilistic model of sequence evolution. These methods generate evolved sequences and reference alignments based on the evolution of the sequence using a probabilistic model to simulate sequence evolution [55]. Since the evolution model totally governs the generated mutations, the accuracy of these benchmarks is limited by how well the accepted model represents the natural evolution. Since the evolution model totally governs the generated mutations, the accuracy of these benchmarks is limited by how well the accepted model represents the natural evolution. The likelihood that, in some cases, the probabilistic model may even skew the estimation if it mimics the sequence relationship model that the tested program uses further complicates the probabilistic model’s creation and selection. These kinds of benchmarks offer pre-established reference alignments that may be used to assess the performance of recently created software. As a result, a scoring system is required to determine how well the tested program aligns with the reference.
Numerous software packages, such as Rose [107], EvolveAGene3 [108], INDELIBE [109], Phylosim [110], Revolver [111], and ALF [112], are capable of conducting simulated sequence evolution. The alignment accuracy, compared to the genuine alignment (known from the simulation), is often evaluated using two metrics: the sum-of-pairs (SP) and the true column (TC) scores [111]. The SP score refers to the fraction of aligned residue pairs that are consistent between the reconstructed and actual alignments, averaged across all pairwise comparisons between individual sequences. The TC score measures the proportion of completely aligned columns replicated in the reconstructed alignment.
Consistency-Based Method
An alternative technique for estimating quality is based on the similarities between alignments produced using various Multiple Sequence Alignment strategies rather than a reference alignment. It is predicated on the notion that two residues are probably accurately aligned if many software programs reliably align them. However, a significant flaw in this approach is that if two residues are regularly aligned incorrectly by separate software programs, the scoring system will accept this error as accurate. This approach employs specific scoring techniques, including the multiple overlap score [113] and the head-or-tail (HoT) score [114,115], in contrast to the estimating methods that rely on reference alignment.
Two popular scoring approaches that quantify the degree of similarity between two alignments by counting common pairs and columns between them are the total column or the column (TC) score and the sum-of-pairs (SP) score [116]. The Wilcoxon signed-rank test [117] and the Friedman rank test [76,118] may be used for alignment accuracy discrimination by providing a p-value, which indicates the likelihood that the performance difference between different methods is due to chance.
Based on the idea that biological sequences lack direction, this consistency test assumes that alignments should remain unchanged regardless of whether the input sequences are provided in the original order or inverted. The overlap measurements mentioned above can be used to quantify the degree of agreement between the alignments produced from the original and reversed sequences. Consistency among aligners and score are two intriguing consistency approaches that may be used easily because they do not assume a reference alignment or sequence of evolution model. Moreover, a set of precise aligners must have great consistency, which makes it attractive. Although most aligners share numerous characteristics and are therefore not “independent”, the consistency requirement nevertheless appeals to the intuitive notion of “independent validation”. The consistency method has a significant flaw in not ensuring accuracy; approaches can be consistently incorrect. More quietly, the selection of aligners within the set impacts consistency. While this can be somewhat alleviated by presenting as many diverse alignments as possible, it is still possible for an accurate alignment to be graded negatively if inaccurate but similar alignments outnumber it.
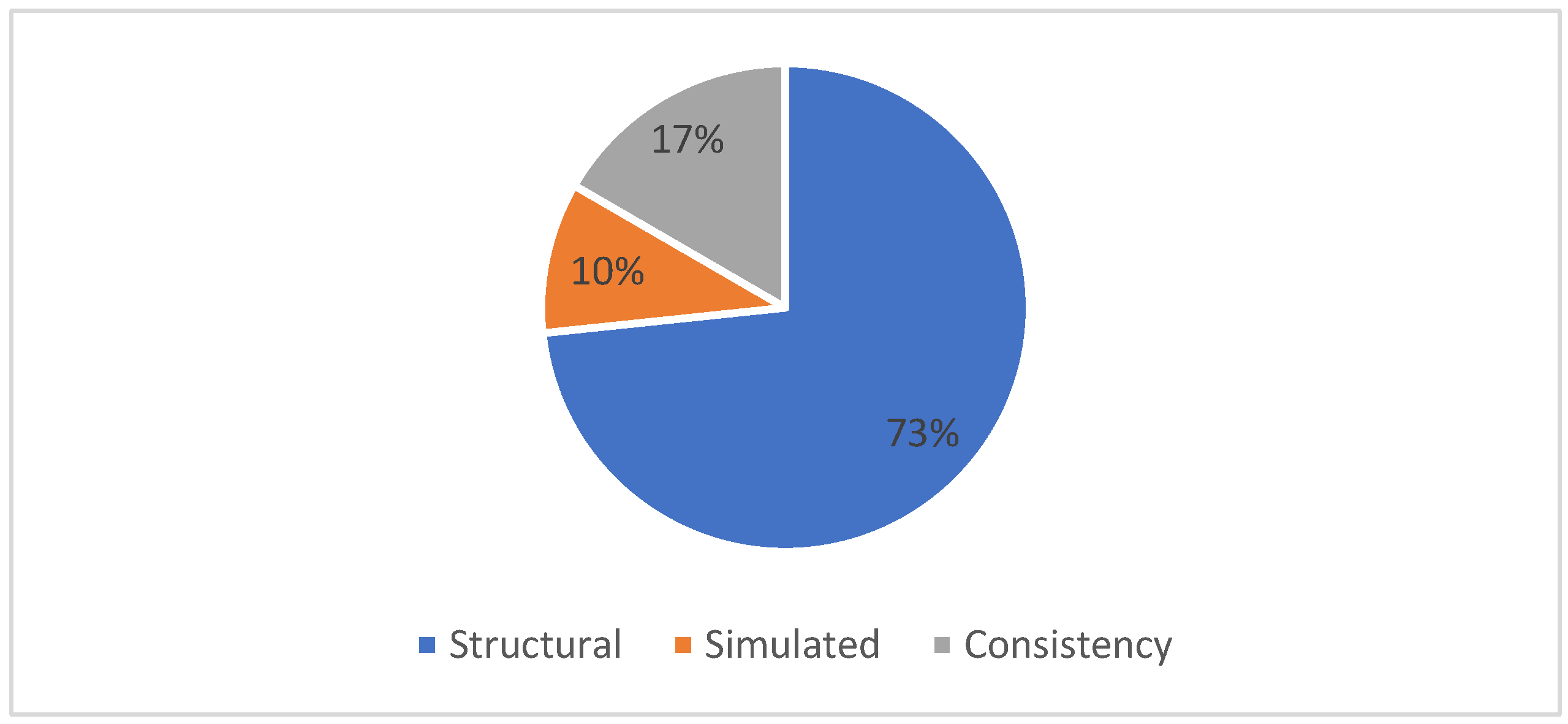
Table 7 shows the performance evaluation measures used for the MSA problem. From Table 7, it can be observed that several performance quality measures are used to validate the effectiveness of the Multiple Sequence Alignment solutions. Based on the published papers from the literature selected for our SLR study, the most popularly used performance quality evaluation measures include structural-based techniques, simulated- based approaches and consistency-based approaches. From Figure 9, it can be observed that structural-based techniques based on the idea that amino acid residues that correspond to the same position in the three-dimensional structure should be aligned are the most popularly used quality measures for MSA with 73%. Next is the consistency predicated on the notion that two residues are probably accurately aligned if many software programs reliably align them with 17%. And finally, the simulation-based techniques use a probabilistic model to simulate sequence evolution and produce evolved sequences and reference alignments with 10%.

Table 7.
The quality estimation of MSA.
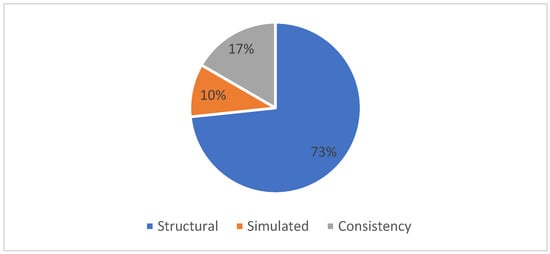
Figure 9.
Distribution of performance quality measures.
5.2.4. RQ4: What Are the Challenges and Open Issues in Bioinspired-Based MSA?
This section addresses RQ4 by delineating the challenges and open issues associated with bioinspired-based Multiple Sequence Alignment (MSA). In recent years, the research community has increasingly relied on bioinspired-based approaches to tackle NP-hard problems [7,69]. These approaches offer the advantage of solving large-scale cases with reduced computational resources, including memory and processing time [24], thereby effectively addressing complex optimization challenges in bioinformatics. While alternative methods exist for optimizing these issues, the versatility of bioinspired-based methods renders them invaluable tools capable of generating “high-quality” solutions within reasonable computing timeframes [41]. An inherent advantage of employing bioinspired-based methods in bioinformatics lies in their ability to effectively address MSA problems, which typically involve large-scale, NP-hard optimization, posing significant constraints on classical optimization techniques [7]. Furthermore, given that data provided by scientists and researchers inherently contain errors, extended bioinspired-based methods such as learnheuristics [123] and simheuristics [124] offer greater adaptability compared to more precise approaches.
Optimizing various objectives is a prevalent task in bioinformatics, rendering the utilization of bioinspired-based methods both appropriate and intrinsic to the field. Notably, the Protein Structure Prediction (PSP) problem and numerous string-related challenges stand out as prominent subjects in bioinformatics research. Moreover, the realm of medical imaging heavily relies on bioinspired-based methods, particularly for tasks such as variable selection and parameter fine-tuning. Over the past few decades, advancements in computational power have facilitated the development of distributed and parallel implementations of bioinspired-based methods. This progress has spurred bioinspired-based methods specialists to introduce increasingly sophisticated and intricate designs, including hybrid and multi-objective techniques.
Bioinspired-based methods are particularly fitting and natural for addressing the multifaceted optimization challenges prevalent in bioinformatics. Notably, within this domain, a subset of string-related difficulties, such as the Protein Structure Prediction (PSP) problem, garners significant attention. These methods extend their applicability to various optimization problems, encompassing alignment challenges and the identification of DNA patterns. Furthermore, the reliance of medical imaging and disease modeling on bioinspired methods for tasks like variable selection and parameter fine-tuning underscores their indispensable role in advancing healthcare research. Moreover, the advent of new high-throughput technologies has led to a surge in available data in bioinformatics, including microarray genomic data, protein and DNA sequences, image-based biomarkers, clinical tests, and bibliographic data. This influx of data introduces novel challenges, necessitating the utilization of knowledge-discovery techniques [125]. Evolved algorithms now enable the exploration of large-scale, real-world data, thereby unlocking the full potential of these methodologies, whereas previous reliance on small-scale benchmarks merely facilitated the testing of fundamental principles [126].
In the forthcoming years, the demand for variable selection in artificial intelligence techniques will progressively necessitate the adoption of bioinspired-based algorithms. Given the sheer volume and diversity of data in bioinformatics, coupled with the imperative for timely solutions and the continuous advancements in computational power, time complexity remains a perennial challenge. Consequently, the landscape of future bioinformatics and bioinspired-based research is multifaceted, offering myriad avenues for exploration and innovation. Some of the overarching open issues in the field can be synthesized as follows:
- The creation of more potent algorithms based on parallel and distributed paradigms and the blending of various algorithms.
- The creation of more resilient algorithms that consider the uncertainty or stochasticity present in bioinformatics (caused by errors in the technology used to collect data or by the characteristics of the data itself).
- The development of frameworks for parameter fine-tuning to take advantage of instance-specific aspects to improve results.
- The application of multi-objective techniques to take into account the various objectives in the majority of issues.
6. Limitations of the Study
Several bioinspired-based techniques for the MSA problem are identified in this SLR. Our goal in creating our protocols is to handle the RQs while optimizing internal and external validity. This argument’s validity is nevertheless subject to many limitations and objections, some of which are discussed in this section.
- Only journal and conference papers that address bioinspired-based methods for solving the MSA problem are included in this SLR. We used our search approach in the study to find and exclude several irrelevant research publications. This ensures that the selected research papers fulfilled the investigation’s needs. However, it is believed that this review would have been improved by including other sources, such as books.
- We limited the scope of our search to English-language materials. The possibility that comparable papers in this field of study exist in languages other than English can lead to linguistic bias. Appreciatively, all of the research papers were written in English. Therefore, we do not have language bias.
- The primary databases were considered while searching through the study articles; nevertheless, other digital libraries containing relevant studies were likely overlooked. To overcome this limitation, we matched the search terms and keywords to a reputable library of research works. However, when looking for the keywords, certain synonyms could be missed. To solve this issue, the SLR methodology has been adjusted to guarantee that no crucial phrases are overlooked.
7. Conclusions and Discussion
In this study, we presented a systematic literature review on bioinspired-based methods for solving the MSA problem that can guide researchers and practitioners in understanding the challenges and new trends in the area. In particular, many bioinspired-based techniques were examined, emphasizing the MSA. The study used the SLR method, which thoroughly analyzes and synthesizes the published articles. We considered studies conducted from 2010 to 2024 to provide more recent and comprehensive developments. After collecting and examining several studies, 45 papers were found after eligibility screening and quality assessment. The results showed that the Genetic Algorithm and Memetic Optimization are the most used bioinspired-based algorithms applied for solving MSA problem with 49%, followed by the Memetic Algorithm representing 13% of the total papers identified for the MSA solutions. Next is the PSO, which accounts for 11% of the total articles identified in the review, followed by the BO and BFO, which represent 5% of each of the total number of bioinspired-based algorithms applied for solving the MSA problem. Finally, we can see that based on the reviewed literature, the Simulated Annealing algorithm is the least utilized algorithm with 2% of the total reviewed papers.
Furthermore, the results of the performance evaluation measures used for the MSA problem were illustrated. Figure 8 shows that several benchmark datasets are used to validate the effectiveness of the Multiple Sequence Alignment solutions. Based on the articles from the literature selected for the proposed SLR study, the most popular benchmark techniques include BAliBASE, SABmark, HOMSTRAD, OXBench, and Prefab. The figure shows that the most commonly applied benchmark datasets are BAliBASE and SABmark, with 64% and 13%, respectively, followed by OXBench and Prefab with 10% each. Finally, HOMSTRAD has been identified as the least benchmark dataset for the MSA with a 3% use case.
Moreover, the proposed review shows the performance evaluation measures used for the MSA problem among the reviewed papers. Figure 9 shows that several performance quality measures were used to validate the effectiveness of the Multiple Sequence Alignment solutions. Based on the published papers from the literature selected for the proposed SLR study, the most popularly used performance quality evaluation measures include structural-based techniques, simulated-based approaches, and consistency-based approaches. Figure 9 shows that structural techniques based on the idea that amino acid residues corresponding to the same position in the three-dimensional structure should be aligned are the most popular quality measures for MSA with 73%. Next is consistency, which is predicated on the notion that two residues are probably accurately aligned if many software programs reliably align them with 17%; finally, the simulation-based techniques, which use a probabilistic model to simulate sequence evolution and produce evolved sequences and reference alignments, come with 10%.
Additionally, we examined the open issues of the study, which include the creation of more potent algorithms based on parallel and distributed paradigms, as well as the blending of various algorithms; the creation of more resilient algorithms that consider the uncertainty or stochasticity present in bioinformatics; the development of frameworks for parameter fine-tuning to take advantage of instance-specific aspects to improve results; and the application of multi-objective techniques to take into account the various objectives in the majority of issues.
Finally, the last section of the proposed systematic literature review (SLR) outlines several study limitations. These limitations include the targeted sources, predominantly journals and conferences. Additionally, the review was limited to English-language publications, potentially excluding valuable contributions from non-English sources. Moreover, the primary databases used for the review may have influenced the scope of the findings. Despite these limitations, acknowledging them provides valuable insights into the constraints of the SLR process. This recognition encourages future research efforts to address these limitations, thereby enhancing the comprehensiveness and inclusivity of systematic literature reviews in MSA methods.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/app14062433/s1, Table S1. List of papers included in the SLR study; Table S2. Overview of the relevant MSA review publications; Table S3. Quality Checklist.
Author Contributions
Writing—original draft and conceptualization, M.K.I.; methodology and formal analysis, M.K.I., U.K.Y., T.A.E.E., and M.N.; writing—review and editing, M.K.I., U.K.Y., T.A.E.E., and M.N.; supervision, U.K.Y.; project administration and funding acquisition, T.A.E.E. All authors have read and agreed to the published version of the manuscript.
Funding
The Deanship of Scientific Research at King Khalid University funded this work through a large group research project under grant number RGP2/52/44.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors extend their appreciation to the Deanship of Scientific Research at King Khalid University for supporting this study through a substantial group research initiative, identified by grant number RGP2/52/44.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Maiolo, L.; Bruno, S.A.; Lucarini, I.; Pecora, A.; De Iacovo, A.; Colace, L. Chemo-Resistive Gas Sensors Based on PbS Colloidal Quantum Dots. In Proceedings of the 2018 IEEE SENSORS, New Delhi, India, 28–31 October 2018; pp. 1–4. [Google Scholar]
- Metsky, H.C.; Matranga, C.B.; Wohl, S.; Schaffner, S.F.; Freije, C.A.; Winnicki, S.M.; Sabeti, P.C. Zika virus evolution and spread in the Americas. Nature 2017, 546, 411–415. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Wang, Y.; Wang, K.; Kang, L.; Peng, F.; Wang, L.; Pang, J. Hybrid genetic algorithm method for efficient and robust evaluation of remaining useful life of supercapacitors. Appl. Energy 2020, 260, 114169. [Google Scholar] [CrossRef]
- Eddy, S.R. A memory-efficient dynamic programming algorithm for optimal alignment of a sequence to an RNA secondary structure. BMC Bioinform. 2002, 3, 18. [Google Scholar] [CrossRef] [PubMed]
- Jakšić, Z.; Devi, S.; Jakšić, O.; Guha, K. A comprehensive review of bio-inspired optimization algorithms including applications in microelectronics and nanophotonics. Biomimetics 2023, 8, 278. [Google Scholar] [CrossRef] [PubMed]
- Chatzou, M.; Magis, C.; Chang, J.M.; Kemena, C.; Bussotti, G.; Erb, I.; Notredame, C. Multiple sequence alignment modeling: Methods and applications. Brief. Bioinform. 2016, 17, 1009–1023. [Google Scholar] [CrossRef] [PubMed]
- Amorim, A.R.; Zafalon GF, D.; de Godoi Contessoto, A.; Valêncio, C.R.; Sato, L.M. Metaheuristics for multiple sequence alignment: A systematic review. Comput. Biol. Chem. 2021, 94, 107563. [Google Scholar] [CrossRef] [PubMed]
- Zambrano-Vega, C.; Nebro, A.J.; García-Nieto, J.; Aldana-Montes, J.F. Comparing multi-objective metaheuristics for solving a three-objective formulation of multiple sequence alignment. Prog. Artif. Intell. 2017, 6, 195–210. [Google Scholar] [CrossRef]
- Calvet, L.; Benito, S.; Juan, A.A.; Prados, F. On the role of metaheuristic optimization in bioinformatics. Int. Trans. Oper. Res. 2023, 30, 2909–2944. [Google Scholar] [CrossRef]
- Chaabane, L.; Khelassi, A.; Terziev, A.; Andreopoulos, N.; Jesus, M.D.; Estrela, V.V. Particle Swarm Optimization with Tabu Search Algorithm (PSO-TS) Applied to Multiple Sequence Alignment Problem. In Advances in Multidisciplinary Medical Technologies—Engineering, Modeling and Findings: Proceedings of the ICHSMT 2019; Springer International Publishing: Berlin/Heidelberg, Germany, 2021; pp. 103–114. [Google Scholar]
- Paruchuri, T.; Kancharla, G.R.; Dara, S. Solving multiple sequence alignment problems by using a swarm intelligent optimization based approach. Int. J. Electr. Comput. Eng. 2023, 13, 1097. [Google Scholar] [CrossRef]
- Mishra, A.; Tripathi, B.K.; Soam, S.S. A genetic algorithm based approach for the optimization of multiple sequence alignment. In Proceedings of the 2020 International Conference on Computational Performance Evaluation (ComPE), Shillong, India, 2–4 July 2020; pp. 415–418. [Google Scholar]
- Dabba, A.; Tari, A.; Zouache, D. Multiobjective artificial fish swarm algorithm for multiple sequence alignment. INFOR Inf. Syst. Oper. Res. 2020, 58, 38–59. [Google Scholar] [CrossRef]
- Yu, Y.; Zhang, C.; Ye, L.; Yang, M.; Zhang, C. A Multi-objective Artificial Bee Colony Algorithm for Multiple Sequence Alignment. In Proceedings of the International Conference on Simulation Tools and Techniques; Springer International Publishing: Cham, Switzerland, 2021; pp. 564–576. [Google Scholar]
- Rubio-Largo, Á.; Vega-Rodríguez, M.A.; González-Álvarez, D.L. Hybrid multi-objective artificial bee colony for multiple sequence alignment. Appl. Soft Comput. 2016, 41, 157–168. [Google Scholar] [CrossRef]
- Makigaki, S.; Ishida, T. Sequence alignment using machine learning for accurate template-based protein structure prediction. Bioinformatics 2020, 36, 104–111. [Google Scholar] [CrossRef]
- Fukuda, H. Cascade and cluster of correlated reactions as causes of stochastic defects in extreme ultraviolet lithography. J. Micro/Nanolithogr. MEMS MOEMS 2020, 19, 024601. [Google Scholar] [CrossRef]
- Bawono, P.; Dijkstra, M.; Pirovano, W.; Feenstra, A.; Abeln, S.; Heringa, J. Multiple sequence alignment. In Bioinformatics: Data, Sequence Analysis, and Evolution; Springer: Berlin/Heidelberg, Germany, 2017; Volume I, pp. 167–189. [Google Scholar]
- Yadav, S.K.; Jha, S.K.; Singh, S.; Dixit, P.; Prakash, S.; Singh, A. Optimizing Multiple Sequence Alignment using Multi-objective Genetic Algorithms. In Proceedings of the 2022 International Conference on Decision Aid Sciences and Applications (DASA) IEEE, Chiangrai, Thailand, 23–25 March 2022; pp. 113–117. [Google Scholar]
- Lalwani, S.; Kumar, R.; Gupta, N. A novel two-level particle swarm optimization approach for efficient multiple sequence alignment. Memetic Comput. 2015, 7, 119–133. [Google Scholar] [CrossRef]
- Gupta, R.; Agarwal, P.; Soni, A.K. MSA-GA: Multiple sequence alignment tool based on genetic approach. Int. J. Soft Comput. Softw. Eng. 2013, 8, 1–11. [Google Scholar]
- Zafalon GF, D.; Gomes, V.Z.; Amorim, A.R.; Valêncio, C.R. A Hybrid Approach using Progressive and Genetic Algorithms for Improvements in Multiple Sequence Alignments. ICEIS 2021, 2, 384–391. [Google Scholar]
- Wang, J.; Yu, L.; Zhang, W.; Gong, Y.; Xu, Y.; Wang, B.; Zhang, D. Irgan: A minimax game for unifying generative and discriminative information retrieval models. In Proceedings of the 40th International ACM SIGIR Conference on Research and Development in Information Retrieval, Tokyo, Japan, 7–11 August 2017; pp. 515–524. [Google Scholar]
- Ali, A.F.; Hassanien, A.E. A Survey of Metaheuristics Methods for Bioinformatics Applications. In Applications of Intelligent Optimization in Biology and Medicine: Current Trends and Open Problems; Springer International Publishing: Cham, Switzerland, 2015; pp. 23–46. [Google Scholar]
- Katoh, K.; Misawa, K.; Kuma, K.I.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Iantorno, S.; Gori, K.; Goldman, N.; Gil, M.; Dessimoz, C. Who watches the watchmen? An appraisal of benchmarks for multiple sequence alignment. Mult. Seq. Alignment Methods 2014, 1079, 59–73. [Google Scholar]
- Rasmussen, T.K.; Krink, T. Improved Hidden Markov Model training for multiple sequence alignment by a particle swarm optimization—Evolutionary algorithm hybrid. Biosystems 2003, 72, 5–17. [Google Scholar] [CrossRef]
- Pei, J.; Grishin, N.V. MUMMALS: Multiple sequence alignment improved by using hidden Markov models with local structural information. Nucleic Acids Res. 2006, 34, 4364–4374. [Google Scholar] [CrossRef]
- Lee, Z.J.; Su, S.F.; Chuang, C.C.; Liu, K.H. Genetic algorithm with ant colony optimization (GA-ACO) for multiple sequence alignment. Appl. Soft Comput. 2008, 8, 55–78. [Google Scholar] [CrossRef]
- Notredame, C.; Higgins, D.G. SAGA: Sequence alignment by genetic algorithm. Nucleic Acids Res. 1996, 24, 1515–1524. [Google Scholar] [CrossRef] [PubMed]
- Paruchuri, T.; Kancharla, G.R.; Dara, S.; Yadav, R.K.; Jadav, S.S.; Dhamercherla, S.; Vidyarthi, A. Nature Inspired Algorithms for Solving Multiple Sequence Alignment Problem: A Review. Arch. Comput. Methods Eng. 2022, 29, 5237–5258. [Google Scholar] [CrossRef]
- Gautam, R.; Kaur, P.; Sharma, M. A comprehensive review on nature inspired computing algorithms for the diagnosis of chronic disorders in human beings. Prog. Artif. Intell. 2019, 8, 401–424. [Google Scholar] [CrossRef]
- Chatterjee, S.; Hasibuzzaman, M.M.; Iftiea, A.; Mukharjee, T.; Nova, S.S. A hybrid genetic algorithm with chemical reaction optimization for multiple sequence alignment. In Proceedings of the 22nd International Conference on Computer and Information Technology (ICCIT), Dhaka, Bangladesh, 18–20 December 2019; pp. 1–6. [Google Scholar]
- Fan, X.; Sayers, W.; Zhang, S.; Han, Z.; Ren, L.; Chizari, H. Review and classification of bio-inspired algorithms and their applications. J. Bionic Eng. 2020, 17, 611–631. [Google Scholar] [CrossRef]
- Rajwar, K.; Deep, K.; Das, S. An exhaustive review of the metaheuristic algorithms for search and optimization: Taxonomy, applications, and open challenges. Artif. Intell. Rev. 2023, 56, 13187–13257. [Google Scholar] [CrossRef] [PubMed]
- Chao, J.; Tang, F.; Xu, L. Developments in algorithms for sequence alignment: A review. Biomolecules 2022, 12, 546. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef]
- Katoh, K.; Kuma, K.I.; Toh, H.; Miyata, T. MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005, 33, 511–518. [Google Scholar] [CrossRef]
- Sievers, F.; Higgins, D.G. Clustal Omega for making accurate alignments of many protein sequences. Protein Sci. 2018, 27, 135–145. [Google Scholar] [CrossRef]
- Chowdhury, B.; Garai, G. A review on multiple sequence alignment from the perspective of genetic algorithm. Genomics 2017, 109, 419–431. [Google Scholar] [CrossRef] [PubMed]
- Almanza-Ruiz, S.H.; Chavoya, A.; Duran-Limon, H.A. Parallel protein multiple sequence alignment approaches: A systematic literature review. J. Supercomput. 2023, 79, 1201–1234. [Google Scholar] [CrossRef]
- Mohammadian, V.; Navimipour, N.J.; Hosseinzadeh, M.; Darwesh, A. Comprehensive and systematic study on the fault tolerance architectures in cloud computing. J. Circuits Syst. Comput. 2020, 29, 2050240. [Google Scholar] [CrossRef]
- Keele, S. Guidelines for Performing Systematic Literature Reviews in Software Engineering; EBSE Technical Report EBSE-2007-01; Software Engineering Group: Staffordshire, UK, 2007. [Google Scholar]
- Genc-Nayebi, N.; Abran, A. A systematic literature review: Opinion mining studies from mobile app store user reviews. J. Syst. Softw. 2017, 125, 207–219. [Google Scholar] [CrossRef]
- Kitchenham, B.; Brereton, O.P.; Budgen, D.; Turner, M.; Bailey, J.; Linkman, S. Systematic literature reviews in software engineering–a systematic literature review. Inf. Softw. Technol. 2009, 51, 7–15. [Google Scholar] [CrossRef]
- Ashtiani, M.N.; Raahemi, B. Intelligent fraud detection in financial statements using machine learning and data mining: A systematic literature review. IEEE Access 2021, 10, 72504–72525. [Google Scholar] [CrossRef]
- Kumar, M. An enhanced algorithm for multiple sequence alignment of protein sequences using genetic algorithm. EXCLI J. 2015, 14, 1232. [Google Scholar]
- Chentoufi, A.; El Fatmi, A.; Bekri, A.; Benhlima, S.; Sabbane, M. Genetic algorithms and dynamic weighted sum method for RNA alignment. In Proceedings of the 2017 Intelligent Systems and Computer Vision (ISCV), Fez, Morocco, 17–19 April 2017; pp. 1–5. [Google Scholar]
- Kaya, M.; Sarhan, A.; Alhajj, R. Multiple sequence alignment with affine gap by using multi-objective genetic algorithm. Comput. Methods Programs Biomed. 2014, 114, 38–49. [Google Scholar] [CrossRef]
- Amorim, A.R.; Visotaky JM, V.; de Godoi Contessoto, A.; Neves, L.A.; De Souza RC, G.; Valêncio, C.R.; Zafalon GF, D. Performance improvement of genetic algorithm for multiple sequence alignment. In Proceedings of the 2016 17th International Conference on Parallel and Distributed Computing, Applications and Technologies (PDCAT), Guangzhou, China, 16–18 December 2016; pp. 69–72. [Google Scholar]
- Naznin, F.; Sarker, R.; Essam, D. Progressive alignment method using genetic algorithm for multiple sequence alignment. IEEE Trans. Evol. Comput. 2012, 16, 615–631. [Google Scholar] [CrossRef]
- Amorim, A.R.; Zafalon GF, D.; Neves, L.A.; Pinto, A.R.; Valêncio, C.R.; Machado, J.M. Improvements in the sensibility of MSA-GA tool using COFFEE objective function. In Proceedings of the 3rd International Conference on Mathematical Modeling in Physical Sciences (IC-MSQUARE 2014), Madrid, Spain, 28–31 August 2014; IOP Publishing: Bristol, UK, 2014; Volume 574, p. 012104. [Google Scholar]
- Rani, R.R.; Ramyachitra, D. Application of genetic algorithm by influencing the crossover parameters for multiple sequence alignment. In Proceedings of the 2017 4th IEEE Uttar Pradesh Section International Conference on Electrical, Computer and Electronics 2017 (UPCON), Mathura, India, 26–28 October 2017; pp. 33–38. [Google Scholar]
- Nayeem, M.A.; Bayzid, M.S.; Rahman, A.H.; Shahriyar, R.; Rahman, M.S. Multi-objective formulation of multiple sequence alignment for phylogeny inference. IEEE Trans. Cybern. 2020, 52, 2775–2786. [Google Scholar] [CrossRef]
- Belattar, K.; Zemali, E.A.; Baouni, S.; Dehni, S. Parallel multiple DNA sequence alignment using genetic algorithm and asynchronous advantage actor critic model. Int. J. Bioinform. Res. Appl. 2022, 18, 460–478. [Google Scholar] [CrossRef]
- Sievers, F.; Barton, G.J.; Higgins, D.G. Multiple sequence alignments. Bioinformatics 2020, 227, 227–250. [Google Scholar]
- Chowdhury, B.; Garai, G. A bi-objective function optimization approach for multiple sequence alignment using genetic algorithm. Soft Comput. 2020, 24, 15871–15888. [Google Scholar] [CrossRef]
- Silva FJ, M.D.; Pérez JM, S.; Pulido JA, G.; Rodriguez, M.A.V. Parallel niche pareto AlineaGA–an evolutionary multi-objective approach on multiple sequence alignment. J. Integr. Bioinform. 2011, 8, 57–72. [Google Scholar] [CrossRef]
- Ortuno, F.M.; Valenzuela, O.; Rojas, F.; Pomares, H.; Florido, J.P.; Urquiza, J.M.; Rojas, I. Optimizing multiple sequence alignments using a genetic algorithm based on three objectives: Structural information, non-gaps percentage and totally conserved columns. Bioinformatics 2013, 29, 2112–2121. [Google Scholar] [CrossRef] [PubMed]
- Nizam, A.; Ravi, J.; Subburaya, K. Cyclic genetic algorithm for multiple sequence alignment. Int. J. Res. Rev. Electr. Comput. Eng. (IJRRECE) 2011, 1, 20. [Google Scholar]
- Naznin, F.; Sarker, R.; Essam, D. Vertical decomposition with genetic algorithm for multiple sequence alignment. BMC Bioinform. 2011, 12, 353. [Google Scholar] [CrossRef]
- Luo, J.; Zhang, L.; Liang, C. A multigroup parallel genetic algorithm for multiple sequence alignment. In Proceedings of the Artificial Intelligence and Computational Intelligence: Third International Conference, AICI 2011, Taiyuan, China, 24–25 September 2011; Proceedings, Part I 3. Springer: Berlin Heidelberg, 2011; pp. 308–316. [Google Scholar]
- Narimani, Z.; Beigy, H.; Abolhassani, H. A new genetic algorithm for multiple sequence alignment. Int. J. Comput. Intell. Appl. 2012, 11, 1250023. [Google Scholar] [CrossRef]
- Zambrano-Vega, C.; Nebro, A.J.; Durillo, J.J.; García-Nieto, J.; Aldana-Montes, J.F. Multiple sequence alignment with multi-objective metaheuristics. a comparative study. Int. J. Intell. Syst. 2017, 32, 843–861. [Google Scholar] [CrossRef]
- Kayed, M.; Elngar, A.A. NestMSA: A new multiple sequence alignment algorithm. J. Supercomput. 2020, 76, 9168–9188. [Google Scholar] [CrossRef]
- Lalwani, S.; Kumar, R.; Deep, K. Multi-objective two-level swarm intelligence approach for multiple RNA sequence-structure alignment. Swarm Evol. Comput. 2017, 34, 130–144. [Google Scholar] [CrossRef]
- Zhan, Q.; Wang, N.; Jin, S.; Tan, R.; Jiang, Q.; Wang, Y. ProbPFP: A multiple sequence alignment algorithm combining hidden Markov model optimized by particle swarm optimization with partition function. BMC Bioinform. 2019, 20, 573. [Google Scholar] [CrossRef] [PubMed]
- Rubio-Largo, Á.; Vega-Rodríguez, M.A.; González-Álvarez, D.L. A hybrid multi-objective memetic metaheuristic for multiple sequence alignment. IEEE Trans. Evol. Comput. 2015, 20, 499–514. [Google Scholar] [CrossRef]
- Ibrahim, M.K.; Yusof, U.K.; Eisa, T.A.E.; Nasser, M. Enhanced Genetic Method for Optimizing Multiple Sequence Alignment. Mathematics 2023, 11, 4578. [Google Scholar] [CrossRef]
- Reddy, G.T.; Reddy MP, K.; Lakshmanna, K.; Rajput, D.S.; Kaluri, R.; Srivastava, G. Hybrid genetic algorithm and a fuzzy logic classifier for heart disease diagnosis. Evol. Intell. 2020, 13, 185–196. [Google Scholar] [CrossRef]
- Su, Y.; Jin, S.; Zhang, X.; Shen, W.; Eden, M.R.; Ren, J. Stakeholder-oriented multi-objective process optimization based on an improved genetic algorithm. Comput. Chem. Eng. 2020, 132, 106618. [Google Scholar] [CrossRef]
- Gondro, C.; Kinghorn, B.P. A simple genetic algorithm for multiple sequence alignment. Genet. Mol. Res. 2007, 6, 964–982. [Google Scholar]
- Notredame, C.; Higgins, D.G.; Heringa, J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000, 302, 205–217. [Google Scholar] [CrossRef]
- Rani, R.R.; Ramyachitra, D. Multiple sequence alignment using multi-objective based bacterial foraging optimization algorithm. Biosystems 2016, 150, 177–189. [Google Scholar] [CrossRef]
- Thompson, J.D.; Koehl, P.; Ripp, R.; Poch, O. BAliBASE 3.0: Latest developments of the multiple sequence alignment benchmark. Proteins Struct. Funct. Bioinform. 2005, 61, 127–136. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Niu, W.J.; Feng, Z.K.; Chen, Y.B.; Min, Y.W.; Liu, S.; Li, B.J. Multireservoir system operation optimization by hybrid quantum-behaved particle swarm optimization and heuristic constraint handling technique. J. Hydrol. 2020, 590, 125477. [Google Scholar] [CrossRef]
- Farshi, T.R.; Drake, J.H.; Özcan, E. A multimodal particle swarm optimization-based approach for image segmentation. Expert Syst. Appl. 2020, 149, 113233. [Google Scholar] [CrossRef]
- Cui, Z.; Zhang, J.; Wu, D.; Cai, X.; Wang, H.; Zhang, W.; Chen, J. Hybrid many-objective particle swarm optimization algorithm for green coal production problem. Inf. Sci. 2020, 518, 256–271. [Google Scholar] [CrossRef]
- Rajagopal, A.; Joshi, G.P.; Ramachandran, A.; Subhalakshmi, R.T.; Khari, M.; Jha, S.; You, J. A deep learning model based on multi-objective particle swarm optimization for scene classification in unmanned aerial vehicles. IEEE Access 2020, 8, 135383–135393. [Google Scholar] [CrossRef]
- Liang, J.; Ge, S.; Qu, B.; Yu, K.; Liu, F.; Yang, H.; Li, Z. Classified perturbation mutation based particle swarm optimization algorithm for parameters extraction of photovoltaic models. Energy Convers. Manag. 2020, 203, 112138. [Google Scholar] [CrossRef]
- Du, K.L.; Swamy, M.N. Search and Optimization by Metaheuristics: Techniques and Algorithms Inspired by Nature; Birkhäuser: Basel, Switzerland, 2016; pp. 1–10. [Google Scholar]
- Wang, C.R.; Zhou, C.L.; Ma, J.W. An improved artificial fish-swarm algorithm and its application in feed-forward neural networks. In Proceedings of the 2005 International Conference on Machine Learning and Cybernetics, Guangzhou, China, 18–21 August 2005; Volume 5, pp. 2890–2894. [Google Scholar]
- Yadav, R.K.; Banka, H. IBBOMSA: An improved biogeography-based approach for multiple sequence alignment. Evol. Bioinform. 2016, 12, EBO–S40457. [Google Scholar] [CrossRef]
- Liu, Y.; Schmidt, B.; Maskell, D.L. MSAProbs: Multiple sequence alignment based on pair hidden Markov models and partition function posterior probabilities. Bioinformatics 2010, 26, 1958–1964. [Google Scholar] [CrossRef]
- Rojas, M.G.; Carballido, J.A.; Olivera, A.C.; Vidal, P.J. A Memetic Cellular Genetic Algorithm for Multiple Sequence Alignment. In Proceedings of the 2020 IEEE Congreso Bienal de Argentina 2020, (ARGENCON), Resistencia, Argentina, 1–4 December 2020; pp. 1–8. [Google Scholar]
- Garg, P. A Comparison between Memetic algorithm and Genetic algorithm for the cryptanalysis of Simplified Data Encryption Standard algorithm. arXiv 2010, arXiv:1004.0574. [Google Scholar]
- Rubio-Largo, Á.; Vega-Rodríguez, M.A.; González-Álvarez, D.L. Parallel H4MSA for multiple sequence alignment. In Proceedings of the Trustcom/BigDataSE/ISPA, Helsinki, Finland, 20–22 August 2015; Volume 3, pp. 242–247. [Google Scholar]
- Rubio-Largo, Á.; Vanneschi, L.; Castelli, M.; Vega-Rodríguez, M.A. A characteristic-based framework for multiple sequence aligners. IEEE Trans. Cybern. 2016, 48, 41–51. [Google Scholar] [CrossRef]
- Zhu, Z.; Zhou, J.; Ji, Z.; Shi, Y.H. DNA sequence compression using adaptive particle swarm optimization-based memetic algorithm. IEEE Trans. Evol. Comput. 2011, 15, 643–658. [Google Scholar] [CrossRef]
- Yao, D.; Jiang, M.; You, X.; Abulizi, A.; Hou, R. An algorithm of multiple sequence alignment based on consensus sequence searched by simulated annealing and star alignment. In Proceedings of the 2015 International Symposium on Bioelectronics and Bioinformatics (ISBB), Beijing, China, 14–17 October 2015; pp. 3–6. [Google Scholar]
- Hussein, A.M.; Abdullah, R.; AbdulRashid, N.; Ali, A.N.B. Protein multiple sequence alignment by basic flower pollination algorithm. In Proceedings of the 2017 8th International Conference on Information Technology (ICIT), Amman, Jordan, 17–18 May 2017; pp. 833–838. [Google Scholar]
- Hussein, A.M.; Abdullah, R.; AbdulRashid, N. Flower pollination algorithm with profile technique for multiple sequence alignment. In Proceedings of the 2019 IEEE Jordan International Joint Conference on Electrical Engineering and Information Technology (JEEIT), Amman, Jordan, 9–11 April 2019; pp. 571–576. [Google Scholar]
- Altwaijry, N.; Almasoud, M.; Almalki, A.; Al-Turaiki, I. Multiple sequence alignment using a multi-objective artificial bee colony algorithm. In Proceedings of the 2020 3rd International Conference on Computer Applications & Information Security (ICCAIS), Riyadh, Saudi Arabia, 19–21 May 2020; pp. 1–6. [Google Scholar]
- Ye, L. A Decomposition and Dominance-Based Multi-objective Artificial Bee Colony Algorithm for Multiple Sequence Alignment. Mob. Inf. Syst. 2022, 2022, 5444055. [Google Scholar]
- Bateman, A.; Coin, L.; Durbin, R.; Finn, R.D.; Hollich, V.; Griffiths-Jones, S.; Eddy, S.R. The Pfam protein families database. Nucleic Acids Res. 2004, 32 (Suppl. 1), D138–D141. [Google Scholar] [CrossRef] [PubMed]
- Lei, X.; Sun, J.; Xu, X.; Guo, L. Artificial bee colony algorithm for solving multiple sequence alignment. In Proceedings of the 2010 IEEE Fifth International Conference on Bio-Inspired Computing: Theories and Applications (BIC-TA), Changsha, Cina, 23–26 September 2010; pp. 337–342. [Google Scholar]
- Zemali, E.A.; Boukra, A. A new hybrid bio-inspired approach to resolve the multiple sequence alignment problem. In Proceedings of the 2016 International Conference on Control, Decision and Information Technologies (CoDIT), Saint Julian’s, Malta, 6–8 April 2016; pp. 108–113. [Google Scholar]
- Manikandan, P.; Ramyachitra, D. Bacterial foraging optimization–genetic algorithm for multiple sequence alignment with multi-objectives. Sci. Rep. 2017, 7, 8833. [Google Scholar] [CrossRef]
- Eusuff, M.; Lansey, K.; Pasha, F. Shuffled frog-leaping algorithm: A memetic meta-heuristic for discrete optimization. Eng. Optim. 2006, 38, 129–154. [Google Scholar] [CrossRef]
- Zhang, X.; Kang, Q.; Cheng, J.; Wang, X. A novel hybrid algorithm based on biogeography-based optimization and grey wolf optimizer. Appl. Soft Comput. 2018, 67, 197–214. [Google Scholar] [CrossRef]
- Talbi, H.; Draa, A. A new real-coded quantum-inspired evolutionary algorithm for continuous optimization. Appl. Soft Comput. 2017, 61, 765–791. [Google Scholar] [CrossRef]
- Mahdad, B.; Srairi, K. Security constrained optimal power flow solution using new adaptive partitioning flower pollination algorithm. Appl. Soft Comput. 2016, 46, 501–522. [Google Scholar] [CrossRef]
- Lassmann, T. Kalign 3: Multiple sequence alignment of large datasets. Bioinformatics 2020, 36, 1928–1929. [Google Scholar] [CrossRef]
- Li, C.; Zhan, G.; Li, Z. News text classification based on improved Bi-LSTM-CNN. In Proceedings of the 9th International Conference on Information Technology in Medicine and Education (ITME), Hangzhou, China, 19–21 October 2018; pp. 890–893. [Google Scholar]
- Gardner, P.P.; Wilm, A.; Washietl, S. A benchmark of multiple sequence alignment programs upon structural RNAs. Nucleic Acids Res. 2005, 33, 2433–2439. [Google Scholar] [CrossRef]
- Cartwright, R.A. DNA assembly with gaps (Dawg): Simulating sequence evolution. Bioinformatics 2005, 21 (Suppl. S3), iii31–iii38. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hall, B.G. Simulating DNA coding sequence evolution with EvolveAGene 3. Mol. Biol. Evol. 2008, 25, 688–695. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, W.; Yang, Z. INDELible: A flexible simulator of biological sequence evolution. Mol. Biol. Evol. 2009, 26, 1879–1888. [Google Scholar] [CrossRef] [PubMed]
- Sipos, B.; Massingham, T.; Jordan, G.E.; Goldman, N. PhyloSim-Monte Carlo simulation of sequence evolution in the R statistical computing environment. BMC Bioinform. 2011, 12, 104. [Google Scholar] [CrossRef] [PubMed]
- Koestler, T.; Haeseler, A.V.; Ebersberger, I. REvolver: Modeling sequence evolution under domain constraints. Mol. Biol. Evol. 2012, 29, 2133–2145. [Google Scholar] [CrossRef] [PubMed]
- Dalquen, D.A.; Anisimova, M.; Gonnet, G.H.; Dessimoz, C. ALF—A simulation framework for genome evolution. Mol. Biol. Evol. 2012, 29, 1115–1123. [Google Scholar] [CrossRef] [PubMed]
- Lassmann, T.; Sonnhammer, E.L. Automatic assessment of alignment quality. Nucleic Acids Res. 2005, 33, 7120–7128. [Google Scholar] [CrossRef] [PubMed]
- Landan, G.; Graur, D. Heads or tails: A simple reliability check for multiple sequence alignments. Mol. Biol. Evol. 2007, 24, 1380–1383. [Google Scholar] [CrossRef]
- Do, C.B.; Mahabhashyam, M.S.; Brudno, M.; Batzoglou, S. ProbCons: Probabilistic consistency-based multiple sequence alignment. Genome Res. 2005, 15, 330–340. [Google Scholar] [CrossRef]
- Thompson, J.D.; Plewniak, F.; Poch, O. A comprehensive comparison of multiple sequence alignment programs. Nucleic Acids Res. 1999, 27, 2682–2690. [Google Scholar] [CrossRef]
- Demšar, J. Statistical comparisons of classifiers over multiple data sets. J. Mach. Learn. Res. 2006, 7, 1–30. [Google Scholar]
- Roshan, U.; Livesay, D.R. Probalign: Multiple sequence alignment using partition function posterior probabilities. Bioinformatics 2006, 22, 2715–2721. [Google Scholar] [CrossRef]
- Kemena, C.; Taly, J.F.; Kleinjung, J.; Notredame, C. STRIKE: Evaluation of protein MSAs using a single 3D structure. Bioinformatics 2011, 27, 3385–3391. [Google Scholar] [CrossRef] [PubMed]
- Dessimoz, C.; Gil, M. Phylogenetic assessment of alignments reveals neglected tree signal in gaps. Genome Biol. 2010, 11, R37. [Google Scholar] [CrossRef] [PubMed]
- Blackburne, B.P.; Whelan, S. Measuring the distance between multiple sequence alignments. Bioinformatics 2012, 28, 495–502. [Google Scholar] [CrossRef] [PubMed]
- Lassmann, T.; Frings, O.; Sonnhammer, E.L. Kalign2: High-performance multiple alignment of protein and nucleotide sequences allowing external features. Nucleic Acids Res. 2009, 37, 858–865. [Google Scholar] [CrossRef] [PubMed]
- Bayliss, C.; Juan, A.A.; Currie, C.S.; Panadero, J. A learnheuristic approach for the team orienteering problem with aerial drone motion constraints. Appl. Soft Comput. 2020, 92, 106280. [Google Scholar] [CrossRef]
- Chica, M.; Juan Pérez, A.A.; Cordon, O.; Kelton, D. Why Simheuristics? Benefits, Limitations, and Best Practices when Combining Metaheuristics with Simulation. Benefits, Limitations, and Best Practices when Combining Metaheuristics with Simulation. 2017. Available online: https://papers.ssrn.com/sol3/papers.cfm?abstract_id=2919208 (accessed on 1 January 2024).
- Talbi, E.G. (Ed.) Hybrid Metaheuristics; Springer: Berlin/Heidelberg, Germany, 2013; Volume 166. [Google Scholar]
- Hughes, J.; Houghten, S.; Mallén-Fullerton, G.M.; Ashlock, D. Recentering and restarting genetic algorithm variations for DNA fragment assembly. In Proceedings of the 2014 IEEE Conference on Computational Intelligence in Bioinformatics and Computational Biology, Honolulu, HI, USA, 21–24 May 2014; pp. 1–8. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).