Biomedical Photoacoustic Imaging Optimization with Deconvolution and EMD Reconstruction
Abstract
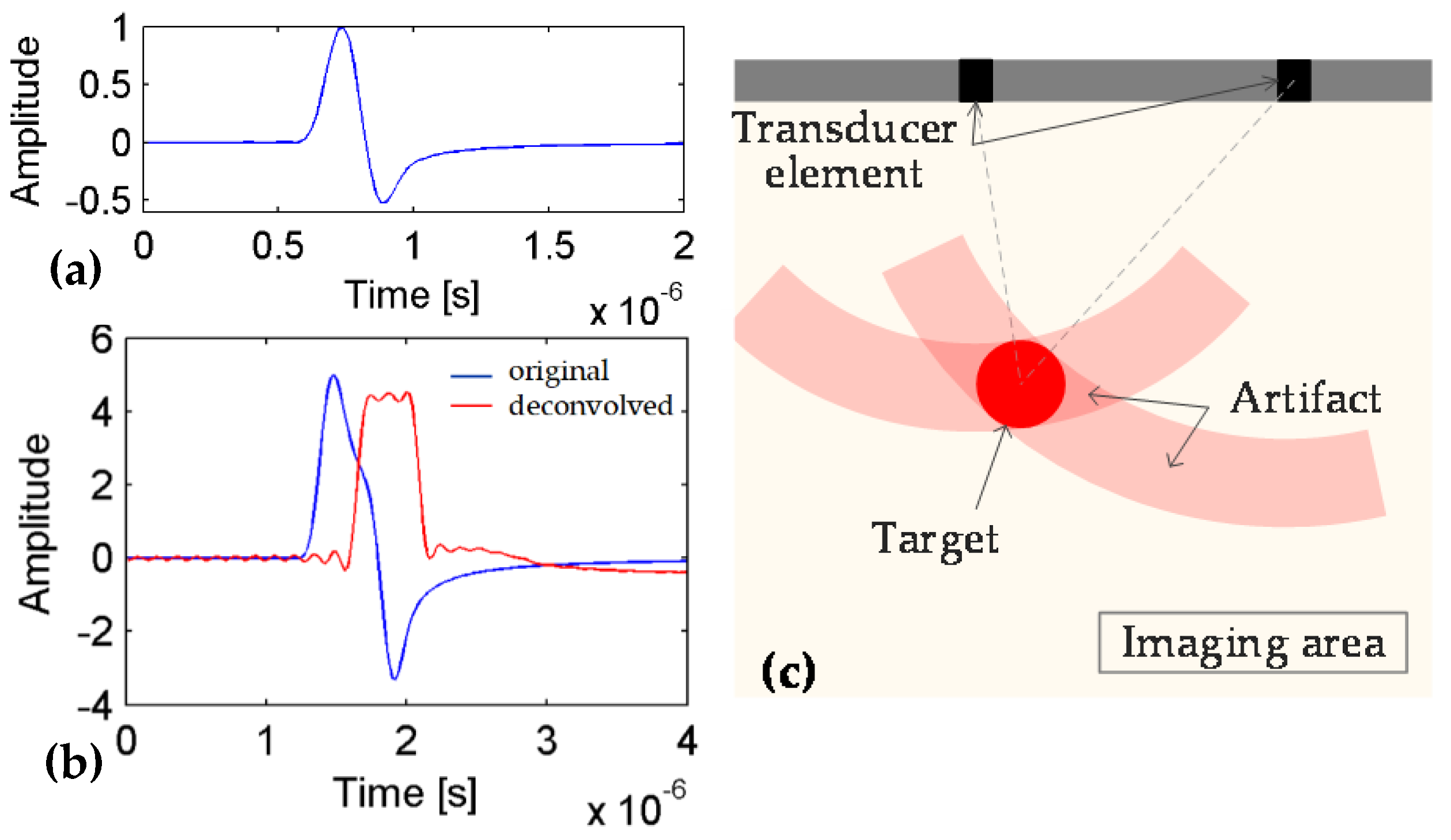
1. Introduction
2. Methods
3. Results
3.1. Numerical Simulations
3.2. Experiments
4. Discussion and Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Xu, M.; Wang, L.V. Photoacoustic imaging in biomedicine. Rev. Sci. Instrum. 2006, 77, 041101. [Google Scholar] [CrossRef]
- Beard, P. Biomedical photoacoustic imaging. Interface Focus 2011, 1, 602–631. [Google Scholar] [CrossRef] [PubMed]
- Ku, G.; Fornage, B.D.; Jin, X.; Xu, M.; Hunt, K.K.; Wang, L.V. Thermoacoustic and photoacoustic tomography of thick biological tissues toward breast imaging. Technol. Cancer Res. Treat. 2005, 4, 559–565. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Pang, Y.; Ku, G.; Xie, X.; Stoica, G.; Wang, L.V. Noninvasive laser-induced photoacoustic tomography for structural and functional in vivo imaging of the brain. Nat. Biotechnol. 2003, 21, 803–806. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Xie, X.; Ku, G.; Wang, L.V.; Stoica, G. Noninvasive imaging of hemoglobin concentration and oxygenation in the rat brain using high-resolution photoacoustic tomography. J. Biomed. Opt. 2006, 11, 024015. [Google Scholar] [CrossRef] [PubMed]
- Oraevsky, A.A.; Andreev, V.A.; Karabutov, A.A.; Esenaliev, R.O. Two-dimensional optoacoustic tomography: Transducer array and image reconstruction algorithm. In Proceedings of the BiOS ‘99 International Biomedical Optics Symposium, San Jose, CA, USA, 14 June 1999; p. 12. [Google Scholar]
- Changhui, L.; Lihong, V.W. Photoacoustic tomography and sensing in biomedicine. Phys. Med. Biol. 2009, 54, R59. [Google Scholar]
- Ermilov, S.A.; Gharieb, R.; Conjusteau, A.; Miller, T.; Mehta, K.; Oraevsky, A.A. Data processing and quasi-3D optoacoustic imaging of tumors in the breast using a linear arc-shaped array of ultrasonic transducers. In Proceedings of the SPIE BiOS, San Jose, CA, USA, 28 February 2008; p. 10. [Google Scholar]
- Ermilov, S.A.; Khamapirad, T.; Conjusteau, A.; Leonard, M.H.; Lacewell, R.; Mehta, K.; Miller, T.; Oraevsky, A.A. Laser optoacoustic imaging system for detection of breast cancer. J. Biomed. Opt. 2009, 14, 024007. [Google Scholar] [CrossRef] [PubMed]
- Kruger, R.A.; Kiser, W.L.; Reinecke, D.R.; Kruger, G.A. Thermoacoustic computed tomography using a conventional linear transducer array. Med. Phys. 2003, 30, 856–860. [Google Scholar] [CrossRef] [PubMed]
- Gamelin, J.; Maurudis, A.; Aguirre, A.; Huang, F.; Guo, P.; Wang, L.V.; Zhu, Q. A real-time photoacoustic tomography system for small animals. Opt. Express 2009, 17, 10489–10498. [Google Scholar] [CrossRef] [PubMed]
- Cai, D.; Li, Z.; Chen, S.-L. In vivo deconvolution acoustic-resolution photoacoustic microscopy in three dimensions. Biomed. Opt. Express 2016, 7, 369–380. [Google Scholar] [CrossRef] [PubMed]
- Ryo, N.; Shin, Y.; Shin-ichiro, U.; Yoshifumi, S. Basic study of improvement of axial resolution and suppression of time side lobe by phase-corrected wiener filtering in photoacoustic tomography. Jpn. J. Appl. Phys. 2018, 57, 07LD11. [Google Scholar] [CrossRef]
- Li, W.; Zhang, L.; Yuan, J.; Liu, X.; Xu, G.; Wang, X.; Carson, P.L. Novel image optimization on photoacoustic tomography. In Proceedings of the 2014 IEEE International Ultrasonics Symposium, Chicago, IL, USA, 3–6 September 2014; pp. 1280–1283. [Google Scholar]
- Cao, M.; Feng, T.; Yuan, J.; Xu, G.; Wang, X.; Carson, P.L. Spread spectrum photoacoustic tomography with image optimization. IEEE Trans. Biomed. Circuits Syst. 2017, 11, 411–419. [Google Scholar] [CrossRef] [PubMed]
- Brunker, J.; Beard, P. Pulsed photoacoustic doppler flowmetry using time-domain cross-correlation: Accuracy, resolution and scalability. J. Acoust. Soc. Am. 2012, 132, 1780–1791. [Google Scholar] [CrossRef] [PubMed]
- Gonzales, R.C.; Woods, R.E. Digital Image Processing; Addison & Wesley Publishing Company: Reading, MA, USA, 1992. [Google Scholar]
- Huang, N.E.; Shen, Z.; Long, S.R.; Wu, M.C.; Shih, H.H.; Zheng, Q.; Yen, N.-C.; Tung, C.C.; Liu, H.H. The empirical mode decomposition and the hilbert spectrum for nonlinear and non-stationary time series analysis. Proc. R. Soc. Lond. Ser. A Math. Phys. Eng. Sci. 1998, 454, 903. [Google Scholar] [CrossRef]
- Zhu, Y.-H.; Yuan, J.; Pinter, S.Z.; Oliver, D.K.; Cheng, Q.; Wang, X.-D.; Tao, C.; Liu, X.-J.; Xu, G.; Carson, P.L. Adaptive optimization on ultrasonic transmission tomography-based temperature image for biomedical treatment. Chin. Phys. B 2017, 26, 064301. [Google Scholar] [CrossRef]
- Treeby, B.E.; Cox, B.T. K-wave: Matlab toolbox for the simulation and reconstruction of photoacoustic wave fields. J. Biomed. Opt. 2010, 15, 021314. [Google Scholar] [CrossRef] [PubMed]






© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, C.; Chen, Y.; Yuan, J.; Zhu, Y.; Cheng, Q.; Wang, X. Biomedical Photoacoustic Imaging Optimization with Deconvolution and EMD Reconstruction. Appl. Sci. 2018, 8, 2113. https://doi.org/10.3390/app8112113
Guo C, Chen Y, Yuan J, Zhu Y, Cheng Q, Wang X. Biomedical Photoacoustic Imaging Optimization with Deconvolution and EMD Reconstruction. Applied Sciences. 2018; 8(11):2113. https://doi.org/10.3390/app8112113
Chicago/Turabian StyleGuo, Chengwen, Yingna Chen, Jie Yuan, Yunhao Zhu, Qian Cheng, and Xueding Wang. 2018. "Biomedical Photoacoustic Imaging Optimization with Deconvolution and EMD Reconstruction" Applied Sciences 8, no. 11: 2113. https://doi.org/10.3390/app8112113
APA StyleGuo, C., Chen, Y., Yuan, J., Zhu, Y., Cheng, Q., & Wang, X. (2018). Biomedical Photoacoustic Imaging Optimization with Deconvolution and EMD Reconstruction. Applied Sciences, 8(11), 2113. https://doi.org/10.3390/app8112113






