Listeria monocytogenes from Marine Fish and the Seafood Market Environment in Northern Greece: Prevalence, Molecular Characterization, and Antibiotic Resistance
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. Microbiological Analysis
2.3. Proteomic Relationship of Listeria monocytogenes Isolates
2.4. Assessment of Biofilm-Forming Ability of Listeria monocytogenes Strains
2.5. Antimicrobial Susceptibility Testing of Listeria monocytogenes Isolates
2.6. Detection of Virulence-Associated Genes in Listeria monocytogenes Isolates
2.7. Statistical Analysis
3. Results
3.1. Prevalence and Molecular Serogroups of Listeria monocytogenes Strains
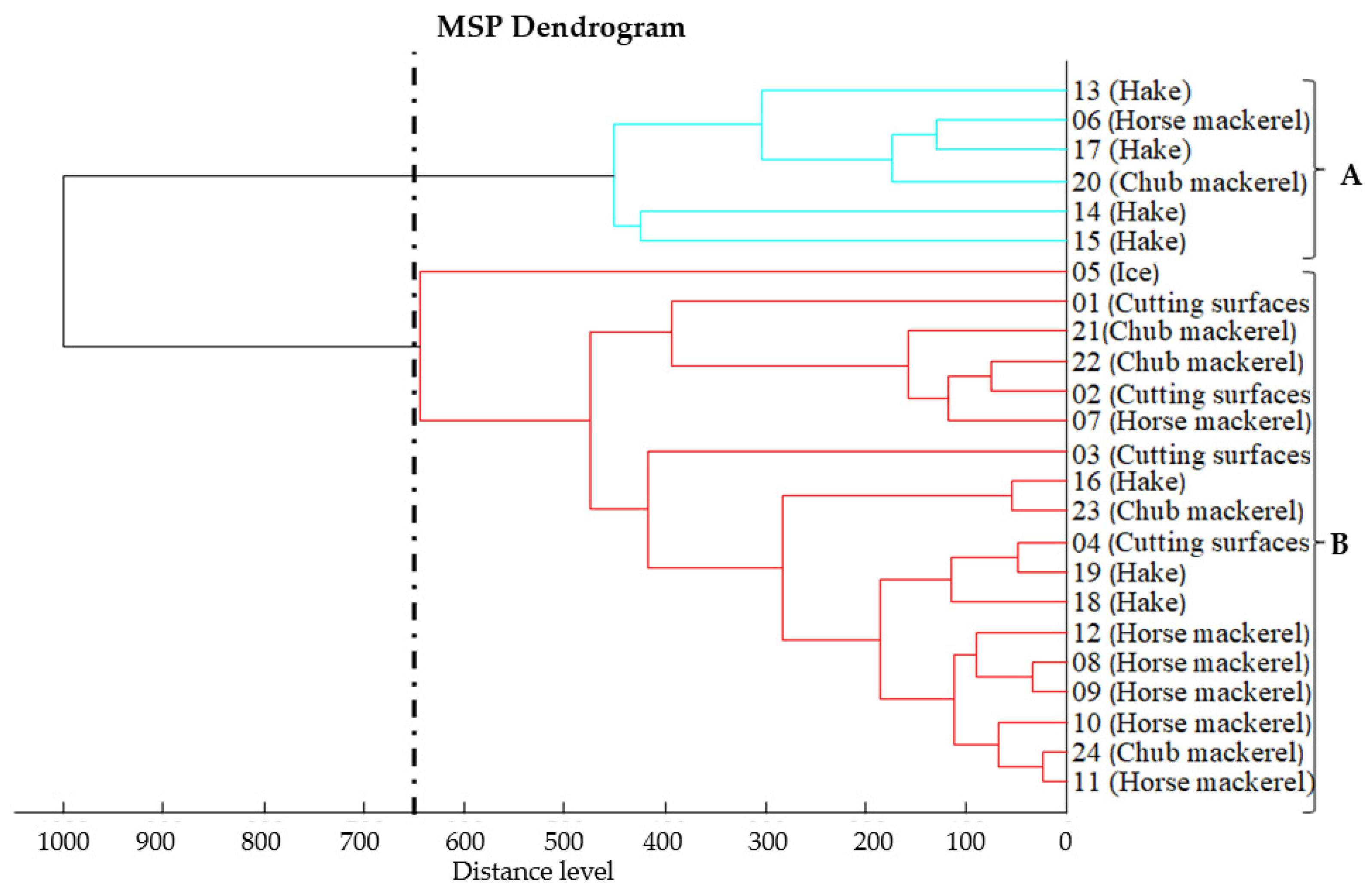
3.2. Proteomic Relatedness of Listeria monocytogenes Isolates
3.3. Assessment of Biofilm-Forming Ability of Listeria monocytogenes Strains
3.4. Antimicrobial Susceptibility Testing of Listeria monocytogenes Strains
3.5. Detection of Virulence-Associated Genes in Listeria monocytogenes Strains
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ariño, A.; Beltrán, J.A.; Herrera, A.; Roncalés, P. Fish and Seafood: Nutritional Value. In Encyclopedia of Human Nutrition; Elsevier: Amsterdam, The Netherlands, 2013; pp. 254–261. [Google Scholar]
- Novoslavskij, A.; Terentjeva, M.; Eizenberga, I.; Valciņa, O.; Bartkevičs, V.; Bērziņš, A. Major Foodborne Pathogens in Fish and Fish Products: A Review. Ann. Microbiol. 2016, 66, 1–15. [Google Scholar] [CrossRef]
- Papadopoulou, C.; Economou, E.; Zakas, G.; Salamoura, C.; Dontorou, C.; Apostolou, J. Microbiological and Pathogenic Contaminants of Seafood in Greece. J. Food Qual. 2007, 30, 28–42. [Google Scholar] [CrossRef]
- Garrity, G.M.; Bell, J.A.; Lilburn, T.G. Taxonomic Outline of The Prokaryotes. In Bergey’s Manual® of Systematic Bacteriology, 2nd ed.; Springer: New York, NY, USA, 2004. [Google Scholar]
- Allerberger, F.; Wagner, M. Listeriosis: A Resurgent Foodborne Infection. Clin. Microbiol. Infect. 2010, 16, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Sibanda, T.; Buys, E.M. Listeria monocytogenes Pathogenesis: The Role of Stress Adaptation. Microorganisms 2022, 10, 1522. [Google Scholar] [CrossRef] [PubMed]
- EFSA and ECDC (European Food Safety Authority and European Centre for Disease Prevention and Control). The European Union One Health 2022 Zoonoses Report. EFSA J. 2023, 21, e8442. [Google Scholar] [CrossRef]
- Bennett John, E.; Dolin, R.; Blaser, M.J. Mandell, Douglas, and Bennett’s Principles and Practice of Infectious Diseases; Elsevier: Philadelphia, PA, USA, 2020. [Google Scholar]
- Dzieciol, M.; Schornsteiner, E.; Muhterem-Uyar, M.; Stessl, B.; Wagner, M.; Schmitz-Esser, S. Bacterial Diversity of Floor Drain Biofilms and Drain Waters in a Listeria monocytogenes Contaminated Food Processing Environment. Int. J. Food Microbiol. 2016, 223, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Ooi, S.T.; Lorber, B. Gastroenteritis Due to Listeria Monocytogenes. Clin. Infect. Dis. 2005, 40, 1327–1332. [Google Scholar] [CrossRef]
- Wu, Y.; Pang, X.; Liu, X.; Wu, Y.; Zhang, X. Functional Genomics Identified Novel Genes Involved in Growth at Low Temperatures in Listeria Monocytogenes. Microbiol. Spectr. 2022, 10, e00710-22. [Google Scholar] [CrossRef]
- Vrinda Menon, K.; Sunil, B.; Latha, C. Prevalence and Antibiotic Resistance Profile of Listeria Spp. Associated with Seafoods from Fish Catchment Areas in Kerala, India. Vet. World 2021, 14, 777–783. [Google Scholar] [CrossRef]
- Zakrzewski, A.J.; Kurpas, M.; Zadernowska, A.; Chajęcka-Wierzchowska, W.; Fraqueza, M.J. A Comprehensive Virulence and Resistance Characteristics of Listeria monocytogenes Isolated from Fish and the Fish Industry Environment. Int. J. Mol. Sci. 2023, 24, 3581. [Google Scholar] [CrossRef]
- Economou, V.; Gousia, P.; Kansouzidou, A.; Sakkas, H.; Karanis, P.; Papadopoulou, C. Prevalence, Antimicrobial Resistance and Relation to Indicator and Pathogenic Microorganisms of Salmonella enterica Isolated from Surface Waters within an Agricultural Landscape. Int. J. Hyg. Environ. Health 2013, 216, 435–444. [Google Scholar] [CrossRef] [PubMed]
- Miettinen, H.; Wirtanen, G. Prevalence and Location of Listeria monocytogenes in Farmed Rainbow Trout. Int. J. Food Microbiol. 2005, 104, 135–143. [Google Scholar] [CrossRef]
- Gozutok, E.; Aydin, A. Presence and Virulence Characterization of Listeria monocytogenes from Fish Samples in the Black Sea, Turkey. Ank. Üniversitesi Vet. Fakültesi Derg. 2022, 69, 387–394. [Google Scholar] [CrossRef]
- Skowron, K.; Wiktorczyk, N.; Grudlewska, K.; Wałecka-Zacharska, E.; Paluszak, Z.; Kruszewski, S.; Gospodarek-Komkowska, E. Phenotypic and Genotypic Evaluation of Listeria monocytogenes Strains Isolated from Fish and Fish Processing Plants. Ann. Microbiol. 2019, 69, 469–482. [Google Scholar] [CrossRef]
- Aalto-Araneda, M.; Korkeala, H.; Lundén, J. Strengthening the Efficacy of Official Food Control Improves Listeria monocytogenes Prevention in Fish-Processing Plants. Sci. Rep. 2018, 8, 13105. [Google Scholar] [CrossRef] [PubMed]
- Tarazi, Y.; Sukhon, S.; Rahbi, A.; Ismail, Z. Molecular Characterization and in Vivo Pathogenicity Study of Listeria monocytogenes Isolated from Fresh and Frozen Local and Imported Fish in Jordan. Open Vet. J. 2021, 11, 517. [Google Scholar] [CrossRef] [PubMed]
- Acciari, V.A.; Torresi, M.; Iannetti, L.; Scattolini, S.; Pomilio, F.; Decastelli, L.; Colmegna, S.; Muliari, R.; Bossù, T.; Proroga, Y.; et al. Listeria monocytogenes in Smoked Salmon and Other Smoked Fish at Retail in Italy: Frequency of Contamination and Strain Characterization in Products from Different Manufacturers. J. Food Prot. 2017, 80, 271–278. [Google Scholar] [CrossRef]
- Nguyen Trang, P.; Thi Anh Ngoc, T.; Masuda, Y.; Hohjoh, K.; Miyamoto, T. Biofilm Formation From Listeria monocytogenes Isolated From Pangasius Fish-Processing Plants. J. Food Prot. 2023, 86, 100044. [Google Scholar] [CrossRef]
- Papaioannou, E.; Giaouris, E.D.; Berillis, P.; Boziaris, I.S. Dynamics of Biofilm Formation by Listeria monocytogenes on Stainless Steel under Mono-Species and Mixed-Culture Simulated Fish Processing Conditions and Chemical Disinfection Challenges. Int. J. Food Microbiol. 2018, 267, 9–19. [Google Scholar] [CrossRef]
- Froese, R.; Pauly, D. (Eds.) FishBase. Available online: www.fishbase.org (accessed on 15 March 2024).
- ISO 11290-1:2017; Microbiology of the Food Chain—Horizontal Method for the Detection and Enumeration of Listeria monocytogenes and of Listeria Spp.—Part 1: Detection Method. International Organization for Standardization: Geneva, Switzerland, 2017. Available online: https://www.iso.org/standard/60313.html (accessed on 22 January 2024).
- Lawrence, L.M.; Gilmour, A. Incidence of Listeria Spp. and Listeria monocytogenes in a Poultry Processing Environment and in Poultry Products and Their Rapid Confirmation by Multiplex PCR. Appl. Environ. Microbiol. 1994, 60, 4600–4604. [Google Scholar] [CrossRef]
- Doumith, M.; Buchrieser, C.; Glaser, P.; Jacquet, C.; Martin, P. Differentiation of the Major Listeria monocytogenes Serovars by Multiplex PCR. J. Clin. Microbiol. 2004, 42, 3819–3822. [Google Scholar] [CrossRef] [PubMed]
- Chachlioutaki, K.; Karavasili, C.; Adamoudi, E.; Tsitsos, A.; Economou, V.; Beltes, C.; Bouropoulos, N.; Katsamenis, O.L.; Doherty, R.; Bakopoulou, A.; et al. Electrospun Nanofiber Films Suppress Inflammation in Vitro and Eradicate Endodontic Bacterial Infection in an E. Faecalis-Infected Ex Vivo Human Tooth Culture Model. ACS Biomater. Sci. Eng. 2022, 8, 2096–2110. [Google Scholar] [CrossRef] [PubMed]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing, 33rd ed.; Clinical and Laboratory Standards Institute: Malvern, PA, USA, 2023; Volume CLSI. [Google Scholar]
- Angelidis, A.S.; Grammenou, A.S.; Kotzamanidis, C.; Giadinis, N.D.; Zdragas, A.G.; Sergelidis, D. Prevalence, Serotypes, Antimicrobial Resistance and Biofilm-Forming Ability of Listeria monocytogenes Isolated from Bulk-Tank Bovine Milk in Northern Greece. Pathogens 2023, 12, 837. [Google Scholar] [CrossRef] [PubMed]
- Kawacka, I.; Pietrzak, B.; Schmidt, M.; Olejnik-Schmidt, A. Listeria monocytogenes Isolates from Meat Products and Processing Environment in Poland Are Sensitive to Commonly Used Antibiotics, with Rare Cases of Reduced Sensitivity to Ciprofloxacin. Life 2023, 13, 821. [Google Scholar] [CrossRef] [PubMed]
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-Resistant, Extensively Drug-Resistant and Pandrug-Resistant Bacteria: An International Expert Proposal for Interim Standard Definitions for Acquired Resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Lawrence, M.L.; Austin, F.W.; Ainsworth, A.J. A Multiplex PCR for Species- and Virulence-Specific Determination of Listeria Monocytogenes. J. Microbiol. Methods 2007, 71, 133–140. [Google Scholar] [CrossRef] [PubMed]
- Rawool, D.B.; Malik, S.V.S.; Shakuntala, I.; Sahare, A.M.; Barbuddhe, S.B. Detection of Multiple Virulence-Associated Genes in Listeria monocytogenes Isolated from Bovine Mastitis Cases. Int. J. Food Microbiol. 2007, 113, 201–207. [Google Scholar] [CrossRef] [PubMed]
- Ben Embarek, P.K. Presence, Detection and Growth of Listeria monocytogenes in Seafoods: A Review. Int. J. Food Microbiol. 1994, 23, 17–34. [Google Scholar] [CrossRef]
- Yücel, N.; Balci, Ş. Prevalence of Listeria, Aeromonas, and Vibrio Species in Fish Used for Human Consumption in Turkey. J. Food Prot. 2010, 73, 380–384. [Google Scholar] [CrossRef]
- Wieczorek, K.; Osek, J. Prevalence, Genetic Diversity and Antimicrobial Resistance of Listeria monocytogenes Isolated from Fresh and Smoked Fish in Poland. Food Microbiol. 2017, 64, 164–171. [Google Scholar] [CrossRef]
- Wang, X.-M.; Lü, X.-F.; Yin, L.; Liu, H.-F.; Zhang, W.-J.; Si, W.; Yu, S.-Y.; Shao, M.-L.; Liu, S.-G. Occurrence and Antimicrobial Susceptibility of Listeria monocytogenes Isolates from Retail Raw Foods. Food Control 2013, 32, 153–158. [Google Scholar] [CrossRef]
- Parihar, V.S.; Barbuddhe, S.B.; Danielsson-Tham, M.L.; Tham, W. Isolation and Characterization of Listeria Species from Tropical Seafoods. Food Control 2008, 19, 566–569. [Google Scholar] [CrossRef]
- Lennox, J.A.; Etta, P.O.; John, G.E.; Henshaw, E.E. Prevalence of Listeria monocytogenes in Fresh and Raw Fish, Chicken and Beef. J. Adv. Microbiol. 2017, 3, 1–7. [Google Scholar] [CrossRef]
- Jamali, H.; Paydar, M.; Ismail, S.; Looi, C.Y.; Wong, W.F.; Radmehr, B.; Abedini, A. Prevalence, Antimicrobial Susceptibility and Virulotyping of Listeria Species and Listeria monocytogenes Isolated from Open-Air Fish Markets. BMC Microbiol. 2015, 15, 144. [Google Scholar] [CrossRef] [PubMed]
- Momtaz, H.; Yadollahi, S. Molecular Characterization of Listeria monocytogenes Isolated from Fresh Seafood Samples in Iran. Diagn. Pathol. 2013, 8, 149. [Google Scholar] [CrossRef] [PubMed]
- Kuzmanovic, J.; Asanin, R.; Baltic, M.; Misic, D.; Dimitrijevic, M.; Stojanovic, M.; Asanin, N.; Kovacevic, I. Presence of Listeria Spp. in Fish Samples, Fish Products and Sea Products. Acta Vet. 2011, 61, 193–203. [Google Scholar] [CrossRef][Green Version]
- Soultos, N.; Abrahim, A.; Papageorgiou, K.; Steris, V. Incidence of Listeria Spp. in Fish and Environment of Fish Markets in Northern Greece. Food Control 2007, 18, 554–557. [Google Scholar] [CrossRef]
- Fallah, A.A.; Saei-Dehkordi, S.S.; Mahzounieh, M. Occurrence and Antibiotic Resistance Profiles of Listeria monocytogenes Isolated from Seafood Products and Market and Processing Environments in Iran. Food Control 2013, 34, 630–636. [Google Scholar] [CrossRef]
- Gudbjörnsdóttir, B.; Suihko, M.-L.; Gustavsson, P.; Thorkelsson, G.; Salo, S.; Sjöberg, A.-M.; Niclasen, O.; Bredholt, S. The Incidence of Listeria monocytogenes in Meat, Poultry and Seafood Plants in the Nordic Countries. Food Microbiol. 2004, 21, 217–225. [Google Scholar] [CrossRef]
- Chen, B.-Y.; Pyla, R.; Kim, T.-J.; Silva, J.L.; Jung, Y.-S. Prevalence and Contamination Patterns of Listeria monocytogenes in Catfish Processing Environment and Fresh Fillets. Food Microbiol. 2010, 27, 645–652. [Google Scholar] [CrossRef]
- Johansson, T.; Rantala, L.; Palmu, L.; Honkanen-Buzalski, T. Occurrence and Typing of Listeria monocytogenes Strains in Retail Vacuum-Packed Fish Products and in a Production Plant. Int. J. Food Microbiol. 1999, 47, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Akkaya, L.; Atabay, H.I.; Gok, V.; Kara, R. Detection of Listeria Species in Fresh Fish and Fish Market Environment by IMS Technique in Turkey. Arch. Lebensmittelhyg. 2011, 62, 16–19. [Google Scholar] [CrossRef]
- Hartemink, R.; Georgsson, F. Incidence of Listeria Species in Seafood and Seafood Salads. Int. J. Food Microbiol. 1991, 12, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Abdollahzadeh, E.; Ojagh, S.M.; Hosseini, H.; Irajian, G.; Ghaemi, E.A. Prevalence and Molecular Characterization of Listeria Spp. and Listeria monocytogenes Isolated from Fish, Shrimp, and Cooked Ready-to-Eat (RTE) Aquatic Products in Iran. LWT 2016, 73, 205–211. [Google Scholar] [CrossRef]
- Vasilev, V.; Japheth, R.; Breuer, R.; Andorn, N.; Ben Abraham, R.; Yoni, Y.; Valinsky, L.; Agmon, V. A Survey of Listeria monocytogenes Strains, Isolated from Ready-to-Eat Foods in Israel over a Period of 10 Years, 1998–2007. Food Control 2010, 21, 1179–1181. [Google Scholar] [CrossRef]
- Siriken, B.; Ayaz, N.D.; Erol, I. Prevalence and Serotype Distribution of Listeria monocytogenes in Salted Anchovy, Raw Anchovy, and Raw Mussel Using IMS-Based Cultivation Technique and PCR. J. Aquat. Food Prod. Technol. 2013, 22, 77–82. [Google Scholar] [CrossRef]
- Davis, J.A.; Jackson, C.R. Comparative Antimicrobial Susceptibility of Listeria monocytogenes, L. Innocua, and L. Welshimeri. Microb. Drug Resist. 2009, 15, 27–32. [Google Scholar] [CrossRef]
- Filiousis, G.; Johansson, A.; Frey, J.; Perreten, V. Prevalence, Genetic Diversity and Antimicrobial Susceptibility of Listeria monocytogenes Isolated from Open-Air Food Markets in Greece. Food Control 2009, 20, 314–317. [Google Scholar] [CrossRef]
- Olaimat, A.N.; Al-Holy, M.A.; Shahbaz, H.M.; Al-Nabulsi, A.A.; Abu Ghoush, M.H.; Osaili, T.M.; Ayyash, M.M.; Holley, R.A. Emergence of Antibiotic Resistance in Listeria monocytogenes Isolated from Food Products: A Comprehensive Review. Compr. Rev. Food Sci. Food Saf. 2018, 17, 1277–1292. [Google Scholar] [CrossRef]
- Athanasios, T.; Anna, P.; John, D.; Antonios, A. Neonatal Meningitis Due to Multi-Resistant Listeria Monocytogenes. J. Antimicrob. Chemother. 1997, 39, 553–554. [Google Scholar]
- Andritsos, N.D.; Mataragas, M. Characterization and Antibiotic Resistance of Listeria monocytogenes Strains Isolated from Greek Myzithra Soft Whey Cheese and Related Food Processing Surfaces over Two-and-a-Half Years of Safety Monitoring in a Cheese Processing Facility. Foods 2023, 12, 1200. [Google Scholar] [CrossRef]
- Takahashi, H.; Miya, S.; Igarashi, K.; Suda, T.; Kuramoto, S.; Kimura, B. Biofilm Formation Ability of Listeria monocytogenes Isolates from Raw Ready-to-Eat Seafood. J. Food Prot. 2009, 72, 1476–1480. [Google Scholar] [CrossRef]
- Meloni, D.; Mazza, R.; Piras, F.; Lamon, S.; Consolati, S.G.; Mureddu, A.; Mazzette, R. The Biofilm Formation Ability of Listeria monocytogenes Isolated from Meat, Poultry, Fish and Processing Plant Environments Is Related to Serotype and Pathogenic Profile of the Strains. Vet. Sci. Dev. 2012, 2, 12. [Google Scholar] [CrossRef][Green Version]
- Nakamura, H.; Takakura, K.-I.; Sone, Y.; Itano, Y.; Nishikawa, Y. Biofilm Formation and Resistance to Benzalkonium Chloride in Listeria monocytogenes Isolated from a Fish Processing Plant. J. Food Prot. 2013, 76, 1179–1186. [Google Scholar] [CrossRef]
| Sample | Number | |
|---|---|---|
| Small fish | European anchovy (Engraulis encrasicolus) | 30 |
| European pilchard (Sardina pilchardus) | 30 | |
| Bogue (Boops boops) | 30 | |
| Total | 90 | |
| Large fish | Horse mackerel (Trachurus mediterraneus) | 25 |
| Chub mackerel (Scomber japonicus) | 25 | |
| Hake (Merlucius merlucius) | 25 | |
| Total | 75 | |
| Environmental samples | Knives | 19 |
| Cutting boards | 19 | |
| Siphons | 15 | |
| Water | 13 | |
| Ice | 19 | |
| Sinks | 19 | |
| Floors | 19 | |
| Total | 123 | |
| Gene | Primer | Concentration | Product (bp) | Target |
|---|---|---|---|---|
| prs | For: GCTGAAGAGATTGCGAAAGAAG | 0.2 μΜ | 370 | Listeria spp. |
| Rev: CAAAGAAACCTTGGATTTGCG | 0.2 μΜ | |||
| ORF 2819 | For: AGCAAAATGCCAAAACTCGT | 1 μΜ | 471 | Listeria monocytogenes serotypes 1/2b, 3b, 4b, 4d, and 4e |
| Rev: CATCACTAAAGCCTCCCATTG | 1 μΜ | |||
| ORF 2110 | For: AGTGGACAATTGATTGGTGAA | 1 μΜ | 597 | Listeria monocytogenes serotypes 4b, 4d, and 4e |
| Rev: CATCCATCCCTTACTTTGGAC | 1 μΜ | |||
| lmo 0737 | For: AGGGCTTCAAGGACTTACCC | 1 μΜ | 691 | Listeria monocytogenes serotypes 1/2a, 1/2c, 3a, and 3c |
| Rev: ACGATTTCTGCTTGCCATTC | 1 μΜ | |||
| lmo 1118 | For: AGGGGTCTTAAATCCTGGAA | 1.5 μΜ | 906 | Listeria monocytogenes serotypes 1/2c and 3c |
| Rev: CGGCTTGTTCGGCATACTTA | 1.5 μΜ |
| Antibiotic | Concentration/Disc | Breakpoint (S *) (mm) |
|---|---|---|
| Amoxicillin/clavulanic acid | 20/10 μg | ≥18 mm |
| Ampicillin | 10 μg | ≥17 mm |
| Ciprofloxacin | 5 μg | ≥21 mm |
| Chloramphenicol | 30 μg | ≥18 mm |
| Clindamycin | 2 μg | ≥21 mm |
| Erythromycin | 15 μg | ≥23 mm |
| Gentamicin | 10 μg | ≥15 mm |
| Meropenem | 10 μg | ≥18 mm |
| Penicillin | 10 IU | ≥29 mm |
| Sulfamethoxazole/trimethoprim | 1.25/23.75 μg | ≥16 mm |
| Tetracycline | 30 μg | ≥19 mm |
| Rifampicin | 5 μg | ≥20 mm |
| Vancomycin | 30 μg | ≥17 mm |
| Gene | Primer | Product (bp) |
|---|---|---|
| inlA | For: ACGAGTAACGGGACAAATGC | 800 |
| Rev: CCCGACAGTGGTGCTAGATT | ||
| inlB | For: TGGGAGAGTAACCCAACCAC | 884 |
| Rev: GTTGACCTTCGATGGTTGCT | ||
| inlC | For: AATTCCCACAGGACACAACC | 517 |
| Rev: CGGGAATGCAATTTTTCACTA | ||
| inlJ | For: TGTAACCCCGCTTACACAGTT | 238 |
| Rev: AGCGGCTTGGCAGTCTAATA | ||
| plcA | For: CTGCTTGAGCGTTCATGTCTCATCCCCC | 1484 |
| Rev: CATGGGTTTCACTCTCCTTCTAC | ||
| prfA | For: CTGTTGGAGCTCTTCTTGGTGAAGCAATCG | 1060 |
| Rev: AGCAACCTCGGTACCATATACTAACTC | ||
| hlyA | For: GCAGTTGCAAGCGCTTGGAGTGAA | 456 |
| Rev: GCAACGTATCCTCCAGAGTGATCG | ||
| iap | For: ACAAGCTGCACCTGTTGCAG | 131 |
| Rev: TGACAGCGTGTGTAGTAGCA | ||
| actA | For: CGCCGCGGAAATTAAAAAAAGA | 839 |
| Rev: ACGAAGGAACCGGGCTGCTAG |
| Isolate | Origin | Molecular Serotype | Virulence-Associated Genes | Antimicrobial Resistance Profile | Biofilm-Forming Ability 1 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| inlA | inlB | inlC | inlJ | plcA | prfA | actA | HlyA | iap | |||||
| 1 | Cutting boards | IVb | + | + | + | + | + | + | + | + | + | CLI, VAN | +++ |
| 2 | Cutting boards | IVb | + | + | + | + | + | + | + | + | + | CLI | ++ |
| 3 | Cutting boards | IIc | + | + | + | + | + | + | + | + | + | CLI, VAN | ++ |
| 4 | Cutting boards | IIc | + | + | + | + | + | + | + | + | + | CLI | ++ |
| 5 | Ice | IIc | + | + | + | + | + | + | + | + | CLI, P, TET, VAN | ++ | |
| 6 | Horse mackerel | IIc | + | + | + | + | + | + | + | + | CLI | ++ | |
| 7 | Horse mackerel | IIc | + | + | + | + | + | + | + | + | + | CLI, VAN | +++ |
| 8 | Horse mackerel | IIc | + | + | + | + | + | + | + | + | + | CLI | +++ |
| 9 | Horse mackerel | IIc | + | + | + | + | + | + | + | + | + | CLI, VAN | ++ |
| 10 | Horse mackerel | IIc | + | + | + | + | + | + | + | + | + | CLI, VAN | ++ |
| 11 | Horse mackerel | IIc | + | + | + | + | + | + | + | + | + | - | ++ |
| 12 | Horse mackerel | IIc | + | + | + | + | + | + | + | + | + | CLI, VAN | ++ |
| 13 | Hake | IIc | + | + | + | + | + | + | + | + | + | CLI | +++ |
| 14 | Hake | IIc | + | + | + | + | + | + | + | + | + | - | +++ |
| 15 | Hake | IIc | + | + | + | + | + | + | + | + | + | CLI, VAN | +++ |
| 16 | Hake | IIc | + | + | + | + | + | + | + | + | + | - | ++ |
| 17 | Hake | IIc | + | + | + | + | + | + | + | + | + | CLI | +++ |
| 18 | Hake | IIc | + | + | + | + | + | + | + | + | + | CLI | ++ |
| 19 | Hake | IIc | + | + | + | + | + | + | + | + | + | CLI | +++ |
| 20 | Chub mackerel | IIc | + | + | + | + | + | + | + | + | + | CLI, VAN | +++ |
| 21 | Chub mackerel | IIc | + | + | + | + | + | + | + | + | + | - | ++ |
| 22 | Chub mackerel | IIc | + | + | + | + | + | + | + | + | + | CLI, VAN | +++ |
| 23 | Chub mackerel | IIc | + | + | + | + | + | + | + | + | + | CLI | ++ |
| 24 | Chub mackerel | IIc | + | + | + | + | + | + | + | + | + | CLI, VAN | ++ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peratikos, P.; Tsitsos, A.; Damianos, A.; Kyritsi, M.A.; Hadjichristodoulou, C.; Soultos, N.; Economou, V. Listeria monocytogenes from Marine Fish and the Seafood Market Environment in Northern Greece: Prevalence, Molecular Characterization, and Antibiotic Resistance. Appl. Sci. 2024, 14, 2725. https://doi.org/10.3390/app14072725
Peratikos P, Tsitsos A, Damianos A, Kyritsi MA, Hadjichristodoulou C, Soultos N, Economou V. Listeria monocytogenes from Marine Fish and the Seafood Market Environment in Northern Greece: Prevalence, Molecular Characterization, and Antibiotic Resistance. Applied Sciences. 2024; 14(7):2725. https://doi.org/10.3390/app14072725
Chicago/Turabian StylePeratikos, Pantelis, Anestis Tsitsos, Alexandros Damianos, Maria A. Kyritsi, Christos Hadjichristodoulou, Nikolaos Soultos, and Vangelis Economou. 2024. "Listeria monocytogenes from Marine Fish and the Seafood Market Environment in Northern Greece: Prevalence, Molecular Characterization, and Antibiotic Resistance" Applied Sciences 14, no. 7: 2725. https://doi.org/10.3390/app14072725
APA StylePeratikos, P., Tsitsos, A., Damianos, A., Kyritsi, M. A., Hadjichristodoulou, C., Soultos, N., & Economou, V. (2024). Listeria monocytogenes from Marine Fish and the Seafood Market Environment in Northern Greece: Prevalence, Molecular Characterization, and Antibiotic Resistance. Applied Sciences, 14(7), 2725. https://doi.org/10.3390/app14072725









