Isolation and Properties of Enterobacter sp. LX3 Capable of Producing Indoleacetic Acid
Abstract
:Featured Application
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation of IAA-Producing Bacteria from Maize Rhizosphere
2.2. Morphological Analysis
2.3. Phenotypic Characterization
2.4. 16S rRNA Gene Sequencing
2.5. Phylogenetic Analysis
2.6. IAA Production
2.7. Plant Growth Promotion Experiment
2.8. IAA Assay
2.9. Chlorophyll Assay
2.10. Cell Growth on Different Carbohydrates
2.11. Cell Growth on Nitrogen Sources
2.12. Statistical Analysis
3. Results
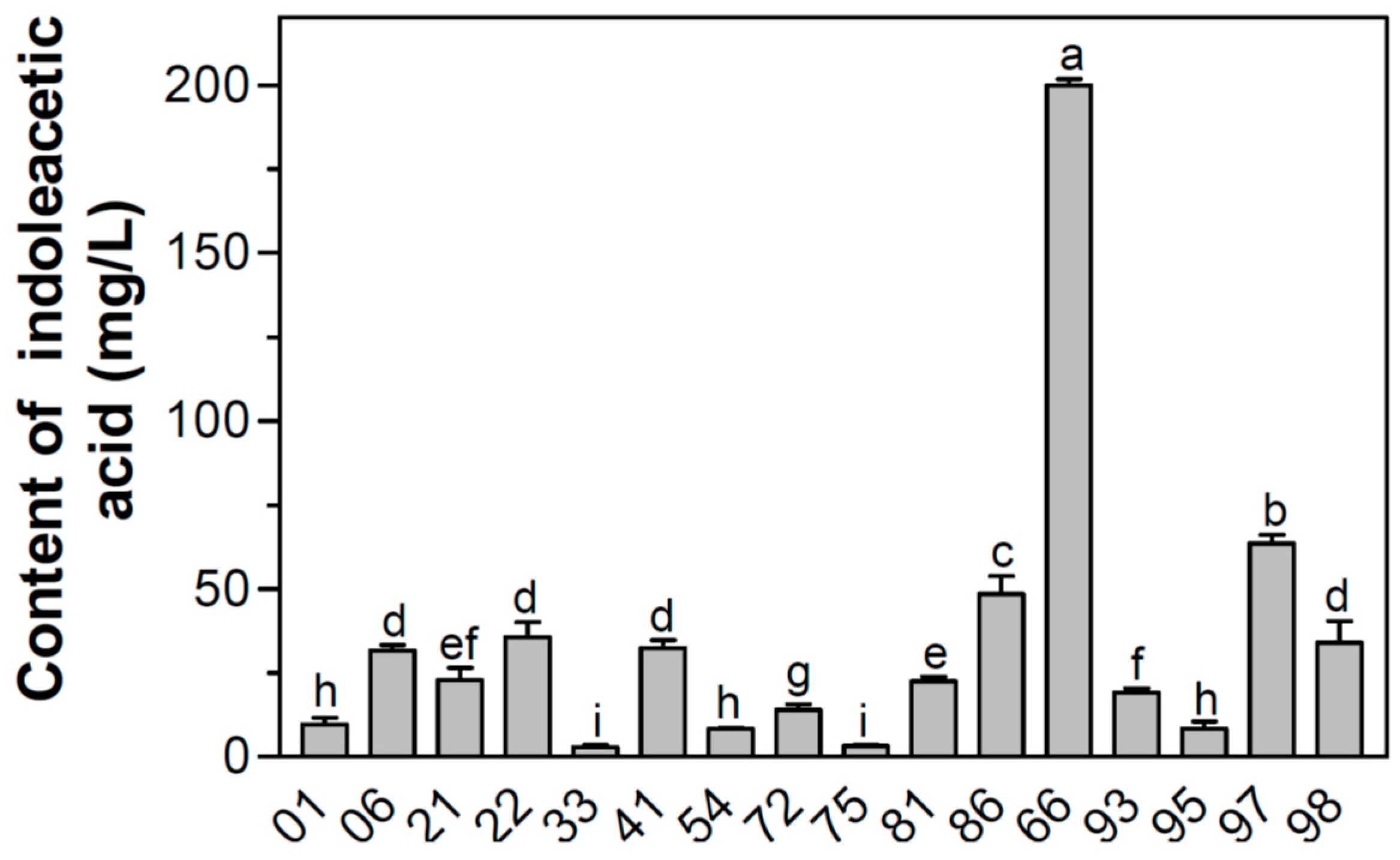
3.1. Screening of IAA-Producing Bacteria and IAA Production
3.2. Phenotypic Properties
3.3. Phenotypic Characteristics
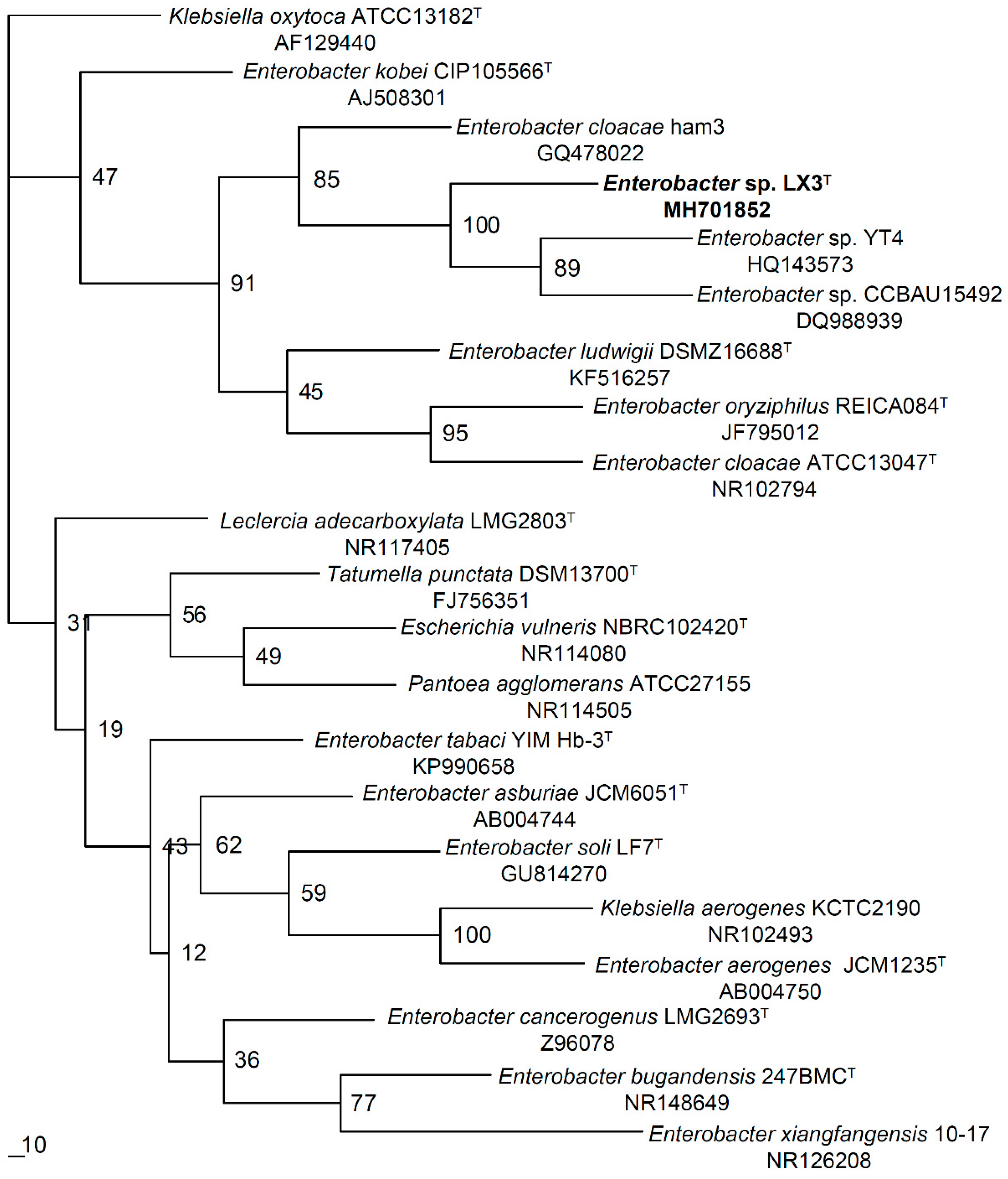
3.4. Phylogenetic Analysis
3.5. Plant Growth Promotion
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Duca, D.; Lorv, J.; Patten, C.L.; Rose, D.; Glick, B.R. Indole-3-acetic acid in plant-microbe interactions. Antonie Van Leeuwenhoek 2014, 106, 85–125. [Google Scholar] [CrossRef] [PubMed]
- Tian, H.; De Smet, I.; Ding, Z. Shaping a root system: Regulating lateral versus primary root growth. Trends Plant Sci. 2014, 19, 426–431. [Google Scholar] [CrossRef] [PubMed]
- Egamberdieva, D.; Wirth, S.J.; Alqarawi, A.A.; Abd_Allah, E.F.; Hashem, A. Phytohormones and beneficial microbes: Essential components for plants to balance stress and fitness. Front. Microbiol. 2017, 8, 2104. [Google Scholar] [CrossRef] [PubMed]
- Ozdal, M.; Ozdal, O.G.; Sezen, A.; Algur, O.F.; Kurbanoglu, E.B. Continuous production of indole-3-acetic acid by immobilized cells of Arthrobacter agilis. 3 Biotech 2017, 7, 23. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Sen, S.K. Exploration of novel rhizospheric yeast isolate as fertilizing soil inoculant for improvement of maize cultivation. J. Sci. Food Agric. 2015, 95, 1491–1499. [Google Scholar] [CrossRef] [PubMed]
- Weselowski, B.; Nathoo, N.; Eastman, A.W.; MacDonald, J.; Yuan, Z.C. Isolation, identification and characterization of Paenibacillus polymyxa CR1 with potentials for biopesticide, biofertilization, biomass degradation and biofuel production. BMC Microbiol. 2016, 16, 244. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.Y.; Zhang, F.; Zhang, D.J.; Srivastava, A.K.; Wu, Q.S.; Zou, Y.N. Mycorrhiza stimulates root-hair growth and IAA synthesis and transport in trifoliate orange under drought stress. Sci. Rep. 2018, 8, 1978. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Defez, R.; Andreozzi, A.; Bianco, C. The Overproduction of indole-3-acetic acid (IAA) in endophytes upregulates nitrogen fixation in both bacterial cultures and inoculated rice plants. Microb. Ecol. 2017, 74, 441–452. [Google Scholar] [CrossRef] [PubMed]
- Egamberdieva, D.; Wirth, S.; Behrendt, U.; Abd_Allah, E.F.; Berg, G. Biochar treatment resulted in a combined effect on soybean growth promotion and a shift in plant growth promoting rhizobacteria. Front. Microbiol. 2016, 7, 209. [Google Scholar] [CrossRef] [PubMed]
- King, E.O.; Ward, M.K.; Raney, D.E. Two simple media for the demonstration of phycocyanin and fluorescin. J. Lab. Clin. Med. 1954, 44, 301–307. [Google Scholar] [PubMed]
- Bric, J.M.; Bostock, R.M.; Silverstone, S.E. Rapid in situ assay for indoleacetic acid production by bacteria immobilized on a nitrocellulose membrane. Appl. Environ. Microbiol. 1991, 57, 535–538. [Google Scholar] [PubMed]
- Murray, R.G.E.; Doetsch, R.N.; Robinow, C.F. Determinative and cytological light microscopy. In Methods for General and Molecular Bacteriology; Gerhardt, P., Murray, R.G.E., Wood, W.A., Krieg, N.R., Eds.; American Society for Microbiology: Washington, DC, USA, 1994; pp. 21–41. [Google Scholar]
- Leifson, E. Staining, shape, and arrangement of bacterial flagella. J. Bcateriol. 1951, 62, 377–389. [Google Scholar]
- Smibert, R.M.; Krieg, N.R. Phenotypic characterization. In Methods for General and Molecular Bacteriology; Gerhardt, P., Murray, R.G.E., Wood, W.A., Krieg, N.R., Eds.; American Society for Microbiology: Washington, DC, USA, 1994; pp. 607–654. ISBN 1-55581-048-9. [Google Scholar]
- Moore, D.D.; Dowhan, D. Preparation and analysis of DNA. In Current Protocols in Molecular Biology; Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., Struhl, R., Eds.; John Wiley & Sons: New York, NY, USA, 1997; Volume 1, pp. 241–245. ISBN 0-471-50338-X. [Google Scholar]
- Jin, Z.; Wang, C.; Li, X. Isolation and some properties of newly isolated oxalate-degrading Pandoraea sp. OXJ-11 from soil. J. App. Microbiol. 2007, 103, 1066–1073. [Google Scholar] [CrossRef] [PubMed]
- Johnson, M.; Zaretskaya, I.; Raytselis, Y.; Merezhuk, Y.; McGinnis, S.; Madden, T.L. NCBI BLAST: A better web interface. Nucl. Acids Res. 2008, 36, W5–W9. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Gibson, T.L.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL-X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. PHYLIP (Phylogeny Inference Package) Version 3.6; Department of Genome Sciences, University of Washington: Seattle, WA, USA, 2005. [Google Scholar]
- Gunning, T.; Cahill, D.M. A soil-free plant growth system to facilitate analysis of plant pathogen interactions in roots. J. Phytopathol. 2009, 157, 497–501. [Google Scholar] [CrossRef]
- Patten, C.L.; Glick, B.R. Role of Pseudomonas putida indoleacetic acid in development of the host plant root system. Appl. Environ. Microbiol. 2002, 68, 3795–3801. [Google Scholar] [CrossRef] [PubMed]
- Arnon, D.I. Copper enzymes in isolated chloroplasts. Polyphenoloxidases in Beta vulgaris. Plant Physiol. 1949, 24, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Grimont, P.A.D.; Grimont, F. Genus XII. Enterobacter. In Bergey’s Manual of Systematic Bacteriology; Brenner, D.J., Krieg, N.R., Staley, J.T., Eds.; Springer: Monroe, MI, USA, 2005; Volume 2, Part B; pp. 661–670. ISBN 978-0-387-24144-9. [Google Scholar]
- Fox, G.E.; Wisotzkey, J.D.; Jurtshuk, P. How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int. J. Syst. Bacteriol. 1992, 42, 166–170. [Google Scholar] [CrossRef] [PubMed]
- Wagner-Döbler, I.; Rheims, H.; Felske, A.; El-Ghezal, A.; Flade-Schröder, D.; Laatsch, H.; Lang, S.; Pukall, R.; Tindall, B.J. Oceanibulbus indolifex, gen. nov., sp. nov., a North Sea Alphaproteo-bacterium producing bioactive metabolites. Int. J. Syst. Evol. Microbiol. 2004, 54, 1177–1184. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Xu, Y.; Lai, X. Antagonistic endophytic bacteria associated with nodules of soybean (Glycine max L.) and plant growth-promoting properties. Braz. J. Microbiol. 2018, 49, 269–278. [Google Scholar] [CrossRef] [PubMed]
- Sun, P.F.; Fang, W.T.; Shin, L.Y.; Wei, J.Y.; Fu, S.F.; Chou, J.Y. Indole-3-acetic acid-producing yeasts in the phyllosphere of the carnivorous plant Drosera indica L. PLoS ONE 2014, 9, e114196. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, M.; Saiz, G.; Dannenmann, M.; Guo, L.; Tao, Y.; Shi, J.; Zuo, Q.; Butterbach-Bahl, K.; Li, G.; et al. Enhancement of root systems improves productivity and sustainability in water saving ground cover rice production system. Field Crops Res. 2017, 213, 186–193. [Google Scholar] [CrossRef]
| Characteristics | Strain LX3 | Characteristics | Strain LX3 |
|---|---|---|---|
| Gram stain | − | Voges-Proskauer reaction | + |
| Straight rod | + | Methyl red | − |
| Endospore formation | − | Nitrate reduction | + |
| Motility | + | Tetrathionate reduction | − |
| Facultative anaerobic | + | Corn oil and tributyrin hydrolysis | − |
| β-Galactosidase | − | Alkaline reaction in Simmons citrate | + |
| Arginine dihydrolase | + | Citrate as sole carbon/energy source | + |
| Lysine decarboxylase | + | Acid and gas production from glucose | + |
| Ornithine decarboxylase | + | Gas production from glucose at 44.5 °C | − |
| Phenylalanine deaminase | − | Acid production from: | |
| Catalase | + | d-Glucose | + |
| Oxidase | − | d-Mannitol | − |
| Urease | + | Inositol | − |
| Acid fast | − | d-Sorbitol | + |
| Gelatin liquefaction | − | l-Rhamnose | + |
| Esculin hydrolysis | + | Sucrose | − |
| Growth at 37 °C | + | Melibiose | + |
| H2S production | − | Arabinose | + |
| Indole production | + | Amygdalin | + |
| Characteristics | Fresh Weight (g) | Root Weight (g) | Plant Height (cm) | Root Length (cm) | Chlorophyll Content (mg/g) |
|---|---|---|---|---|---|
| Control | 0.34 ± 0.013 | 0.20 ± 0.007 | 10.83 ± 0.17 | 12.83 ± 0.21 | 0.74 ± 0.011 |
| Strain LX3 | 0.41 ± 0.012 b | 0.22 ± 0.005 a | 13.17 ± 0.15 b | 15.16 ± 0.17 b | 0.80 ± 0.013 a |
| Characteristic | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lysine decarboxylase | + | − | − | − | − | − | − | N | − | − | + | − | − |
| β-Galactosidase | − | + | + | + | + | + | + | N | + | + | − | + | + |
| Arginine dihydrolase | + | + | N | + | + | + | + | N | + | + | − | + | − |
| Urea hydrolysis | + | − | + | − | − | − | − | N | − | − | − | − | − |
| H2S production | − | − | − | − | − | − | − | + | − | − | − | − | − |
| Voges-Proskauer reaction | + | − | − | + | + | + | + | + | − | + | + | + | − |
| Indole production | + | − | − | − | − | − | − | + | − | − | − | − | + |
| Acid production from: | |||||||||||||
| Inositol | − | − | − | − | − | + | − | − | − | − | + | − | N |
| d-Mannitol | − | + | + | + | + | + | + | N | + | + | + | + | + |
| Sucrose | − | + | + | − | + | + | + | + | + | + | + | + | |
| d-Sorbitol | + | + | + | − | + | + | + | N | + | + | + | + | − |
| Melibiose | + | + | − | − | + | + | − | − | + | + | + | + | + |
| l-Rhamnose | + | + | − | + | + | + | − | N | + | + | + | + | + |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chi, Q.; Tang, W.; Liu, L.; Meng, J.; Dong, X.; Chen, W.; Li, X. Isolation and Properties of Enterobacter sp. LX3 Capable of Producing Indoleacetic Acid. Appl. Sci. 2018, 8, 2108. https://doi.org/10.3390/app8112108
Chi Q, Tang W, Liu L, Meng J, Dong X, Chen W, Li X. Isolation and Properties of Enterobacter sp. LX3 Capable of Producing Indoleacetic Acid. Applied Sciences. 2018; 8(11):2108. https://doi.org/10.3390/app8112108
Chicago/Turabian StyleChi, Qingshan, Wenzhu Tang, Lu Liu, Jun Meng, Xiaoli Dong, Wenfu Chen, and Xianzhen Li. 2018. "Isolation and Properties of Enterobacter sp. LX3 Capable of Producing Indoleacetic Acid" Applied Sciences 8, no. 11: 2108. https://doi.org/10.3390/app8112108
APA StyleChi, Q., Tang, W., Liu, L., Meng, J., Dong, X., Chen, W., & Li, X. (2018). Isolation and Properties of Enterobacter sp. LX3 Capable of Producing Indoleacetic Acid. Applied Sciences, 8(11), 2108. https://doi.org/10.3390/app8112108




