From Genomes to Applications: Comparative Analysis of Aeribacillus pallidus Reveals a Thermophilic Chassis for Biotechnology
Abstract
1. Introduction
2. Materials and Methods
2.1. Whole Genome Sequence and Accession Numbers of Nucleotide Sequence
2.2. Genome Annotation and Functional Categorization of A. pallidus Strains
2.3. Prediction of CAZymes and Secondary Metabolite Biosynthetic Gene Clusters
2.4. Whole-Genome Phylogenomic Clustering of Aeribacillus Strains
2.5. Pan-Genome and Core-Genome Analysis
3. Results
3.1. General Genomic Properties of A. pallidus Strains
3.2. Comparative Functional Annotation and COG Categories of A. pallidus Strains
3.3. Thermal Stress Response
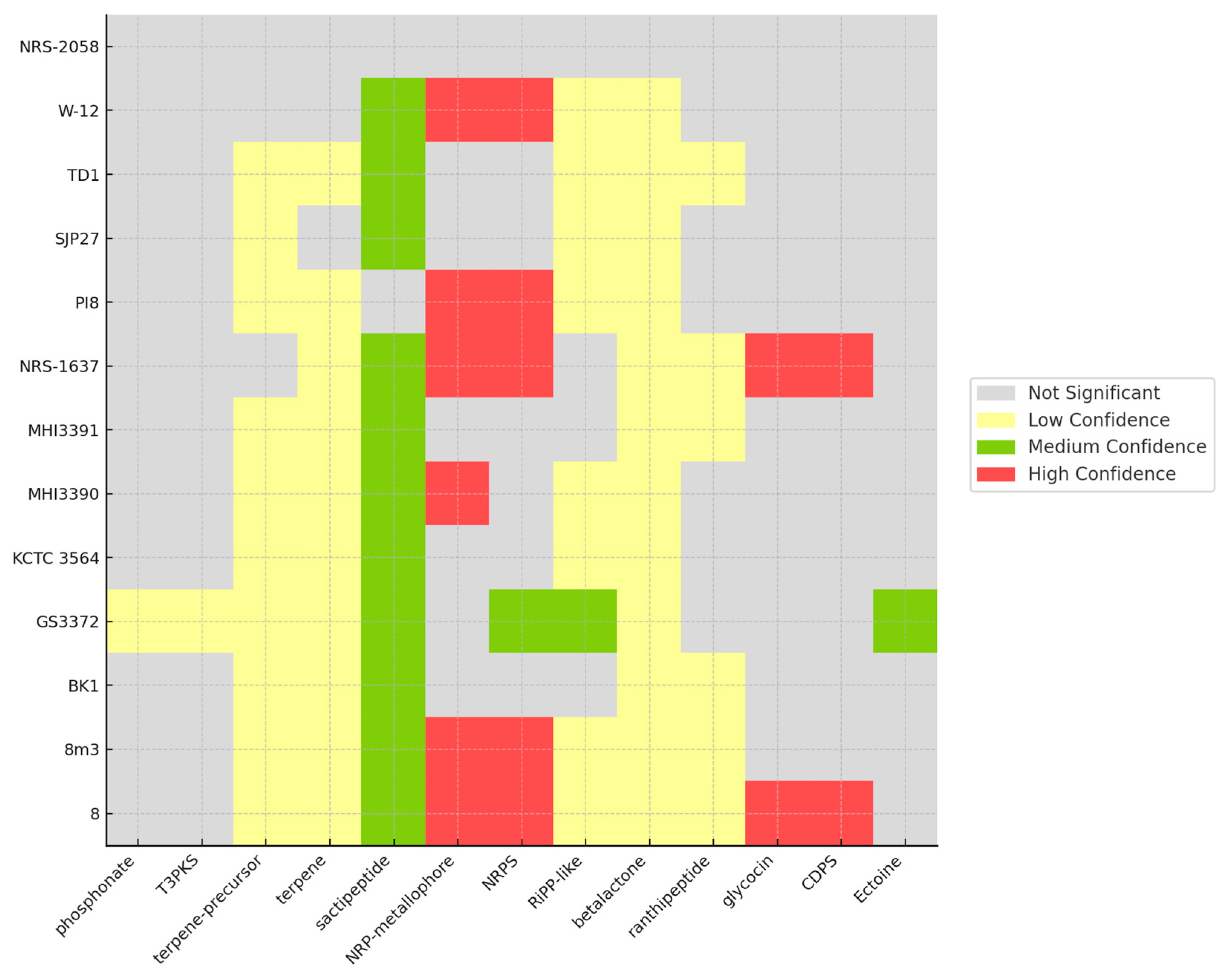
3.4. CAZyme and Secondary Metabolite Identification
3.5. Phylogenomic Clustering Reveals Genetic Relationships Among Aeribacillus Strains
3.6. Pan/Core Genome Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Scholz, T.; Demharter, W.; Hensel, R.; Kandler, O. Bacillus pallidus sp. nov., a new thermophilic species from sewage. Syst. Appl. Microbiol. 1987, 9, 91–96. [Google Scholar] [CrossRef]
- Banat, I.M.; Marchant, R.; Rahman, T.J. Geobacillus debilis sp. nov., a novel obligately thermophilic bacterium isolated from a cool soil environment, and reassignment of Bacillus pallidus to Geobacillus pallidus comb. nov. Int. J. Syst. Evol. Microbiol. 2004, 54, 2197–2201. [Google Scholar] [CrossRef]
- Minana-Galbis, D.; Pinzon, D.L.; Loren, J.G.; Manresa, A.; Oliart-Ros, R.M. Reclassification of Geobacillus pallidus (Scholz et al. 1988) Banat et al. 2004 as Aeribacillus pallidus gen. nov., comb. nov. Int. J. Syst. Evol. Microbiol. 2010, 60, 1600–1604. [Google Scholar] [CrossRef]
- Yasawong, M.; Areekit, S.; Pakpitchareon, A.; Santiwatanakul, S.; Chansiri, K. Characterization of thermophilic halotolerant Aeribacillus pallidus TD1 from Tao dam hot spring, Thailand. Int. J. Mol. Sci. 2011, 12, 5294–5303. [Google Scholar] [CrossRef]
- Timilsina, P.M.; Pandey, G.R.; Shrestha, A.; Ojha, M.; Karki, T.B. Purification and characterization of a noble thermostable algal starch liquefying alpha-amylase from Aeribacillus pallidus BTPS-2 isolated from geothermal spring of Nepal. Biotechnol. Rep. 2020, 28, e00551. [Google Scholar] [CrossRef]
- Yildirim, V.; Baltaci, M.O.; Ozgencli, I.; Sisecioglu, M.; Adiguzel, A.; Adiguzel, G. Purification and biochemical characterization of a novel thermostable serine alkaline protease from Aeribacillus pallidus C10: A potential additive for detergents. J. Enzym. Inhib. Med. Chem. 2017, 32, 468–477. [Google Scholar] [CrossRef]
- Ktata, A.; Krayem, N.; Aloulou, A.; Bezzine, S.; Sayari, A.; Chamkha, M.; Karray, A. Purification, biochemical and molecular study of lipase producing from a newly thermoalkaliphilic Aeribacillus pallidus for oily wastewater treatment. J. Biochem. 2020, 167, 89–99. [Google Scholar] [CrossRef]
- López López, M.J.; Jurado Rodríguez, M.d.M.; López González, J.A.; Estrella González, M.J.; Martínez Gallardo, M.R.; Toribio Gallardo, A.J.; Suárez Estrella, F. Characterization of Thermophilic Lignocellulolytic Microorganisms in Composting. Front. Microbiol. 2021, 12, 697480. [Google Scholar] [CrossRef]
- Filippidou, S.; Jaussi, M.; Junier, T.; Wunderlin, T.; Jeanneret, N.; Regenspurg, S.; Li, P.-E.; Lo, C.-C.; Johnson, S.; McMurry, K. Genome sequence of Aeribacillus pallidus strain GS3372, an endospore-forming bacterium isolated in a deep geothermal reservoir. Genome Announc. 2015, 3, 00981-15. [Google Scholar] [CrossRef]
- Kita, K.; Yoshida, S.; Masuo, S.; Nakamura, A.; Ishikawa, S.; Yoshida, K.-i. Genes encoding a novel thermostable bacteriocin in the thermophilic bacterium Aeribacillus pallidus PI8. J. Appl. Microbiol. 2023, 134, lxad293. [Google Scholar] [CrossRef]
- Lücking, G.; Albrecht, K.; Märtlbauer, E.; Schauer, K. Draft genome sequences of two thermophilic, spore-forming Aeribacillus pallidus strains isolated from dairy products. Microbiol. Resour. Announc. 2024, 13, e00896-23. [Google Scholar] [CrossRef]
- Stavridou, E.; Karapetsi, L.; Nteve, G.M.; Tsintzou, G.; Chatzikonstantinou, M.; Tsaousi, M.; Martinez, A.; Flores, P.; Merino, M.; Dobrovic, L. Landscape of microalgae omics and metabolic engineering research for strain improvement: An overview. Aquaculture 2024, 587, 740803. [Google Scholar] [CrossRef]
- Davey, J.W.; Hohenlohe, P.A.; Etter, P.D.; Boone, J.Q.; Catchen, J.M.; Blaxter, M.L. Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nat. Rev. Genet. 2011, 12, 499–510. [Google Scholar] [CrossRef]
- Yildiz, S.Y.; Radchenkova, N.; Arga, K.Y.; Kambourova, M.; Toksoy Oner, E. Genomic analysis of Brevibacillus thermoruber 423 reveals its biotechnological and industrial potential. Appl. Microbiol. Biotechnol. 2015, 99, 2277–2289. [Google Scholar] [CrossRef]
- Yaşar Yıldız, S. Genomic insights into Thermomonas hydrothermalis: Potential applications in industrial biotechnology. World J. Microbiol. Biotechnol. 2025, 41, 30. [Google Scholar] [CrossRef]
- Yasar Yildiz, S.; Finore, I.; Leone, L.; Romano, I.; Lama, L.; Kasavi, C.; Nicolaus, B.; Toksoy Oner, E.; Poli, A. Genomic analysis provides new insights into biotechnological and industrial potential of Parageobacillus thermantarcticus M1. Front. Microbiol. 2022, 13, 923038. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef]
- Olson, R.D.; Assaf, R.; Brettin, T.; Conrad, N.; Cucinell, C.; Davis, J.J.; Dempsey, D.M.; Dickerman, A.; Dietrich, E.M.; Kenyon, R.W. Introducing the bacterial and viral bioinformatics resource center (BV-BRC): A resource combining PATRIC, IRD and ViPR. Nucleic Acids Res. 2023, 51, D678–D689. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Szklarczyk, D.; Heller, D.; Hernández-Plaza, A.; Forslund, S.K.; Cook, H.; Mende, D.R.; Letunic, I.; Rattei, T.; Jensen, L.J. eggNOG 5.0: A hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019, 47, D309–D314. [Google Scholar] [CrossRef]
- Vallenet, D.; Engelen, S.; Mornico, D.; Cruveiller, S.; Fleury, L.; Lajus, A.; Rouy, Z.; Roche, D.; Salvignol, G.; Scarpelli, C. MicroScope: A platform for microbial genome annotation and comparative genomics. Database 2009, 2009, bap021. [Google Scholar] [CrossRef]
- Zheng, J.; Ge, Q.; Yan, Y.; Zhang, X.; Huang, L.; Yin, Y. dbCAN3: Automated carbohydrate-active enzyme and substrate annotation. Nucleic Acids Res. 2023, 51, W115–W121. [Google Scholar] [CrossRef]
- Medema, M.H.; Blin, K.; Cimermancic, P.; De Jager, V.; Zakrzewski, P.; Fischbach, M.A.; Weber, T.; Takano, E.; Breitling, R. antiSMASH: Rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 2011, 39, W339–W346. [Google Scholar] [CrossRef]
- Konstantinidis, K.T.; Tiedje, J.M. Genomic insights that advance the species definition for prokaryotes. Proc. Natl. Acad. Sci. USA 2005, 102, 2567–2572. [Google Scholar] [CrossRef]
- Ondov, B.D.; Treangen, T.J.; Melsted, P.; Mallonee, A.B.; Bergman, N.H.; Koren, S.; Phillippy, A.M. Mash: Fast genome and metagenome distance estimation using MinHash. Genome Biol. 2016, 17, 132. [Google Scholar] [CrossRef]
- Vallenet, D.; Calteau, A.; Dubois, M.; Amours, P.; Bazin, A.; Beuvin, M.; Burlot, L.; Bussell, X.; Fouteau, S.; Gautreau, G. MicroScope: An integrated platform for the annotation and exploration of microbial gene functions through genomic, pangenomic and metabolic comparative analysis. Nucleic Acids Res. 2020, 48, D579–D589. [Google Scholar] [CrossRef]
- Blondel, V.D.; Guillaume, J.-L.; Lambiotte, R.; Lefebvre, E. Fast unfolding of communities in large networks. J. Stat. Mech. Theory Exp. 2008, 2008, P10008. [Google Scholar] [CrossRef]
- Deb, S. Pan-genome evolution and its association with divergence of metabolic functions in Bifidobacterium genus. World J. Microbiol. Biotechnol. 2022, 38, 231. [Google Scholar] [CrossRef]
- Khan, Z.; Shahwar, D. Role of heat shock proteins (HSPs) and heat stress tolerance in crop plants. In Sustainable Agriculture in the Era of Climate Change; Springer: Berlin/Heidelberg, Germany, 2020; pp. 211–234. [Google Scholar]
- Luo, D.; Wu, Z.; Bai, Q.; Zhang, Y.; Huang, M.; Huang, Y.; Li, X. Universal stress proteins: From gene to function. Int. J. Mol. Sci. 2023, 24, 4725. [Google Scholar] [CrossRef]
- Gragerov, A.; Nudler, E.; Komissarova, N.; Gaitanaris, G.A.; Gottesman, M.E.; Nikiforov, V. Cooperation of GroEL/GroES and DnaK/DnaJ heat shock proteins in preventing protein misfolding in Escherichia coli. Proc. Natl. Acad. Sci. USA 1992, 89, 10341–10344. [Google Scholar] [CrossRef]
- Hu, C.; Yang, J.; Qi, Z.; Wu, H.; Wang, B.; Zou, F.; Mei, H.; Liu, J.; Wang, W.; Liu, Q. Heat shock proteins: Biological functions, pathological roles, and therapeutic opportunities. MedComm 2022, 3, e161. [Google Scholar] [CrossRef]
- Mayer, M.; Bukau, B. Hsp70 chaperones: Cellular functions and molecular mechanism. Cell. Mol. Life Sci. 2005, 62, 670. [Google Scholar] [CrossRef]
- Eelager, M.P.; Masti, S.P.; Chougale, R.B.; Dalbanjan, N.P.; Kumar, S.P. Noni (Morinda citrifolia) leaf extract incorporated methylcellulose active films: A sustainable strategy for browning inhibition in apple slice packaging. Int. J. Biol. Macromol. 2024, 269, 132270. [Google Scholar] [CrossRef]
- Katikaridis, P.; Bohl, V.; Mogk, A. Resisting the heat: Bacterial disaggregases rescue cells from devastating protein aggregation. Front. Mol. Biosci. 2021, 8, 681439. [Google Scholar] [CrossRef]
- Queraltó, C.; Álvarez, R.; Ortega, C.; Díaz-Yáñez, F.; Paredes-Sabja, D.; Gil, F. Role and regulation of Clp proteases: A target against gram-positive bacteria. Bacteria 2023, 2, 21–36. [Google Scholar] [CrossRef]
- Michel, A.; Agerer, F.; Hauck, C.R.; Herrmann, M.; Ullrich, J.; Hacker, J.r.; Ohlsen, K. Global regulatory impact of ClpP protease of Staphylococcus aureus on regulons involved in virulence, oxidative stress response, autolysis, and DNA repair. J. Bacteriol. 2006, 188, 5783–5796. [Google Scholar] [CrossRef]
- Jensen, C.; Fosberg, M.J.; Thalsø-Madsen, I.; Bæk, K.T.; Frees, D. Staphylococcus aureus ClpX localizes at the division septum and impacts transcription of genes involved in cell division, T7-secretion, and SaPI5-excision. Sci. Rep. 2019, 9, 16456. [Google Scholar] [CrossRef]
- Rohrwild, M.; Coux, O.; Huang, H.; Moerschell, R.P.; Yoo, S.J.; Seol, J.H.; Chung, C.H.; Goldberg, A.L. HslV-HslU: A novel ATP-dependent protease complex in Escherichia coli related to the eukaryotic proteasome. Proc. Natl. Acad. Sci. USA 1996, 93, 5808–5813. [Google Scholar] [CrossRef]
- Kebe, N.M.; Samanta, K.; Singh, P.; Lai-Kee-Him, J.; Apicella, V.; Payrot, N.; Lauraire, N.; Legrand, B.; Lisowski, V.; Mbang-Benet, D.-E. The HslV protease from Leishmania major and its activation by C-terminal HslU peptides. Int. J. Mol. Sci. 2019, 20, 1021. [Google Scholar] [CrossRef]
- Zarzecka, U.; Modrak-Wójcik, A.; Figaj, D.; Apanowicz, M.; Lesner, A.; Bzowska, A.; Lipinska, B.; Zawilak-Pawlik, A.; Backert, S.; Skorko-Glonek, J. Properties of the HtrA protease from bacterium Helicobacter pylori whose activity is indispensable for growth under stress conditions. Front. Microbiol. 2019, 10, 961. [Google Scholar] [CrossRef]
- Yi, L.; Liu, B.; Nixon, P.J.; Yu, J.; Chen, F. Recent advances in understanding the structural and functional evolution of FtsH proteases. Front. Plant Sci. 2022, 13, 837528. [Google Scholar] [CrossRef]
- Kirthika, P.; Lloren, K.K.S.; Jawalagatti, V.; Lee, J.H. Structure, substrate specificity and role of lon protease in bacterial pathogenesis and survival. Int. J. Mol. Sci. 2023, 24, 3422. [Google Scholar] [CrossRef]
- Laksanalamai, P.; Robb, F.T. Small heat shock proteins from extremophiles: A review. Extremophiles 2004, 8, 1–11. [Google Scholar] [CrossRef]
- Najar, I.N.; Thakur, N. A systematic review of the genera Geobacillus and Parageobacillus: Their evolution, current taxonomic status and major applications. Microbiology 2020, 166, 800–816. [Google Scholar] [CrossRef]
- Gelfand, D.H. Taq DNA polymerase. In PCR Technology: Principles and Applications for DNA Amplification; Springer: Berlin/Heidelberg, Germany, 1989; pp. 17–22. [Google Scholar]
- Keay, L.; Moser, P.W.; Wildi, B.S. Proteases of the genus Bacillus. II. Alkaline proteases. Biotechnol. Bioeng. 1970, 12, 213–249. [Google Scholar] [CrossRef]
- Cordeiro, C.A.M.; Martins, M.L.L.; Luciano, A.B. Production and properties of alpha-amylase from thermophilic Bacillus sp. Braz. J. Microbiol. 2002, 33, 57–61. [Google Scholar] [CrossRef]


| Strain | DDBJ/EMBL/GenBank Accession Number |
|---|---|
| NRS-2058 | JBCNBW010000001.1 |
| KCTC3564 | CP017703.1 |
| 8m3 | LWBR01000001.1 |
| W-12 | QURG01000001.1 |
| 8 | LVHY01000001.1 |
| TD1 | SFCD01000001.1 |
| BK1 | CP160301.1 |
| PI8 | AP022323.1 |
| MHI3390 | JAVLRY010000001.1 |
| NRS-1637 | JARTFV010000001.1 |
| GS3372 | JYCD01000002.1 |
| SJP27 | JBFQFV010000001.1 |
| MHI3391 | JAVLRZ010000001.1 |
| Genome | Size (bp) | GC Content (%) | N50 | L50 | Number of Contigs (with PEGs) | Number of Subsystems | Number of Coding Sequences | Number of RNAs |
|---|---|---|---|---|---|---|---|---|
| NRS-2058 | 3,245,679 | 39.0 | 58,619 | 18 | 109 | 284 | 3481 | 75 |
| W-12 | 3,839,138 | 38.9 | 99,755 | 12 | 140 | 304 | 4283 | 37 |
| TD1 | 3,748,965 | 38.8 | 45,881 | 26 | 207 | 300 | 4300 | 53 |
| SJP27 | 3,377,776 | 39.2 | 47,336 | 24 | 209 | 293 | 3933 | 69 |
| PI8 | 3,833,114 | 39.0 | - | 1 | 1 | 304 | 4229 | 104 |
| NRS-1637 | 3,863,896 | 38.7 | 71,699 | 16 | 120 | 302 | 4385 | 88 |
| MHI3391 | 3,782,925 | 39.1 | 39,514 | 27 | 171 | 301 | 4326 | 87 |
| MHI3390 | 3,911,438 | 39.0 | 281,598 | 4 | 66 | 307 | 4413 | 87 |
| KCTC3564 | 4,089,457 | 39.3 | - | 1 | 1 | 300 | 4535 | 104 |
| GS3372 | 4,985,863 | 57.4 | 66,220 | 25 | 185 | 302 | 5592 | 72 |
| BK1 | 3,935,118 | 39.2 | - | 1 | 1 | 301 | 4293 | 105 |
| 8m3 | 3,818,610 | 38.9 | 136,754 | 9 | 79 | 306 | 4245 | 68 |
| 8 | 3,903,800 | 38.8 | 85,252 | 15 | 169 | 301 | 4478 | 95 |
| NRS-2058 | W-12 | TD1 | SJP27 | PI8 | NRS-1637 | MHI3391 | MHI3390 | KCTC3564 | GS3372 | BK1 | 8m3 | 8 | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| COG Category | COG Category Description | CDS | ||||||||||||
| Cellular Processes and Signaling | ||||||||||||||
| D | Cell cycle control, cell division, chromosome partitioning | 49 | 43 | 50 | 48 | 45 | 46 | 47 | 47 | 46 | 67 | 44 | 42 | 48 |
| M | Cell wall/membrane/envelope biogenesis | 157 | 154 | 153 | 118 | 138 | 153 | 155 | 175 | 184 | 216 | 163 | 158 | 150 |
| N | Cell motility | 73 | 75 | 73 | 61 | 73 | 75 | 70 | 71 | 70 | 86 | 70 | 72 | 71 |
| O | Post-translational modification, protein turnover, chaperones | 104 | 118 | 113 | 116 | 119 | 117 | 120 | 120 | 121 | 132 | 121 | 118 | 116 |
| T | Signal transduction mechanisms | 116 | 147 | 161 | 124 | 145 | 149 | 149 | 151 | 150 | 224 | 148 | 146 | 148 |
| U | Intracellular trafficking, secretion, and vesicular transport | 55 | 48 | 49 | 46 | 51 | 56 | 52 | 54 | 59 | 85 | 47 | 51 | 55 |
| V | Defense mechanisms | 42 | 56 | 67 | 52 | 65 | 68 | 57 | 74 | 64 | 68 | 59 | 50 | 71 |
| W | Extracellular structures | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 |
| Z | Cytoskeleton | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 |
| Information Storage and Processing | ||||||||||||||
| A | RNA processing and modification | 0 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 |
| B | Chromatin structure and dynamics | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| J | Translation, ribosomal structure and biogenesis | 189 | 169 | 176 | 169 | 170 | 170 | 169 | 172 | 172 | 201 | 169 | 169 | 170 |
| K | Transcription | 188 | 268 | 272 | 240 | 264 | 264 | 267 | 264 | 270 | 389 | 257 | 270 | 269 |
| L | Replication, recombination and repair | 216 | 275 | 239 | 377 | 329 | 317 | 337 | 307 | 578 | 251 | 463 | 280 | 421 |
| Metabolism | ||||||||||||||
| C | Energy production and conversion | 144 | 245 | 248 | 205 | 234 | 237 | 224 | 239 | 229 | 254 | 226 | 246 | 237 |
| E | Amino acid transport and metabolism | 278 | 302 | 309 | 251 | 295 | 300 | 292 | 296 | 282 | 465 | 279 | 295 | 299 |
| F | Nucleotide transport and metabolism | 100 | 95 | 101 | 93 | 99 | 97 | 106 | 99 | 100 | 111 | 96 | 95 | 98 |
| G | Carbohydrate transport and metabolism | 150 | 213 | 224 | 178 | 222 | 228 | 214 | 222 | 217 | 294 | 206 | 215 | 231 |
| H | Coenzyme transport and metabolism | 113 | 146 | 153 | 135 | 146 | 137 | 137 | 152 | 155 | 182 | 135 | 146 | 137 |
| I | Lipid transport and metabolism | 110 | 125 | 131 | 83 | 123 | 125 | 100 | 121 | 100 | 154 | 102 | 122 | 126 |
| P | Inorganic ion transport and metabolism | 171 | 262 | 250 | 201 | 240 | 243 | 239 | 270 | 221 | 295 | 246 | 263 | 243 |
| Q | Secondary metabolites biosynthesis, transport and catabolism | 53 | 79 | 72 | 49 | 73 | 74 | 56 | 78 | 72 | 105 | 62 | 72 | 74 |
| Poorly Characterized | ||||||||||||||
| S | Function unknown | 1068 | 1036 | 1004 | 923 | 985 | 1049 | 1035 | 1042 | 1073 | 1463 | 994 | 1024 | 1059 |
| NRS-2058 | W-12 | TD1 | SJP27 | PI8 | NRS-1637 | MHI3391 | MHI3390 | KCTC3564 | GS3372 | BK1 | 8m3 | 8 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Heat shock protein 60 kDa family chaperone GroEL | |||||||||||||
| Heat shock protein 10 kDa family chaperone GroES | |||||||||||||
| RNA polymerase sigma factor RpoD | |||||||||||||
| RNA polymerase heat shock sigma factor SigI | |||||||||||||
| Chaperone protein DnaK | |||||||||||||
| Chaperone protein DnaJ | |||||||||||||
| Heat shock protein GrpE | |||||||||||||
| Chaperone protein HtpG | |||||||||||||
| ClpCP protease substrate adapter protein MecA | |||||||||||||
| ATP-dependent Clp protease, ATP-binding subunit ClpC | |||||||||||||
| ATP-dependent Clp protease proteolytic subunit ClpP (EC 3.4.21.92) | |||||||||||||
| ATP-dependent Clp protease ATP-binding subunit ClpX | |||||||||||||
| Putative membrane-bound ClpP-class protease associated with aq_911 | |||||||||||||
| ATP-dependent Clp protease, ATP-binding subunit ClpE | |||||||||||||
| Chaperone protein ClpB (ATP-dependent unfoldase) | |||||||||||||
| 33 kDa chaperonin HslO | |||||||||||||
| ATP-dependent protease subunit HslV (EC 3.4.25.2) | |||||||||||||
| ATP-dependent hsl protease ATP-binding subunit HslU | |||||||||||||
| Serine protease, DegP/HtrA, do-like (EC 3.4.21.-) | |||||||||||||
| HflK protein | |||||||||||||
| HflC protein | |||||||||||||
| Lon-like protease with PDZ domain | |||||||||||||
| Small heat shock protein |
| HMM Profile | CAZyme Classes/Associated Module | NRS-2058 | W-12 | TD1 | SJP27 | PI8 | NRS-1637 | MHI3391 | MHI3390 | KCTC3564 | GS3372 | BK1 | 8m3 | 8 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AA1 | Auxiliary Activity Family 1 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 2 | 4 | 1 | 1 | 0 |
| AA3 | Auxiliary Activity Family 3 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| AA4 | Auxiliary Activity Family 4 | 0 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 5 | 3 | 4 | 4 | 4 |
| AA6 | Auxiliary Activity Family 6 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| CBM20 | Carbohydrate-binding Module Family 20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| CBM34 | Carbohydrate-binding Module Family 34 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| CBM48 | Carbohydrate-binding Module Family 48 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| CBM50 | Carbohydrate-binding Module Family 50 | 4 | 3 | 3 | 3 | 3 | 3 | 4 | 4 | 3 | 1 | 3 | 3 | 3 |
| CBM68 | Carbohydrate-binding Module Family 68 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| CBM96 | Carbohydrate-binding Module Family 96 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| CE1 | Carbohydrate Esterase Family 1 | 1 | 2 | 3 | 1 | 2 | 2 | 1 | 3 | 1 | 5 | 2 | 2 | 2 |
| CE4 | Carbohydrate Esterase Family 4 | 4 | 5 | 5 | 4 | 4 | 4 | 4 | 5 | 4 | 10 | 4 | 4 | 4 |
| CE7 | Carbohydrate Esterase Family 7 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| CE9 | Carbohydrate Esterase Family 9 | 1 | 1 | 1 | 1 | 2 | 1 | 2 | 2 | 1 | 1 | 2 | 1 | 1 |
| CE14 | Carbohydrate Esterase Family 14 | 3 | 2 | 1 | 1 | 2 | 1 | 1 | 2 | 1 | 4 | 2 | 2 | 1 |
| GH1 | Glycosyl Hydrolase Family 1 | 0 | 2 | 2 | 0 | 2 | 2 | 2 | 2 | 2 | 0 | 2 | 2 | 2 |
| GH3 | Glycosyl Hydrolase Family 3 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| GH4 | Glycosyl Hydrolase Family 4 | 0 | 2 | 2 | 4 | 2 | 2 | 2 | 2 | 2 | 0 | 2 | 2 | 0 |
| GH13_2 | Glycosyl Hydrolase Family 13/Subf 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| GH13_9 | Glycosyl Hydrolase Family 13/Subf 9 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| GH13_14 | Glycosyl Hydrolase Family 13/Subf 14 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| GH13_20 | Glycosyl Hydrolase Family 13/Subf 20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| GH13_29 | Glycosyl Hydrolase Family 13/Subf 29 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| GH13_31 | Glycosyl Hydrolase Family 13/Subf 31 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| GH13_45 | Glycosyl Hydrolase Family 13/Subf 45 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 0 | 1 | 1 | 1 |
| GH15 | Glycosyl Hydrolase Family 15 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 |
| GH18 | Glycosyl Hydrolase Family 18 | 3 | 2 | 3 | 2 | 4 | 3 | 3 | 3 | 4 | 4 | 3 | 2 | 3 |
| GH20 | Glycosyl Hydrolase Family 20 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| GH23 | Glycosyl Hydrolase Family 23 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 |
| GH25 | Glycosyl Hydrolase Family 25 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| GH31_1 | Glycosyl Hydrolase Family 31/Subf 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| GH31_2 | Glycosyl Hydrolase Family 31/Subf 2 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| GH32 | Glycosyl Hydrolase Family 32 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| GH38 | Glycosyl Hydrolase Family 38 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| GH57 | Glycosyl Hydrolase Family 57 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 2 | 2 | 1 | 0 | 0 | 2 |
| GH73 | Glycosyl Hydrolase Family 73 | 1 | 1 | 1 | 1 | 1 | 2 | 0 | 0 | 0 | 0 | 1 | 1 | 2 |
| GH84 | Glycosyl Hydrolase Family 84 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| GH109 | Glycosyl Hydrolase Family 109 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| GH130_4 | Glycosyl Hydrolase Family 130/Subf 4 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| GH170 | Glycosyl Hydrolase Family 170 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 1 |
| GH171 | Glycosyl Hydrolase Family 171 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | |
| GH176 | Glycosyl Hydrolase Family 176 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| GH177 | Glycosyl Hydrolase Family 177 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| GH179 | Glycosyl Hydrolase Family 179 | 0 | 2 | 2 | 1 | 2 | 2 | 1 | 3 | 1 | 1 | 1 | 3 | 2 |
| GH188 | Glycosyl Hydrolase Family 188 | 0 | 5 | 6 | 5 | 3 | 7 | 5 | 6 | 6 | 5 | 5 | 6 | 7 |
| GT1 | Glycosyl Transferase Family 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| GT2 | Glycosyl Transferase Family 2 | 6 | 0 | 0 | 0 | 1 | 1 | 3 | 2 | 3 | 12 | 3 | 0 | 1 |
| GT4 | Glycosyl Transferase Family 4 | 7 | 7 | 7 | 1 | 5 | 6 | 8 | 5 | 3 | 8 | 6 | 5 | 6 |
| GT5 | Glycosyl Transferase Family 5 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 5 |
| GT8 | Glycosyl Transferase Family 8 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 0 |
| GT27 | Glycosyl Transferase Family 27 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| GT28 | Glycosyl Transferase Family 28 | 3 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| GT35 | Glycosyl Transferase Family 35 | 1 | 0 | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 5 | 4 | 4 | 4 |
| GT51 | Glycosyl Transferase Family 51 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 4 | 5 | 5 | 1 |
| GT61 | Glycosyl Transferase Family 51 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 |
| GT100 | Glycosyl Transferase Family 100 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| GT108 | Glycosyl Transferase Family 108 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| GT111 | Glycosyl Transferase Family 111 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| GT118 | Glycosyl Transferase Family 118 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| GT119 | Glycosyl Transferase Family 119 | 6 | 1 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 |
| GT121 | Glycosyl Transferase Family 121 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 3 | 0 | 0 | 1 | 1 | 1 |
| PL12 | Polysaccharide Lyase Family 12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| PL12_1 | Polysaccharide Lyase Family 12/Subf 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| PL12_3 | Polysaccharide Lyase Family 12/Subf 3 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| SLH | Surface layer (S-layer) Homology | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 |
| Organism | CDS | CDS (Without Artefact Fam.) | Pan CDS | Core CDS | Var CDS | Strain Specific CDS | Core CDS (%) | Var CDS (%) | Strain Spe. CDS (%) |
|---|---|---|---|---|---|---|---|---|---|
| A. alveayuensis 24KAM51 | 7047 | 6666 | 6666 | 2179 | 4487 | 1404 | 32.688 | 67.312 | 21.062 |
| A. alveayuensis DSM 19092 | 3359 | 3161 | 3161 | 1103 | 2058 | 361 | 34.894 | 65.106 | 11.42 |
| A. composti B-14742 | 3980 | 3755 | 3755 | 1175 | 2580 | 102 | 31.292 | 68.708 | 2.716 |
| A. composti B-14745 | 3630 | 3418 | 3418 | 1138 | 2280 | 164 | 33.294 | 66.706 | 4.798 |
| A. composti HB-1 | 4026 | 3765 | 3765 | 1161 | 2604 | 80 | 30.837 | 69.163 | 2.125 |
| A. composti KCTC 33824 | 4006 | 3781 | 3781 | 1158 | 2623 | 70 | 30.627 | 69.373 | 1.851 |
| A. composti NRS-1511 | 4099 | 3873 | 3873 | 1177 | 2696 | 88 | 30.39 | 69.61 | 2.272 |
| A. composti NRS-1512 | 3933 | 3715 | 3715 | 1178 | 2537 | 96 | 31.709 | 68.291 | 2.584 |
| A. composti NRS-1630 | 4153 | 3903 | 3903 | 1172 | 2731 | 20 | 30.028 | 69.972 | 0.512 |
| A. composti NRS-1631 | 4140 | 3885 | 3885 | 1173 | 2712 | 7 | 30.193 | 69.807 | 0.18 |
| A. composti NRS-1632 | 4075 | 3836 | 3836 | 1169 | 2667 | 57 | 30.474 | 69.526 | 1.486 |
| A. composti NRS-1633 | 3941 | 3706 | 3706 | 1166 | 2540 | 76 | 31.462 | 68.538 | 2.051 |
| A. composti NRS-2045 | 3953 | 3733 | 3733 | 1177 | 2556 | 36 | 31.53 | 68.47 | 0.964 |
| A. kexueae KCTC 33881 | 3488 | 3325 | 3325 | 1097 | 2228 | 949 | 32.992 | 67.008 | 28.541 |
| A. pallidus 8 | 4273 | 3946 | 3946 | 1172 | 2774 | 50 | 29.701 | 70.299 | 1.267 |
| A. pallidus 8m3 | 4046 | 3806 | 3806 | 1188 | 2618 | 59 | 31.214 | 68.786 | 1.55 |
| A. pallidus BK1 | 4122 | 3860 | 3860 | 1147 | 2713 | 65 | 29.715 | 70.285 | 1.684 |
| A. pallidus GS3372 | 5519 | 5298 | 5298 | 1223 | 4075 | 3125 | 23.084 | 76.916 | 58.985 |
| A. pallidus KCTC3564 | 4524 | 4200 | 4200 | 1161 | 3039 | 289 | 27.643 | 72.357 | 6.881 |
| A. pallidus MHI3390 | 4199 | 3945 | 3945 | 1188 | 2757 | 185 | 30.114 | 69.886 | 4.689 |
| A. pallidus MHI3391 | 4078 | 3837 | 3837 | 1143 | 2694 | 154 | 29.789 | 70.211 | 4.014 |
| A. pallidus NRS-1637 | 4159 | 3898 | 3898 | 1172 | 2726 | 16 | 30.067 | 69.933 | 0.41 |
| A. pallidus NRS-2058 | 3450 | 3277 | 3277 | 1084 | 2193 | 991 | 33.079 | 66.921 | 30.241 |
| A. pallidus PI8 | 4000 | 3735 | 3735 | 1158 | 2577 | 93 | 31.004 | 68.996 | 2.49 |
| A. pallidus SJP27 | 3648 | 3451 | 3451 | 1117 | 2334 | 160 | 32.367 | 67.633 | 4.636 |
| A. pallidus TD1 | 4037 | 3817 | 3817 | 1192 | 2625 | 172 | 31.229 | 68.771 | 4.506 |
| A. pallidus W-12 | 4071 | 3857 | 3857 | 1190 | 2667 | 89 | 30.853 | 69.147 | 2.307 |
| Aeribacillus sp. FSL K6-1121 | 4423 | 4141 | 4141 | 1176 | 2965 | 156 | 28.399 | 71.601 | 3.767 |
| Aeribacillus sp. FSL K6-1305 | 4170 | 3894 | 3894 | 1157 | 2737 | 80 | 29.712 | 70.288 | 2.054 |
| Aeribacillus sp. FSL K6-2211 | 4365 | 4061 | 4061 | 1151 | 2910 | 114 | 28.343 | 71.657 | 2.807 |
| Aeribacillus sp. FSL K6-2833 | 4233 | 3947 | 3947 | 1165 | 2782 | 86 | 29.516 | 70.484 | 2.179 |
| Aeribacillus sp. FSL K6-2848 | 4567 | 4251 | 4251 | 1171 | 3080 | 159 | 27.546 | 72.454 | 3.74 |
| Aeribacillus sp. FSL K6-3256 | 3976 | 3713 | 3713 | 1175 | 2538 | 30 | 31.646 | 68.354 | 0.808 |
| Aeribacillus sp. FSL K6-8210 | 4348 | 4038 | 4038 | 1155 | 2883 | 107 | 28.603 | 71.397 | 2.65 |
| Aeribacillus sp. FSL K6-8394 | 3911 | 3682 | 3682 | 1124 | 2558 | 213 | 30.527 | 69.473 | 5.785 |
| Aeribacillus sp. FSL M8-0235 | 4012 | 3749 | 3749 | 1136 | 2613 | 153 | 30.301 | 69.699 | 4.081 |
| Aeribacillus sp. FSL M8-0254 | 4169 | 3940 | 3940 | 1144 | 2796 | 266 | 29.036 | 70.964 | 6.751 |
| Aeribacillus sp. FSL W8-0870 | 4343 | 4046 | 4046 | 1178 | 2868 | 81 | 29.115 | 70.885 | 2.002 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yıldız, S.Y.; Radchenkova, N. From Genomes to Applications: Comparative Analysis of Aeribacillus pallidus Reveals a Thermophilic Chassis for Biotechnology. Appl. Sci. 2025, 15, 10866. https://doi.org/10.3390/app152010866
Yıldız SY, Radchenkova N. From Genomes to Applications: Comparative Analysis of Aeribacillus pallidus Reveals a Thermophilic Chassis for Biotechnology. Applied Sciences. 2025; 15(20):10866. https://doi.org/10.3390/app152010866
Chicago/Turabian StyleYıldız, Songül Yaşar, and Nadja Radchenkova. 2025. "From Genomes to Applications: Comparative Analysis of Aeribacillus pallidus Reveals a Thermophilic Chassis for Biotechnology" Applied Sciences 15, no. 20: 10866. https://doi.org/10.3390/app152010866
APA StyleYıldız, S. Y., & Radchenkova, N. (2025). From Genomes to Applications: Comparative Analysis of Aeribacillus pallidus Reveals a Thermophilic Chassis for Biotechnology. Applied Sciences, 15(20), 10866. https://doi.org/10.3390/app152010866






