Enzymatic Analysis of Chitin Deacetylases on Crystalline Chitin with Varied Molecular Weights: Insights from Active Pocket Characteristic Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Phylogenetic Analysis and Active-Site Characterization of CDAs
2.3. Heterologous Expression and Purification of Recombinant CDAs
2.4. Enzyme Assay of CDAs
2.5. Optimization of Reaction Conditions for Recombinant Proteins
2.6. Catalytic Performance on Chitin Substrates
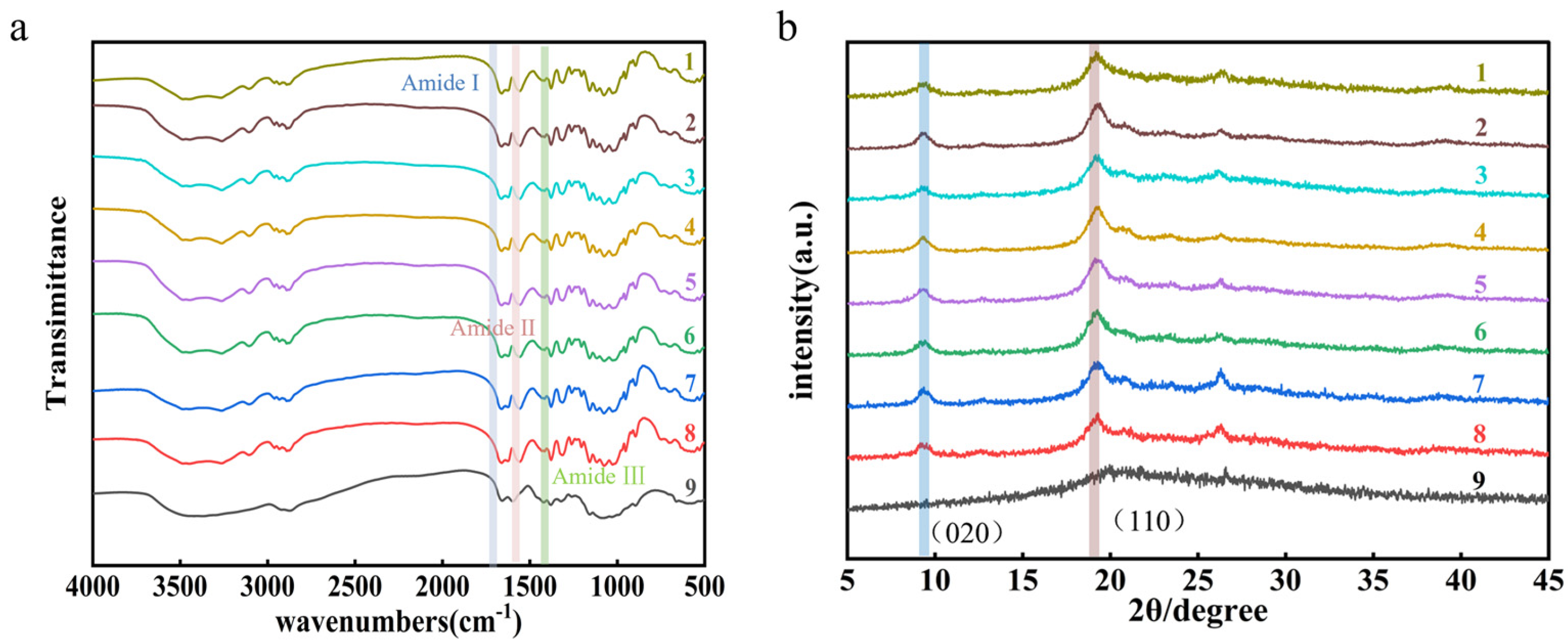
2.7. Substrate Characterization
2.7.1. Molecular Weight Determination of Substrates
2.7.2. FT-IR Analysis
2.7.3. X-Ray Diffraction (XRD) Analysis
2.8. Statistical Analysis
3. Results and Discussion
3.1. Bioinformatics Analysis of CDAs
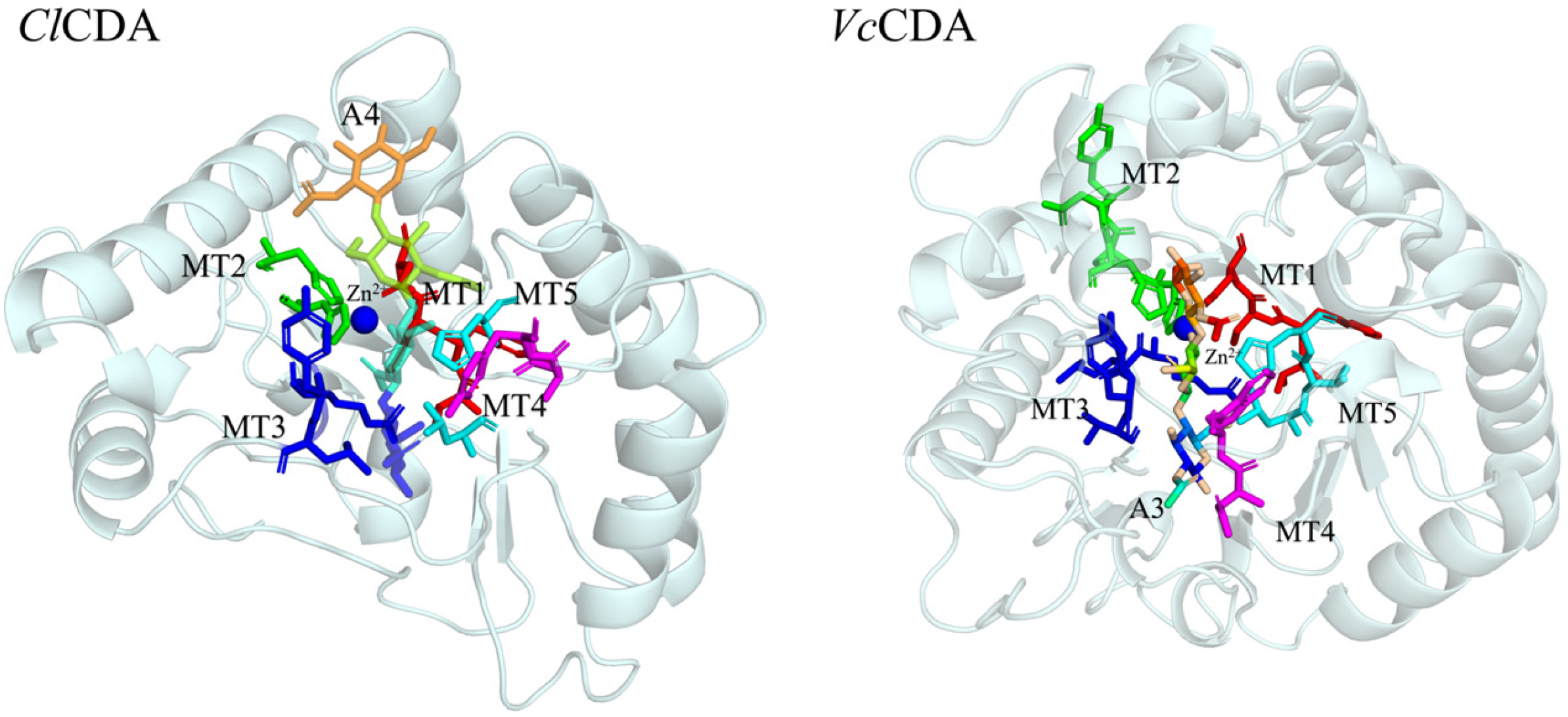
3.1.1. Phylogenetic Analysis and Multiple Sequence Alignment
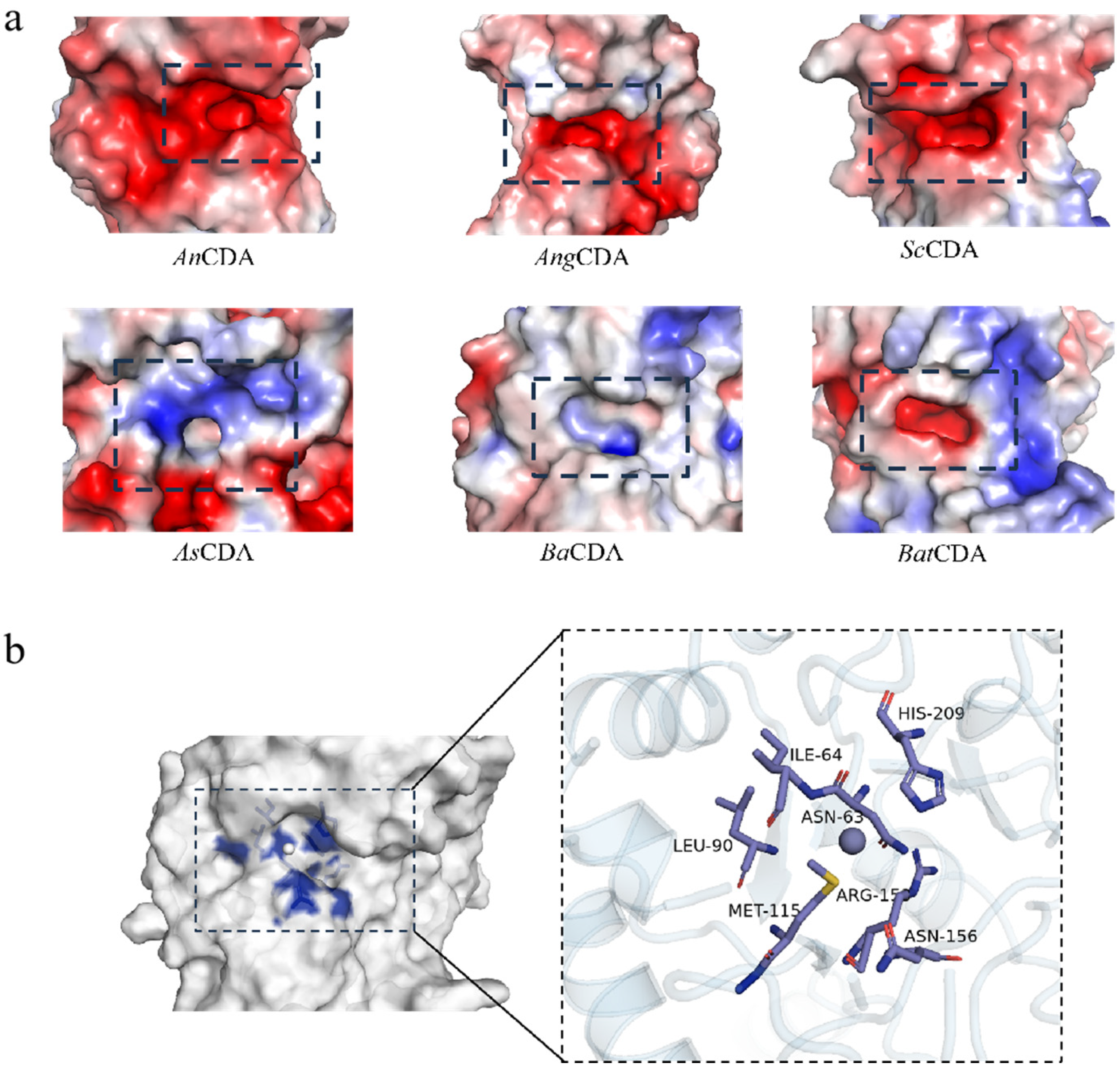
3.1.2. Charge Distribution and Geometric Parameters of the Active Pocket
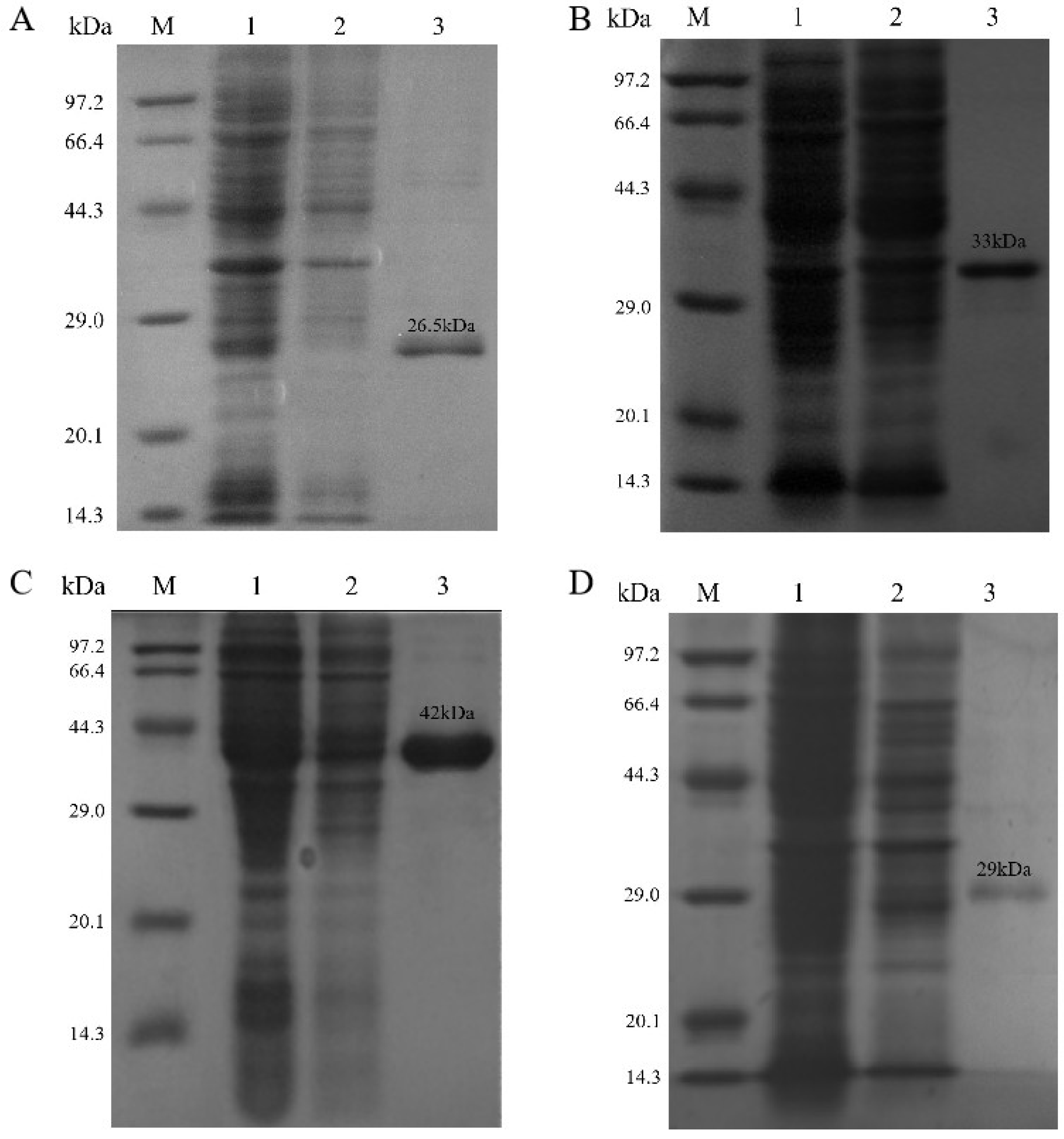
3.2. Heterologous Expression and Purification of Recombinant CDAs
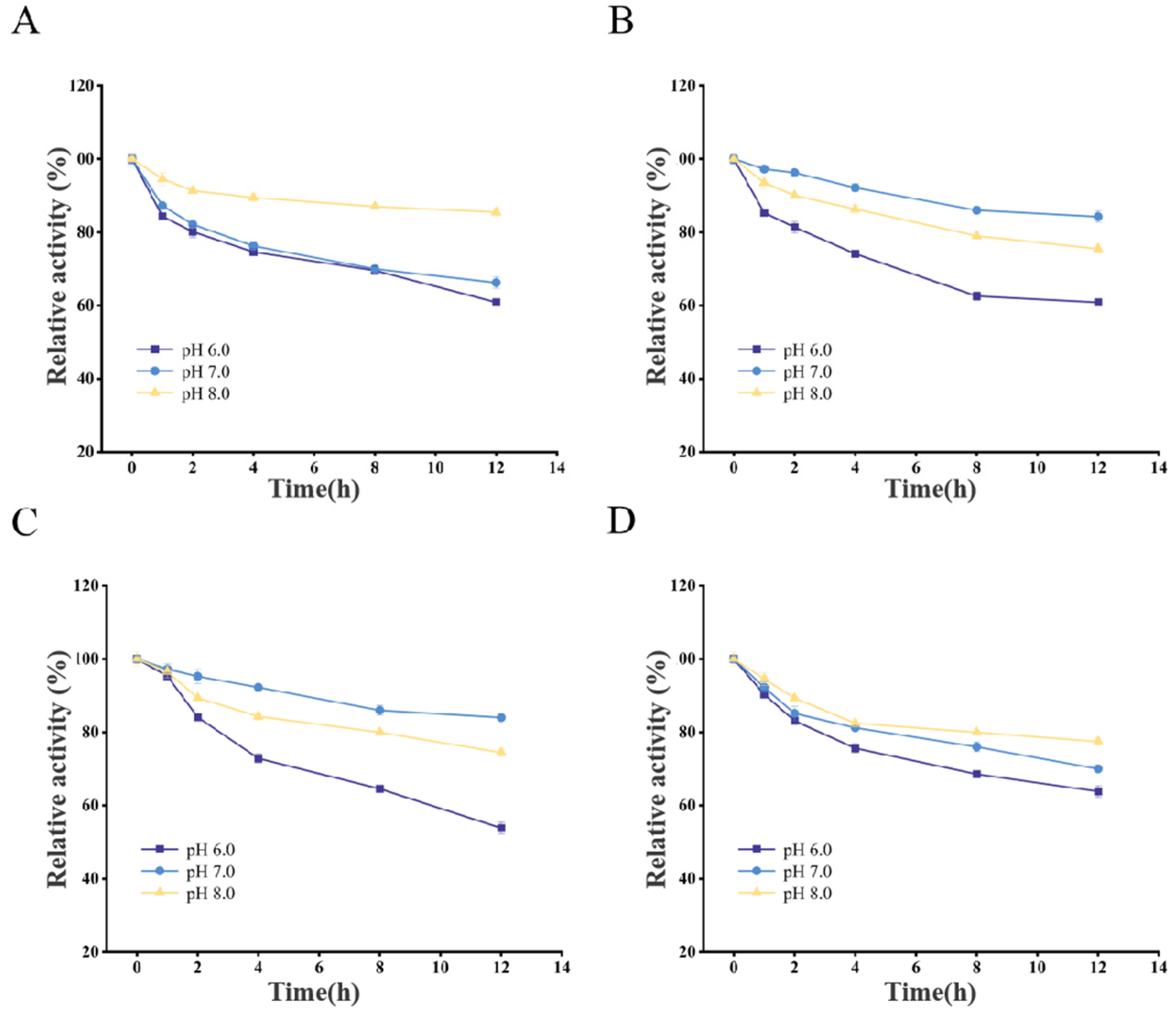
3.3. Optimal Reaction Conditions for Recombinant CDAs
3.4. Recombinant Enzymes Process Chitin Substrates with Diverse Molecular Parameters
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CDA | Chitin deacetylase |
| DD | Degree of deacetylation |
| GlcNAc | N-Acetyl-D-glucosamine |
| MT1–MT5 | Motif 1–motif5 |
| △DD | Percentage of acetyls removed |
References
- Kim, S.-K. Chitin, Chitosan, Oligosaccharides and Their Derivatives: Biological Activities and Applications; CRC Press: Boca Raton, FL, USA, 2010. [Google Scholar] [CrossRef]
- Mohan, K.; Ganesan, A.R.; Ezhilarasi, P.; Kondamareddy, K.K.; Rajan, D.K.; Sathishkumar, P.; Rajarajeswaran, J.; Conterno, L. Green and eco-friendly approaches for the extraction of chitin and chitosan: A review. Carbohydr. Polym. 2022, 287, 119349. [Google Scholar] [CrossRef]
- Xu, Y.; Han, J.; Lin, H. Fabrication and characterization of a self-crosslinking chitosan hydrogel under mild conditions without the use of strong bases. Carbohydr. Polym. 2017, 156, 372–379. [Google Scholar] [CrossRef]
- Gonçalves, C.; Ferreira, N.; Lourenço, L. Production of low molecular weight chitosan and chitooligosaccharides (COS): A review. Polymers 2021, 13, 2466. [Google Scholar] [CrossRef] [PubMed]
- Pan, H.; Zhao, T.; Xu, L.; Shen, Y.; Wang, L.; Ding, Y. Preparation of novel chitosan derivatives and applications in functional finishing of textiles. Int. J. Biol. Macromol. 2020, 153, 971–976. [Google Scholar] [CrossRef] [PubMed]
- Friedman, M.; Juneja, V.K. Review of antimicrobial and antioxidative activities of chitosans in food. J. Food Prot. 2010, 73, 1737–1761. [Google Scholar] [CrossRef]
- Lin, S.; Qin, Z.; Chen, Q.; Fan, L.; Zhou, J.; Zhao, L. Efficient immobilization of bacterial GH family 46 chitosanase by carbohydrate-binding module fusion for the controllable preparation of chitooligosaccharides. J. Agric. Food. Chem. 2019, 67, 6847–6855. [Google Scholar] [CrossRef]
- Pokhrel, S.; Yadav, P.N. Functionalization of chitosan polymer and their applications. J. Macromol. Sci. Part A 2019, 56, 450–475. [Google Scholar] [CrossRef]
- El Knidri, H.; El Khalfaouy, R.; Laajeb, A.; Addaou, A.; Lahsini, A. Eco-friendly extraction and characterization of chitin and chitosan from the shrimp shell waste via microwave irradiation. Process Saf. Environ. Prot. 2016, 104, 395–405. [Google Scholar] [CrossRef]
- Nawaz, H.; Mannan, R.; Nishan, U. Modification of physical and functional characteristics of chitin extracted from microwave-treated Nelumbo nucifera rhizome flour. Polym. Polym. Compos. 2021, 29, S1257–S1267. [Google Scholar] [CrossRef]
- Giraldo, J.D.; García, Y.; Vera, M.; Garrido-Miranda, K.A.; Andrade-Acuna, D.; Marrugo, K.P.; Rivas, B.L.; Schoebitz, M. Alternative processes to produce chitin, chitosan, and their oligomers. Carbohydr. Polym. 2024, 332, 121924. [Google Scholar] [CrossRef]
- Hossbach, J.; Busswinkel, F.; Kranz, A.; Wattjes, J.; Cord-Landwehr, S.; Moerschbacher, B.M. A chitin deacetylase of Podospora anserina has two functional chitin binding domains and a unique mode of action. Carbohydr. Polym. 2018, 183, 1–10. [Google Scholar] [CrossRef]
- Qing, L.; Gao, J.; Du, L.; Liu, Y.; Guo, N.; Sun, J.; Dong, H.; Mao, X. Chitinase and deacetylase-based chitin-degrading bacteria: One-pot cascade bioconversion of chitin to chitooligosaccharides. J. Agric. Food. Chem. 2024, 72, 23937–23946. [Google Scholar] [CrossRef] [PubMed]
- Liang, B.; Song, W.; Xing, R.; Liu, S.; Yu, H.; Li, P. The source, activity influencing factors and biological activities for future development of chitin deacetylase. Carbohydr. Polym. 2023, 321, 121335. [Google Scholar] [CrossRef]
- Naqvi, S.; Moerschbacher, B.M. The cell factory approach toward biotechnological production of high-value chitosan oligomers and their derivatives: An update. Crit. Rev. Biotechnol. 2017, 37, 11–25. [Google Scholar] [CrossRef] [PubMed]
- Kaczmarek, M.B.; Struszczyk-Swita, K.; Xiao, M.; Szczęsna-Antczak, M.; Antczak, T.; Gierszewska, M.; Steinbüchel, A.; Daroch, M. Polycistronic expression system for Pichia pastoris composed of chitino-and chitosanolytic enzymes. Front. Bioeng. Biotechnol. 2021, 9, 710922. [Google Scholar] [CrossRef] [PubMed]
- Aragunde, H.; Biarnés, X.; Planas, A. Substrate recognition and specificity of chitin deacetylases and related family 4 carbohydrate esterases. Int. J. Mol. Sci. 2018, 19, 412. [Google Scholar] [CrossRef]
- Lindner, S.; Bonin, M.; Hellmann, M.J.; Moerschbacher, B.M. Three intertwining effects guide the mode of action of chitin deacetylase de-and N-acetylation reactions. Carbohydr. Polym. 2025, 347, 122725. [Google Scholar] [CrossRef]
- Blair, D.E.; Schüttelkopf, A.W.; MacRae, J.I.; van Aalten, D.M. Structure and metal-dependent mechanism of peptidoglycan deacetylase, a streptococcal virulence factor. Proc. Natl. Acad. Sci. USA 2005, 102, 15429–15434. [Google Scholar] [CrossRef]
- Blair, D.E.; Hekmat, O.; Schüttelkopf, A.W.; Shrestha, B.; Tokuyasu, K.; Withers, S.G.; van Aalten, D.M. Structure and mechanism of chitin deacetylase from the fungal pathogen Colletotrichum lindemuthianum. Biochemistry 2006, 45, 9416–9426. [Google Scholar] [CrossRef]
- Nakamura, A.M.; Nascimento, A.S.; Polikarpov, I. Structural diversity of carbohydrate esterases. Biotechnol. Res. Innov. 2017, 1, 35–51. [Google Scholar] [CrossRef]
- Mapuranga, J.; Chang, J.; Li, H.; Zhang, Y.; Li, R.; Song, L.; Zhang, N.; Yang, W. The molecular structure, biological roles, and inhibition of plant pathogenic fungal chitin deacetylases. Front. Plant Sci. 2024, 14, 1335646. [Google Scholar] [CrossRef]
- Yang, X.; Gao, H.; Yan, J.; Zhou, J.; Shi, L. Intramolecular chaperone-assisted dual-anchoring activation (ICDA): A suitable preorganization for electrophilic halocyclization. Chem. Sci. 2024, 15, 6130–6140. [Google Scholar] [CrossRef]
- Taher, M.; Dubey, K.D.; Mazumdar, S. Computationally guided bioengineering of the active site, substrate access pathway, and water channels of thermostable cytochrome P450, CYP175A1, for catalyzing the alkane hydroxylation reaction. Chem. Sci. 2023, 14, 14316–14326. [Google Scholar] [CrossRef]
- Nie, Y.; Wang, S.; Xu, Y.; Luo, S.; Zhao, Y.; Xiao, R.; Montelione, G.; Hunt, J.; Szyperski, T. Enzyme engineering based on X-ray structures and kinetic profiling of substrate libraries: Alcohol dehydrogenases for stereospecific synthesis of a broad range of chiral alcohols. ACS Catal. 2018, 8, 5145–5152. [Google Scholar] [CrossRef]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.; Clamp, M.; Barton, G.J. Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef]
- Jurrus, E.; Engel, D.; Star, K.; Monson, K.; Brandi, J.; Felberg, L.E.; Brookes, D.H.; Wilson, L.; Chen, J.; Liles, K. Improvements to the APBS biomolecular solvation software suite. Protein Sci. 2018, 27, 112–128. [Google Scholar] [CrossRef]
- Graef, J.; Ehrt, C.; Rarey, M. Binding site detection remastered: Enabling fast, robust, and reliable binding site detection and descriptor calculation with DoGSite3. J. Chem. Inf. Model. 2023, 63, 3128–3137. [Google Scholar] [CrossRef] [PubMed]
- Schmidtke, P.; Le Guilloux, V.; Maupetit, J.; Tufféry, P. Fpocket: Online tools for protein ensemble pocket detection and tracking. Nucleic Acids Res. 2010, 38, W582–W589. [Google Scholar] [CrossRef]
- Sun, Y.; Zhang, J.; Wu, S.; Wang, S. Statistical optimization for production of chitin deacetylase from Rhodococcus erythropolis HG05. Carbohydr. Polym. 2014, 102, 649–652. [Google Scholar] [CrossRef]
- Delezuk, J.A.d.M.; Cardoso, M.B.; Domard, A.; Campana-Filho, S.P. Ultrasound-assisted deacetylation of beta-chitin: Influence of processing parameters. Polym. Int. 2011, 60, 903–909. [Google Scholar] [CrossRef]
- Poirier, M.; Charlet, G. Chitin fractionation and characterization in N, N-dimethylacetamide/lithium chloride solvent system. Carbohydr. Polym. 2002, 50, 363–370. [Google Scholar] [CrossRef]
- Kasaai, M.R. A review of several reported procedures to determine the degree of N-acetylation for chitin and chitosan using infrared spectroscopy. Carbohydr. Polym. 2008, 71, 497–508. [Google Scholar] [CrossRef]
- Kenawy, E.-R.; Ali, S.S.; Al-Etewy, M.; Sun, J.; Wu, J.; El-Zawawy, N. Synthesis, characterization and biomedical applications of a novel Schiff base on methyl acrylate-functionalized chitosan bearing p-nitrobenzaldehyde groups. Int. J. Biol. Macromol. 2019, 122, 833–843. [Google Scholar] [CrossRef] [PubMed]
- Xiao, M.; Chen, D.; Liu, S.; Chen, A.; Fang, A.; Tian, B.; Yu, Y.; Bi, C.; Kang, Z.; Yang, Y. A chitin deacetylase Ps CDA2 from Puccinia striiformis f. sp. tritici confers disease pathogenicity by suppressing chitin-triggered immunity in wheat. Mol. Plant Pathol. 2023, 24, 1467–1479. [Google Scholar] [CrossRef] [PubMed]
- Christodoulidou, A.; Bouriotis, V.; Thireos, G. Two sporulation-specific chitin deacetylase-encoding genes are required for the ascospore wall rigidity of Saccharomyces cerevisiae. J. Biol. Chem. 1996, 271, 31420–31425. [Google Scholar] [CrossRef] [PubMed]
- Davis, L.L.; Bartnicki-Garcia, S. Chitosan synthesis by the tandem action of chitin synthetase and chitin deacetylase from Mucor rouxii. Biochemistry 1984, 23, 1065–1073. [Google Scholar] [CrossRef]
- Tsigos, I.; Martinou, A.; Kafetzopoulos, D.; Bouriotis, V. Chitin deacetylases: New, versatile tools in biotechnology. Trends Biotechnol. 2000, 18, 305–312. [Google Scholar] [CrossRef]
- Pawaskar, G.M.; Raval, K.; Rohit, P.; Shenoy, R.P.; Raval, R. Cloning, expression, purification and characterization of chitin deacetylase extremozyme from halophilic Bacillus aryabhattai B8W22. 3 Biotech 2021, 11, 515. [Google Scholar] [CrossRef]
- Zhao, Y.; Park, R.-D.; Muzzarelli, R.A. Chitin deacetylases: Properties and applications. Mar. Drugs 2010, 8, 24–46. [Google Scholar] [CrossRef] [PubMed]
- Tuveng, T.R.; Rothweiler, U.; Udatha, G.; Vaaje-Kolstad, G.; Smalås, A.; Eijsink, V.G. Structure and function of a CE4 deacetylase isolated from a marine environment. PLoS ONE 2017, 12, e0187544. [Google Scholar] [CrossRef]
- Yamada, M.; Kurano, M.; Inatomi, S.; Taguchi, G.; Okazaki, M.; Shimosaka, M. Isolation and characterization of a gene coding for chitin deacetylase specifically expressed during fruiting body development in the basidiomycete Flammulina velutipes and its expression in the yeast Pichia pastoris. FEMS Microbiol. Lett. 2008, 289, 130–137. [Google Scholar] [CrossRef]
- Geoghegan, I.A.; Gurr, S.J. Chitosan mediates germling adhesion in Magnaporthe oryzae and is required for surface sensing and germling morphogenesis. PLoS Pathog. 2016, 12, e1005703. [Google Scholar] [CrossRef]
- Yang, G.; Wang, Y.; Fang, Y.; An, J.; Hou, X.; Lu, J.; Zhu, R.; Liu, S. A novel potent crystalline chitin decomposer: Chitin deacetylase from Acinetobacter schindleri MCDA01. Molecules 2022, 27, 5345. [Google Scholar] [CrossRef]
- Abd El-Ghany, M.N.; Hamdi, S.A.; Zahran, A.K.; Abou-Taleb, M.A.; Heikel, A.M.; Abou El-Kheir, M.T.; Farahat, M.G. Characterization of novel cold-active chitin deacetylase for green production of bioactive chitosan. AMB Express 2025, 15, 5. [Google Scholar] [CrossRef]
- Zhu, X.-Y.; Zhao, Y.; Zhang, H.-D.; Wang, W.-X.; Cong, H.-H.; Yin, H. Characterization of the specific mode of action of a chitin deacetylase and separation of the partially acetylated chitosan oligosaccharides. Mar. Drugs 2019, 17, 74. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Gay, L.M.; Tuveng, T.R.; Agger, J.W.; Westereng, B.; Mathiesen, G.; Horn, S.J.; Vaaje-Kolstad, G.; Van Aalten, D.M.; Eijsink, V.G. Structure and function of a broad-specificity chitin deacetylase from Aspergillus nidulans FGSC A4. Sci. Rep. 2017, 7, 1746. [Google Scholar] [CrossRef]
- Wujieti, B.; Feng, X.; Liu, E.; Li, D.; Hao, M.; Zhou, L.; Cui, W. A theoretical study on the activity and selectivity of IDO/TDO inhibitors. Phys. Chem. Chem. Phys. 2024, 26, 16747–16764. [Google Scholar] [CrossRef]
- Distaso, M.; Cea-Rama, I.; Coscolín, C.; Chernikova, T.N.; Tran, H.; Ferrer, M.; Sanz-Aparicio, J.; Golyshin, P.N. The mobility of the cap domain is essential for the substrate promiscuity of a family IV esterase from sorghum rhizosphere microbiome. Appl. Environ. Microbiol. 2023, 89, e01807–e01822. [Google Scholar] [CrossRef] [PubMed]
- Qin, C.; Li, H.; Xiao, Q.; Liu, Y.; Zhu, J.; Du, Y. Water-solubility of chitosan and its antimicrobial activity. Carbohydr. Polym. 2006, 63, 367–374. [Google Scholar] [CrossRef]
- Li, K.; Liang, Y.; Fang, J.; Peng, J.; Tan, M. Chitin deacetylase from Bacillus aryabhattai TCI-16: Heterologous expression, characterization, and deacetylation performance. J. Agric. Food Chem. 2024, 72, 9268–9278. [Google Scholar] [CrossRef] [PubMed]


| Source of Strain | GenBank Accession Numbers | Name | Source of Strain | GenBank Accession Numbers | Name |
|---|---|---|---|---|---|
| Acinetobacter schindleri | MW295944.1 | AsCDA | Pestalotiopsis sp. | A0A1L3THR9 | PesCDA |
| Bacillus licheniformis | Q65LW7 | BlCDA | Rhizopus nigricans | AY779045 | RnCDA |
| Bacillus aryabhattai TCI-16 | WP_028412810.1 | BatCDA | Mucor rouxii | P50325 | MrCDA |
| Aspergillus nidulans FGSC A4 | Q5AQQ0 | AnCDA | Colletotrichum lindemuthianum | Q6DWK3 | ClCDA |
| Bacillus aryabhattai B8W22 | WP_028412720.1 | BaCDA | Podospora anserina | B2AAQ0 | PaCDA |
| Arthrobacter | 5LFZ | ArCE4 | Rhizopus circinans | A0A9P6XCG4 | RcCDA |
| Arthrobacter sp. Jub115 | WP_133834943 | ArCDA | Rhizopus stolonifer | A0A367KRZ9 | RsCDA |
| Coprinopsis cinerea | A8P2Z0 | CcCDA | Streptomyces bacillaris | WP_112490308 | SbCDA |
| Flammulina velutipes | BAE92728 | FvCDA | Streptomyces griseoinacarnatus RB7AG | QFX82172 | SgCDA |
| Aspergillus flavus | Q2UI04 | AfCDA | Saccharomyces cerevisiae S288C | Q06703 | ScCDA |
| Puccinia striiformis f. sp. tritici | A0A0L0W1E0 | PgtCDA | Aspergillus niger | A2QZC8 | AngCDA |
| Name | Hydrophobicity Score | Volume [Å3] | Surface Area [Å2] | Surface-to-Volume (S/V) Ratio | Substrate and the Removed Acetyl Group (%) |
|---|---|---|---|---|---|
| AsCDA | 37.933 | 172.130 | 391.39 | 2.274 | Chitin (63%) |
| BlCDA | 23.923 | 150.020 | 317.250 | 2.115 | Glycol chitin |
| BatCDA | 25.867 | 152.288 | 355.315 | 2.333 | Colloidal chitin (44%) |
| BaCDA | 31.375 | 142.096 | 300.068 | 2.112 | Glycol chitin |
| ArCE4 | 11.583 | 151.550 | 344.330 | 2.272 | Chitin(8%) |
| ArCDA | 10.300 | 135.170 | 307.300 | 2.273 | β-chitin (2.99U/mg) |
| PaCDA | 11.750 | 154.624 | 356.668 | 2.307 | Chitin (3%) |
| RcCDA | 25.250 | 121.860 | 329.730 | 2.706 | Glycol chitin, no activity against chitin |
| ClCDA | 21.571 | 152.576 | 363.734 | 2.384 | / |
| MrCDA | 16.231 | 139.780 | 368.790 | 2.638 | Glycol chitin |
| RnCDA | 23.471 | 177.660 | 423.640 | 2.385 | Glycol chitin |
| PesCDA | 18.538 | 154.624 | 358.381 | 2.318 | Colloidal chitin (7%) |
| CcCDA | 9.750 | 148.480 | 354.772 | 2.389 | No activity against chitin |
| AnCDA | 9.077 | 123.900 | 339.230 | 2.738 | Chitin (0.5%) |
| RsCDA | 22.818 | 112.640 | 262.530 | 2.331 | Colloidal chitin (27.5%) |
| SbCDA | 27.750 | 150.530 | 375.170 | 2.492 | Colloidal chitin, no activity against chitin |
| SgCDA | 17.182 | 101.410 | 223.190 | 2.201 | Colloidal chitin |
| ScCDA | 16.182 | 112.640 | 296.820 | 2.280 | Shrimp shell chitin (11%) |
| AngCDA | 6.923 | 132.100 | 346.600 | 2.624 | No activity against chitin |
| FvCDA | 10.846 | 118.272 | 286.080 | 2.419 | Glycol chitin |
| AfCDA | 8.750 | 144.380 | 332.850 | 2.305 | Glycol chitin |
| PgtCDA | 21.733 | 112.130 | 292.240 | 2.606 | No activity against chitin |
| Substrate | Molecular Weight (kDa) | Deacetylation (%) | Crystallinity (%) | AnCDA (△DD) | AsCDA (△DD) | BaCDA (△DD) | ScCDA (△DD) |
|---|---|---|---|---|---|---|---|
| Commercial chitin | 563.42 ± 1.89 | 8.70 ± 2.66 | 76.49 ± 1.23 | 2.24 ± 0.50 | 33.47 ± 1.44 | 21.52 ± 1.03 | 7.41 ± 0.81 |
| Substrate a | 346.74 ± 3.12 | 18.06 ± 1.18 | 79.97 ± 1.87 | 3.10 ± 0.44 | 38.26 ± 0.68 | 25.43 ± 0.92 | 14.08 ± 0.96 |
| Substrate b | 322.67 ± 4.23 | 19.21 ± 0.92 | 73.97 ± 1.35 | 3.19 ± 0.92 | 37.32 ± 0.97 | 28.66 ± 0.87 | 16.37 ± 1.28 |
| Substrate c | 278.72 ± 2.38 | 19.35 ± 0.78 | 78.78 ± 1.87 | 5.84 ± 2.66 | 42.93 ± 1.21 | 29.88 ± 1.24 | 21.77 ± 0.79 |
| Substrate d | 164.40 ± 5.23 | 16.19 ± 0.89 | 80.03 ± 2.01 | 9.11 ± 0.89 | 47.58 ± 1.04 | 34.41 ± 1.03 | 20.44 ± 0.84 |
| Substrate e | 64.80 ± 1.98 | 16.99 ± 1.21 | 76.00 ± 1.48 | 16.42 ± 1.20 | 52.46 ± 0.91 | 37.57 ± 1.23 | 30.45 ± 1.18 |
| Substrate f | 33.36 ± 2.23 | 16.70 ± 0.93 | 81.07 ± 1.29 | 21.51 ± 1.03 | 49.49 ± 1.23 | 35.57 ± 1.27 | 28.48 ± 1.21 |
| Substrate g | 14.46 ± 3.68 | 19.72 ± 1.37 | 77.14 ± 0.89 | 28.43 ± 1.30 | 59.54 ± 1.53 | 49.08 ± 1.34 | 42.18 ± 1.68 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, K.; Yang, S.; Cai, J. Enzymatic Analysis of Chitin Deacetylases on Crystalline Chitin with Varied Molecular Weights: Insights from Active Pocket Characteristic Analysis. Appl. Sci. 2025, 15, 10721. https://doi.org/10.3390/app151910721
Chen K, Yang S, Cai J. Enzymatic Analysis of Chitin Deacetylases on Crystalline Chitin with Varied Molecular Weights: Insights from Active Pocket Characteristic Analysis. Applied Sciences. 2025; 15(19):10721. https://doi.org/10.3390/app151910721
Chicago/Turabian StyleChen, Kaige, Shengyu Yang, and Jun Cai. 2025. "Enzymatic Analysis of Chitin Deacetylases on Crystalline Chitin with Varied Molecular Weights: Insights from Active Pocket Characteristic Analysis" Applied Sciences 15, no. 19: 10721. https://doi.org/10.3390/app151910721
APA StyleChen, K., Yang, S., & Cai, J. (2025). Enzymatic Analysis of Chitin Deacetylases on Crystalline Chitin with Varied Molecular Weights: Insights from Active Pocket Characteristic Analysis. Applied Sciences, 15(19), 10721. https://doi.org/10.3390/app151910721




