1. Introduction
1.1. Plant Electrical Signals
Since 1873, action potential in
Venus flytrap has been first measured by Burden-Sanderson, and the plant electrophysiology has been studied over the past 140 years [
1]. A large number of experimental data are recorded by different experiment methods [
2]. In fact, plant electrical signals is a response to stimulation by environment and involves in many processes in plant physiological activities [
3,
4,
5,
6,
7], e.g., photosynthesis [
8,
9], respiration [
10,
11], transpiration [
12], ATP content variation and heat tolerance.
Owing to different measurement methods, there are three data types of plant electrical signals, including text data, binary data, and image data. In general, text data is used in the traditional intracellular and extracellular recording and patch clamp measurement. In
Table 1, the size of text data is approximately 70 MB/h (RM6240BD, Chengdu Instrument factory, China) at a given sample frequency [
13]. For many vascular plants, the frequency of action potentials and variation potentials is less than 10 Hz. The data files are always saved as the text format for good readability in most recording systems. Once the amount of electrodes reaches dozens and the sample frequency is higher than one kilohertz, the data can be saved as a binary data format. In
Table 1, the size of multi-electrode arrays (MEA) data is 10 GB–60 GB/h saved as a binary data format (Alpha MED Scientific Inc., Ibaraki Osaka, Japan) [
14]. For optical recording method, which measures the plant cells membrane potential change through fluorescence intensity by using voltage-sensitive dye imaging, the size of image data is about 15 GB–70 GB/h at a given image resolution, sample frequency and optical channel number [
15].
1.2. The Analysis Methods of Plant Electrophysiological Data
Three types of analysis methods for plant electrical signals are listed in
Table 2. Although there have been many reports describing various analysis methods for plant electrical signals, the lack of shareable standard plant electrical signals data limits the reproducibility in experiments to verify these results and algorithms.
1.3. Multi-Source Data Sharing, Management and Analysis Techniques
Compared to single-source data analysis, multi-source can not only validate interdependence experimental results to obtain more reliable and accurate results, but it also eliminate contradictions and complement each other [
26,
27]. Data sharing improves the research exchange with other organizations and enable more researchers to use and utilize existing data resources. In addition, it can reduce the cost of data management and the work of data collection [
28].
Although desktop-based offline analysis techniques for plant electrical signals have been improved, there are still challenges in analysis of plant electrophysiological data, e.g., overlapped noise in the raw recordings, variation of response waveform owing to different stimulations, varieties, and ongoing plant electrophysiological data collection. Therefore, online computing and analysis can promote in-depth research on plant electrical signals interpretation and data mining for finding useful information of crop stress tolerance.
For these reasons, we retrieved the electronic resources, i.e., Ei Compendex, Web of Science, PubMed, Google Scholar, IEEE Xplore, ACM Digital Library, ScienceDirect, Springer Link, Google etc. Through reviewing literatures and the retrieving key words are plant electrical signal, membrane potential, action potential (AP), variation potential (VP), sharing, data storage, large scale data, and online analysis. In 2014, Spain, Britain and the PLEASED team pioneered the web-based data storage and popular science for plant electrical signals aided by EU’s Seventh Framework Program, in which they opened several electrophysiological data set stored in business Mega cloud storage. It inspires us to do the in-depth research for data sharing, especially for online analysis.
Currently, there is no public web-based plant electrophysiological platform for data sharing, online computing and analysis. Luckily, the abundant data analysis and sharing technologies for animal and human body electrophysiological data can enlighten us.
With respect to current important electrophysiological data sharing platforms for animal and human body, we have summarized in a review paper [
29]. Many of them are active and allow users to create datasets on web page and access datasets online through a web browser or custom desktop software. The electrophysiological data include electrical signals, experimental metadata, annotation data and image data. Hence, efficient semi-structure and unstructured data management is a key challenge.
To realize electrophysiological data sharing and analysis for experimental and clinical research for humans, many web platforms have been developed for human electrophysiological data management in the past years. Among these systems, PhysioNet has the powerful functions like rich data types [
30] (PhysioBank includes EEG, ECG, EMG etc.), a large number of offline analysis tools (PhysioTookit), basic online data browsing function (Lightwave [
31], PhysioBank ATM) and scientific research cooperation among different teams (PhysioNetWorks). The similar platforms also contain INCF (International Neuroinformatics Coordinating Facility), NEO [
32], G-Node (German Neuroinformatics Node) [
33], IEEG-Portal (The International Epilepsy Electrophysiology Portal) [
34] and CARMEN (The Code Analysis Repository & Modelling for E-Neuroscience) [
35,
36]. Cloudwave is a visualization and analysis platform of electrophysiology (EEG, ECG) [
37]. It is a part of the Prevention and Risk Identification of SUDEP Mortality (PRISM) project, using highly scalable open source cloud computing infrastructure European Data Format (EDF) and relational database for data storage [
38]. However, Cloudwave focuses on the EEG/ECG visualization and data storage model, it does not involve the algorithms in data analysis.
For data management and storage, Hadoop [
39], typically as a distributed storage and parallel instance, is an open source, scalable distributed computing, and storage platform. NeuroPigPen is a scalable toolkit to manage large volumes of electrophysiological signals data by Apache Pig and Apache Hadoop [
40]. Ngu and Huh proposed a Hadoop framework-based paralleled B+−tree system to deal with the management of big data [
41]. In the aspect of big data analysis, the analysis of big data can extract meaningful information and even mine the hidden information in big data [
42]. It is also worth further research on how to process with big data quickly and effectively.
1.4. Challenges
Two key challenges in plant electrical signals research are sharing experimental data and scientific computing. In the big data era, for data-intensive scientific discovery, scientific research areas require efficient analysis methods and storage capabilities to cope with large-scale data [
43]. Using the large-scale datasets and analysis tools, researchers can interpret data from various perspectives, which may verify the hypothesis or make a new scientific discovery. However, ongoing massive datasets from different labs may use various recording methods with different experimental protocols or procedures. Thus, the data sharing, not only can help guide beginners who study plant electrical signals but also provide a reference for experts in this research field.
The challenges of web-based data management include:
- (1)
How to design reasonable data storage solutions to deal with structured and unstructured plant electrophysiological data, which data size is from tens of KB to several GB? With the rapid increase in data scale, there is a demand to design a reasonable storage schema to achieve rapid query and calculation; a unified data interface allows users to operate data and metadata for the upper application [
44].
- (2)
How to design data visualization and analysis workflow, e.g., versatile classification algorithms, feature extraction methods, and visual methods, to assist researchers to understand the data and reveal new knowledge?
- (3)
How to design a standardized plant electrical data storage model and file format? As shown in
Table 1, the file format, data size, and data storage types of the plant electrical signals data are various. Although human electrophysiological data format can be a reference, there are still many differences in experimental protocols, data scale, and noise processing in plant electrical signals.
- (4)
How to select the efficient computing framework? When the size of a single file or dataset reaches several GB, the efficiency of standalone processing is very low. It is necessary to select an efficient distributed computing framework to realize real-time online calculation of plant electrophysiological data and improve the efficiency of data processing.
Fortunately, Hadoop [
39] is an open source, scalable distributed computing, and storage platform. The main components of it include Hadoop Distributed File System and distributed computing framework MapReduce. Hadoop has good scalability, fault tolerance, and supports for large-scale datasets, but it is not good at high-performance I/O, real-time computing, or iterative computation [
45]. To our knowledge, Spark is a memory-based distributed computing framework, which aims to provide an interactive data analysis and be used for complex multi-pass algorithms (such as iterative analysis). By storing data in memory when calculating, Spark is more than 40 times faster than Hadoop, and can interactively query large-scale datasets at sub-second level [
46]. In addition, there are some other large data processing engines including Microsoft Dryad, Storm, Tez, and Flink [
47]. HBase, an open source, non-relational, distributed database, is part of Hadoop project and runs on the top of HDFS. It supports random, real-time read/write operation on large datasets.
Therefore, we proposed a system architecture and developed a web-based porotype system to support data sharing, online analysis, and visualization for ongoing increasing plant electrophysiological multi-source data. To cope with the increasing of semi-structured, unstructured data scale, a distributed data storage and management solution was implemented using a non-relational (NoSQL) database HBase that runs on Hadoop. Motivated by online analysis, we also designed parallel computing algorithms for large datasets based on Spark, namely parallelization process of the fluorescent images analysis, plant electrical signals feature extraction and classification methods.
The rest of this paper is organized as follows. In
Section 2, the architecture of the proposed system is described;
Section 3 presents the detail of the implementation of the proposed system; In
Section 4, we evaluate the system, including the test of system performance, result analysis, and future work; Finally,
Section 5 is conclusions.
2. System Architecture
Besides storage of plant electrophysiological data, it is also necessary to achieve a web-based real-time online computation. In fact, since the increasing demand for storage and computing from the growing amount of data, there are both computing-intensive and data-intensive tasks in the process of plant electrophysiological data analysis. Therefore, parallel computing or distributed computing is considered. Here, we use the HDFS and HBase database for the multi-source data storage. These datasets can be distributed in multiple computer nodes, making them more suitable for the distributed computing. Furthermore, for the online computing, we choose Spark to conduct the complex computing tasks.
Web-based application analysis requires not only rapid response but also an easily comprehensible friendly visualization, and then it supports easy understanding and in-depth scientific analysis. For small-scale data visualization on the web platform, it is easy to meet real-time requirements. However, when the data is processed on a large scale, the network transmission delay and the load on the browser will increase, even result in a frustrating experience in data visualization. Therefore, the visualization of a large amount of data is a challenge.
In addition, data annotations enable users to mark important information on visualization results. On one hand, it facilitates multi-user communication and sharing research ideas. On the other hand, expert annotations, standardized experimental data enlighten people to develop new analysis methods and are collected for training use.
For these reasons, we proposed a system architecture of the plant electrophysiological data sharing and online computing platform based on Hadoop and Spark for potential large-scale data storage and computing. The plant electrophysiological data sharing architecture is shown in
Figure 1.
The first layer is the data storage layer. Plant electrophysiological data include intracellular and extracellular recording data, MEA data, fluorescence image sequences, patch clamps data, and ion flow data. The size and type of these data are different. Therefore, the original files and experimental metadata are stored in the HDFS, HBase and MySQL database respectively. To facilitate the application of the upper application, a unified data management interface is necessary, including the read, deletion, update and adding operation on experimental data and metadata.
The second layer is the computing layer, which implements complex computation tasks by Spark, including the extraction of plant electrical signals from the fluorescence image sequence, pseudo-color map generation, the waveform extraction of the action potentials and the algorithm of the template matching.
The third layer is the application layer, which is implemented by Struts2, Hibernate, and other libraries. It is used to run relative simple computing tasks, read HDFS, HBase and MySQL data by calling the computing layer task, and interact with a remote client through the HTTP protocol using JSON.
The fourth layer is the user layer. That is the interface layer of the web user, which completes data uploading, management, online computing, analysis and other functions by providing the web access interface.
Figure 2 shows a functional schematic of the web system. In this context, it supports uploading of plant electrical signals and image datasets measured by the four kinds of recording technologies to Hadoop cluster for data storage, management, and analysis. Online visualization of analysis results from the different datasets can be smoothly finished by the friendly web interface. The basic data workflow includes: (1) Users get multi-source plant electrophysiological datasets by three classic technologies in
Table 1. (2) By web browser, users can create and upload datasets by the web interface. The uploaded datasets and metadata will be stored in Hadoop cluster. (3) After datasets creation, users can use our online data visualization and analysis tools to explore and share data.
3. Implementation of The Proposed System
Based on the needs of plant electrophysiological data sharing and analysis platform, we implemented the system prototype. For computing framework, Spark can replace MapReduce in Hadoop to improve efficiency.
Hadoop mainly includes two important parts: HDFS (Hadoop distributed file system) and MapReduce computing framework. Raw data is stored in HDFS. Hadoop computing tasks are assigned by the task scheduler to DataNodes, and then MapReduce completes the data segmentation, mapping, shuffle, reduce steps, and results collection.
HBase, a distributed column-oriented database, runs on the top of HDFS. Compared with other NoSQL Databases, such as Cassandra and MongoDB, HBase is easier to integrate with Hadoop and Spark. In our application, we used HBase to manage the small files. It supports random, real-time read/write operation on large datasets.
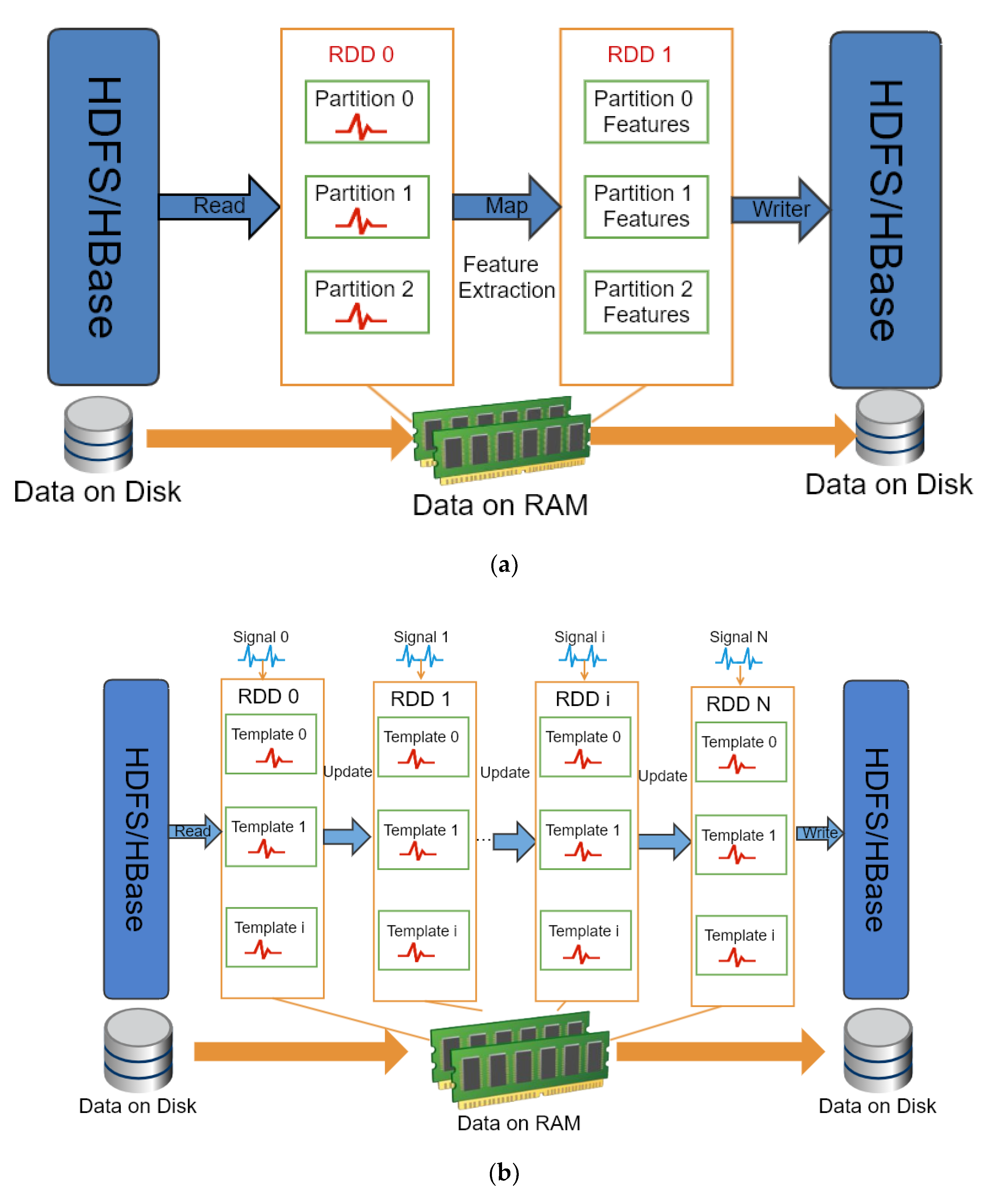
Spark is a memory-based distributed computing framework, which is more efficient than MapReduce and ensures high reliability and scalability at the same time. Both Spark and Flink are efficient big data computing frameworks. They both can integrate with Hadoop platform. The main advantages of Flink are streaming computing, iterative computing and memory management. Considering that our computing is still in the batch mode and Spark is more mature, Spark is a suitable candidate for parallel processing image and text data in our system. Spark uses the Resilient Distributed Datasets (RDD) as the core data units for transformation operation and computation in memory when memory is sufficient. However, MapReduce reads the hard disk data by the split and partition, and the intermediate results of the calculation are written into a disk. Spark organizes tasks with directed acyclic graphs. The RDD can generate new RDDs, and each RDD can perform the corresponding computational tasks. It is suitable for efficient iterative computation, interactive query and stream processing. Therefore, Spark is more suitable for iterative computation compared to MapReduce. After Spark 2.0, Spark also provides the Dataframe and Dataset API, both of which are the high-level API for processing structured data. In our system, we used RDD to process the unstructured data, such as image data and other plant electrical signals data. It can operate data in the low-level API to achieve more flexible data processing.
3.1. Data Storage
3.1.1. Metadata
Metadata is the data about data, which is used to describe the characteristics and attributes of other data for experimental protocol, data interpretation, management, preservation, retrieval and sharing. In different scenarios, the metadata has completely different meanings.
In this work, there are two types of metadata, i.e., the file metadata of the distributed file system HDFS and the plant electrophysiological metadata. HDFS metadata is used to describe the file and directory information of the HDFS file system and data block information. The plant electrophysiological metadata belongs to the scientific data metadata, which is mainly used to describe the experimental object, the experimental conditions, the stimulus way, the experimental data, the experimental target and the record information. The electrophysiological metadata of plants are shown in
Table 3.
(1) File Metadata
The Hadoop Distributed File System (HDFS) includes a single NameNode and Secondary NameNode, and a number of DataNodes. NameNode manages the namespace of the entire file system, and provides metadata information for the user to access files, the byte size of the metadata of a file is 224 + 2 × length of filename, the byte size of the directory is 264 + 2 × length of filename, and its each piece (includes all copy blocks) of metadata requires 152 + 72 × the number of copies. DataNode is used to store the files in the form of data block. In Hadoop1.x, the default data block size is 64 MB, which is the smallest storage unit for NameNode management. Files are divided into the data blocks according to the setting, and the data blocks and backups will be distributed evenly in the cluster. Too many small files can overburden the metadata of NameNode. HDFS is more suitable for large files storage. HBase can more efficiently manage and store the small files.
HBase, a distributed column-oriented database, runs on the top of HDFS. Therefore, it could be used as a direct input/output source of MapReduce, combining with Hadoop seamlessly [
48]. Regions are the basic elements of availability and distribution for a table. Each table in HBase can be split into multiple regions by row. As a region reaches the size threshold, it will split again. These different regions will be stored in multiple Region Servers by load balancing. In each region, data is organized into multiple Stores. A Store corresponds to a Column Family for a given region in a table. For small files, HBase can merge them into Stores. Hence, HBase has a more efficient way to store large amount of small files.
(2) Experimental Metadata
Metadata is important for reproducible experiments and the reliability and applicability of experimental data. The experimental data without metadata is almost meaningless for the researchers.
Therefore, different metadata information is required to describe different plant electrical signals recording methods and experimental protocols. Considering the characteristics of plant electrophysiological experiments, we established a two-level metadata description. The first level is the dataset metadata, and the second level is the experimental metadata of the respective record file. The information about the dataset and three kinds of plant electrophysiological methods are shown in
Table 3.
Here, we use MySQL to store the two levels of dataset and experiment metadata as well as annotation information. Enhanced entity relationship (EER) design diagram is shown in (
Supplementary Material Figure S1). In web applications, users often read a small number of experiment information (often one row or several rows from MySQL table) in each operation. Hence, it is more suitable to store experiment metadata in MySQL than HBase. However, as the data scale increases, we will also consider integrating key features with raw data in HBase to support data analysis tasks at scale.
3.1.2. Storage Model and Retrieval Method
(1) Storage Model
In this study, we use a hybrid data storage model for online applications, as shown in
Figure 3. Plant electrophysiological data and their metadata are separated. Semi-structured and unstructured plant electrophysiological data are stored in HDFS and HBase. Key features of raw data will also integrate with raw data in HBase, and plant electrophysiological metadata is stored in MySQL. The main file formats in the current system are shown in
Table 4. The intracellular and extracellular recording and the MEA recording data have different binary file formats according to different data acquisition software. To facilitate the experimental analysis, the raw plant electrical signals data is converted to a commonly readable CSV format for reading and analysis. CSV files will be stored in HDFS or HBase in a compressed format. Spark or MapReduce can read these CSV files in an efficient way for computing. For optical recording, the raw TIFF format images need to be converted to JPEG or PNG files for visualization in a browser.
When user creates a dataset, system will save the experimental metadata of dataset into the MySQL, which will auto-generate a unique ID for the dataset. Next, when user uploads files to the dataset, the unique ID combines with the filename to generate the unique RowKey for each file in the HBase. Then, save the file into HBase. The unique ID can be used to search all the files belong to the special dataset in HBase.
Large files are defined as data size larger than 64MB, such as MEA files, are stored directly in HDFS. However, for the plant electrical signals recording, there are a large number of small files (the file size is less than the size of the HDFS data block). Therefore, HBase is used to store small files. In the addition, the extracted features are also stored in the HBase for the batch analysis tasks [
49].
(2) Retrieve
When the dataset is created, users can retrieve it by the dataset metadata, e.g., the dataset name, creator, creation time, and even whether dataset is open to the public.
Retrieval function is implemented through the back-end of system, and fuzzy query is performed by calling MySQL database. For the dataset query, the main SQL statements are as follows:
Select datasetid, name from Dataset where name like ‘%$var_1$%’ and Author like ‘$%var_2$%’ and createtime like ‘%$var_3$%’ and open like ‘%$var_4$%’ and species like ‘%$var_5$%’ and recordMethod like ‘%$var_6$%’ and stimulation like ‘%$var_7$%’ and sigtype like ‘%$var_4$%’.
3.2. Plant Electrical Data Online Analysis Method
3.2.1. Web-Based Electrical Signals Extracting Method for Fluorescence Images
Plant electrical signals can be obtained by optical mapping methods in a non-contact method. Optical mapping technology can obtain plant fluorescence image sequence in a certain period. The plant electrical signals can be extracted through the image processing. The previous work of our research group was based on the local desktop system conditions. The detail of the extraction algorithm [
18] of plant electrical signals in a single area of the image is shown in the
Supplementary Materials Method 1.
To carry out the online computing of a large amount of data, the extraction process of the plant electrical signals includes following two steps: (1) obtaining the time series from fluorescence images using voltage-sensitive dye; (2) fitting calculation is carried out to correct bleaching. In other words, the main calculation includes gray value extraction and curve fitting processing. In addition, besides the calculation of a single area, the entire image can be divided equally and calculated the gray value of each small area, which can obtain a number of the fluorescence time curve from series of continuous fluorescence images, and then get the corresponding plant electrical signals by fitting each fluorescence time curve. Single and multi-ROI of plant electrical signals extraction methods can be done through the distributed computing. Since the image time series is stored in HBase, distributed computing can be implemented using MapReduce. For enhancing the speed of calculation, we used Spark. The detail of plant electrical signals extraction for images datasets based on Spark is showed in Algorithm 1:
| Algorithm 1: Parallel plant electrical signals extraction for fluorescence images datasets based on Spark |
Spark Master:Set parameters (cores number, memory and others) of Spark context. Create the SparkContext object SC. Calling the newAPIHadoopRDD method of SC, and read images from the HBase
Spark Slaves (Parallel)- 3.
For each slave node, read image from HBase, and convert it to PairRDD<key = rowKey, value = the byte array of image> - 4.
If single-region analysis: - 5.
compute the average gray value in the user specified region - 6.
if multi-ROI analysis: - 7.
For each region, compute its average gray value. - 8.
Transform to PairRDD<key=rowKey, value= the average gray value (single value or array)>
Spark Master:- 9.
Collect all results to local machine by calling collect() method of RDD. - 10.
if single-region analysis: - 11.
transform average signals to double array. - 12.
if multi-ROI analysis: - 13.
make a transposition for the signals matrix. - 14.
Call the parallel method to transform signals transposition to RDD
Spark Slaves(Parallel):- 15.
For each slave node. read each signal. - 16.
if single-region analysis: - 17.
Based on the parameters, fitting the signals RF(t) by the exponent functions to get fitting signals F(t). Then minus the F(t) by RF(t), we can get the electrical signals S(t) = RF(t) − F(t). - 18.
if multi-ROI analysis: - 19.
For each region, fitting the signals RF(t) by the exponent functions to get fitting signals F(t). Then minus the F(t) by RF(t), we can get the electrical signals S(t) = RF(t) − F(t).
Spark Master:- 20.
Use collect () method to collect electrical signals from each slave node. Save electrical signals in HBase.
|
Figure 4 indicates the extraction process of the electrical signals from plant fluorescence image based on MapReduce. In Mapper1, it reads each image, and obtains time series by calculating the gray scale of the corresponding area of each image, and then stores them directly to the disk after sorting. The second stage Mapper2, when the user inputs the fitting parameters, each fluorescence time series is smoothed and fitted respectively, the fitting curve and fitting parameters are restored in the disk after sorting. The results of the calculation are cached to the web server as a copy for same calculation request. If it exists, the results in the cache are read and sent to the client to reduce the burden of the server and improve the efficiency of the response.
To improve the computing performance, we used Spark to extract the electrical signals from plant fluorescence image series. After extracting the gray value of each image, it was necessary to transpose the fluorescence data matrix to obtain the fluorescence time series of different areas. The extraction process of the electrical signals of the plant fluorescence images based on Spark is shown in
Figure 5.
3.2.2. Online Feature Extraction and Classification of Plant Electrical Signals
In order to analyze the basic characteristics of plant electrical signals, feature extraction functions are integrated on the web page for obtaining 12 features of plant electrical signals, i.e., waveform duration, magnitude, waveform rise slope, falling slope, area, mean, standard deviation, slope, skewness, Hjorth activity, Hjorth mobility, Hjorth complexity. The skewness feature can represent the left or right skewness of waveform shape. Kurtosis is used for indicating the length of “tail” in waveform. Hjorth parameters (activity, mobility and complexity) represent the variance, mean frequency, and change of frequency of the signal respectively. A detailed description of these features can be found in our published desktop version of the research work [
16,
18].
To reduce the load on the web server, the calculation of these 12 parameters was implemented on the web browser side via JavaScript scripts.
Waveform extraction algorithm is described in our previous work [
18], the computational complexity of a single electrical signals is O(N2). Therefore, a small amount of electrical signals waveform extraction can be executed directly on the web server. However, distributed computing is used when the electrical signals data is on a large scale.
Using Spark to process the electrical signals extraction and classification are shown in the
Figure 6a. Firstly, reading signals in each channel by SparkContext. Then get the derivative for each signal. For each differenced signal, search all possible peak or valley positions as well as their start and end position. The detail of algorithm is showed in Algorithm 2. After that, the results are stored in HDFS. The web server reads the calculation results and transfers them to the web browser to display each possible AP waveform from start and end position in the form of annotations. As shown in
Figure 6b, on-line template matching algorithm is applied for classification. The detail of algorithm is showed in Algorithm 3.
Spark provides some functionalities, e.g., SparkContext, PairRDD, newAPIHadoopRDD and collect. The SparkContext object is the entry point of Spark program. The SparkContext is used to connect to Spark cluster, create RDD and other operations. The newAPIHadoopRDD is a method of SparkContext object that is used by Spark to read HBase data and convert it to RDD object. The PairRDD is a key-value pair of RDD object. The API of collect is a type of RDD operation that converts RDD into List object in Java.
| Algorithm 2: Parallel extraction of all AP-like signals based on Spark |
Spark Master:Set parameters (CPU, memory and others) of Spark context. Create the SparkContext object SC. Calling the newAPIHadoopRDD method of SC, read CSV file from the HBase
Spark Slave(Parallel):- 3.
For each slave node, read each signal from HBase transform to PairRDD<key = filename, value = raw signal>. - 4.
then apply the waveform extraction algorithm to each signal. - 5.
all extracted AP-like signals will be saved into a new PairRDD <key = filename + channel name, value = AP-like signal>
Spark Master:- 6.
Use collect() method to collect all AP-like signals from each slave node. Then save them into HBase.
|
| Algorithm 3: Parallel classification of AP signals based on Spark |
Spark Master:Set parameters (CPU, memory and others) of Spark context. Create the SparkContext object SC. By Calling the newAPIHadoopRDD method of SC, read template signals from the HBase transform it to a PairRDD<key = template number, value = the template signals >as a broadcast variable.
Spark Slave(Parallel):- 3.
Read each AP-like signal from HBase as PairRDD<key = filename + channel name, value = AP-like signal>. - 4.
For each AP-like signal compare with each template get the highest similarity coefficient, new PairRDD<key = template number, value = the highest similarity coefficient>. - 5.
if the highest similarity coefficient is higher than 0.91. - 6.
the AP-like waveform is a AP. - 7.
if the highest similarity coefficient is low than 0.91. - 8.
the AP-like waveform is not a AP.
Spark Master:- 9.
Use collect() method the collect all the result. add the new AP to the templates in HBase.
|
3.2.3. The Visualization of Plant Electrical Signals
(1) The Visualization of Time Series
We can store intracellular and extracellular recording electrical signals and MEA recording electrical signals in HDFS and HBase database. Next, in order to facilitate the analysis of electrical signals, we provide the visualization function of the web-based plant electrical signals. For avoiding delay problem caused by reading a large amount of data to the client browser, the data are segmented and then transferred to the client browser. The main visualization process flow of the algorithm is shown in
Supplementary Materials Method 2.
Figure 7 shows the data transfer between the web browser and the web server. Browser fetches new data continuously from the web server according to the requests of the user.
(2) The Visualization of Temporal and Spatial Distribution of Plant Electrical Signals
The temporal and spatial visualization of plant electrical signals mainly refers to the temporal and spatial visualization of optical mapping data or MEA data. It’s used to discover and understand the changes in the electrophysiological signals. The spatial-temporal dataset contains time and location information. Besides time and location information, other attributes are also included such as the amplitude information of the plant electrical signals. For different datasets, there are different in accuracy of time and location information. For example, generally, the sampling interval of the optical mapping data is 0.2 s or 1 s, and the time accuracy of the MEA data is 0.05 ms. And the different objective lens resolution determines the size of the fluorescence image field, and the size of the selected image area can also be set. The MEA electrode recording area in the 64 channels with MED64 is usually fixed at 3.55 mm by 3.55 mm. Position and time scale are factors that directly affect visualization.
This visualization method requires to read the time series of all areas once and then display on a fluorescent image. When the sampling rate of the time series is low, the size of the displayed data is small and the browser load is light to render easily in a short time and the data transmit in a fast speed. However, when the time series sampling rate is very high, such as MEA data, the dataset is large, and then the sampling rate must be reduced in the background, only a small amount of time series are sent to the client for the visualization.
In the visualization, the plant electrical signals need to be drawn. Generally, the number of time points of optical mapping are less than 1000; but the sampling rate of MEA recording data is very high, which results in a sharp increase in the number of time points. Therefore, in order to save the drawing time, the sampling rate needs to be reduced.
3.3. Data Access and Download
User can download their own datasets (private and public) and all the public datasets. (1) The logged-in user can search and edit their own datasets and download the Zip files of the datasets in the “Dashboard” page. (2) Each user can download the public datasets in the “Public Dataset” page.
4. Results and Discussion
4.1. Data
The optical recording and MEA recording data are from sunflower at 2–3 weeks, and the details of the method have been published in the literature [
14,
15]. The original format of fluorescence image of optical recording is TIFF format. The electrical signals induced by electrical stimulation are from cucumber at 3–4 weeks. The detailed recording method can refer to our previous work [
18] and the format of the original file is CSV. All types of electrophysiological data are stored in HDFS and HBase.
4.2. Web-Based Plant Electrical Signals Analysis and Visualization
4.2.1. Plant Electrical Signals Extraction and Visualization based Fluorescence Images
In
Figure 8, the four figures show the results of extracting plant electrical signals from multiple regions in fluorescence images. The figure on the top-left is the average gray value of selecting ROIs from the time-series images in a given time range. The top-right figure shows the smoothed gray curves. Users can set the smooth parameter for the moving average filter algorithm. The bottom-left figure is the gray curves after fitting with three functions. The bottom-right figure is the final plant electrical signals curves.
Dygraph.js is used to visualize the signals curves. Multiple curves can be displayed simultaneously in the same axis. The value of each data point can be viewed when moving the mouse on it, and the figures can be zoomed in and out.
Furthermore,
Figure 9 is a spatiotemporal visualization of the plant electrical signals distribution on the fluorescence image. Each fluorescence image equally divided into 20 × 27 regions. Users can set the region size for different datasets. Each region in the image has a value for the plant electrical signals extracted from the time-series images. Next, the plant electrical signals variation in each region can be observed. When the mouse is moved to the curve, the corresponding amplitude will be displayed.
4.2.2. Visualization of Multi-Channel Plant Electrical Signals
Figure 10a shows the visualization of the 4-channel plant electrical signal. It’s implemented by Dygraph.js. By setting each axis size and grid color, multiple Dygraph diagrams are generated automatically according to the number of channels. Users can set the size of each view. For quick display, the sampling rate for the raw data is set to 1 Hz, which reduces the time to read, transmit and display.
Figure 10b displays 4-channel plant electrical signals in a view that can be zoomed in and out and display the signals value.
Data annotations are important for describing the experimental data and experimental procedures in detail. Commonly, the comments of expert users are more valuable. Thus, allowing users to add comments can assist the establishment of standardized datasets and the primary users to better understand the experimental data. As shown in
Figure 11, users can add annotations directly to the plant electrical signals curves. They can delete and save the annotations too. Here, we provide an annotation function, which allows users to add annotations to the plant electrical signals curves directly to indicate the start time of the stimulation artifacts, the start and end position of the AP and other information (
Figure 12).
4.3. Web-Based Feature Extraction and Classification of Plant Electrical Signals
Figure 12 shows the feature extraction web page. Firstly, user can select the start position (0 in the box in the figure) and the end position (1 in the box in the figure) of the plant electrical signals and then click the feature calculation button in the figure to calculate the 12 feature parameters.
In order to provide automatic waveform recognition, the waveform extraction and template matching algorithm are implemented on the web platform for extracting the induced AP-like waveforms from the original signals with artifacts and background noise and determining whether the waveform is AP waveform [
18]. The waveform extraction algorithm running on Spark can extract AP waveforms from 272 signals in 17 s, and a total of 357 possible AP waveforms are extracted.
After extracting the AP-like waveforms of the plant electrical signals, it is still necessary to determine whether it is an AP waveform. Our system integrates the template-matching algorithm using the template library stored in HBase. In brief, we initially selected eight standard action potential signals as templates. Next, each AP-like waveform was compared with the templates in
Figure 13 to calculate the similarity.
Figure 14 displays typical examples of observed waveforms. The similarity-based algorithm allows us to determine the classification and update the template library. The details of algorithm were described in our previous works [
18]. In the present work, we focused on spark-based parallel computing for the previous classification algorithm running on a desktop-based system.
The dataset is the same as our previous work [
18]. We tested 329 waveforms including 96 non-AP waveforms and 233 AP waveforms with template matching algorithm. In this work, we compared different thresholds and selected the threshold which showed the best performance. Here, Classification_Threshold is the value of similarity for identifying an AP-like waveform and deciding whether it is added as a new one to the template library. Moreover, Update_Threshold is the value of similarity for deciding whether an AP-like waveform merges with the old template to update the template. Using this proposed method, the 329 waveforms were identified and classified. However, the selection of Classification_Threshold is still prior knowledge-based. For example, if setting of Classification_Threshold is too high to few new templates added in the library, even leading to a low accuracy. When the Classification_Threshold = 0.91 and Update_threshold = 0.95, the classification accuracy is highest and up to 96%. Although the template library can be updated using template-matching algorithm, parallel template matching is not achieved completely through the Spark. When updating the template, the classification accuracy could up to 96%. Without updating of templates, the accuracy of the classification of plant electrical signals is only 89%. Each signal should be processed in serially because of the update of the template. In order to solve the parallelization of the template-matching algorithm, we can consider the multiple iterations, which updates part of the templates in each iteration and finally makes the classification accuracy and calculation time to achieve a better balance.
4.4. System Stress Test
By using Jmeter 3.1 to run the stress test for the system, we set 50, 100, 200, 400, 800, and 1000 users respectively to test the system. Through HTTP requests, it can test the various aspects performance of system, e.g., database I/O rate, waveform extraction rate, template-matching rate. Each user runs for the five iterations. The configuration parameters of test environment are shown in
Table 5, and the test results are shown in
Table 6.
Num: Numbers of user.
Sample: Total number of requests.
Average: Average response time. Unit: ms.
Median: median value of response time, Unit: ms.
90% Line: 90 percent requests’ response time less than this value, Unit: ms.
Min: Minimal response time, Unit: ms.
Max: Maximal response time, Unit: ms.
Throughput: Request number per unit time.
Received: The amount of data received from the server per unit of time, Unit: KB/s.
Send: The amount of data sent per unit of time, Unit: KB/S
Multiple threads are used to simulate the concurrent connections of users in Jmeter. i.e., the number of threads represents number of users. In general, the 90% response time of users is longer than 2 s when the number of concurrent users is 400, the user experience is bad. Especially when the number of users is up to 800 and 1000, the most response time is longer than 10 s. Taking into account all indices, i.e., the average response time, the median response time, 90% Line, when the number of concurrent users is approximately 100, the user has a good experience. However, when the number of concurrent is 200 or 400, the user experience becomes bad, some requests will have a high delay. When the number of concurrent is up to 800, the request response time becomes longer, and performance is degradative. By analyzing the single HTTP request, it is found that the request time for database read, waveform extraction, and template matching has a significant increase relative to other requests when the load increases. To improve the performance of the system in the future, waveform extraction algorithm and template matching algorithm can be optimized to reduce the time complexity of the algorithm and the query in the database can be speeded up by using the extended index. Moreover, the technology of load balancing and increasing the number of servers will reduce the pressure on the single server. The database can also use read/write splitting technology for performance improvement.
4.5. Spark Parallel Performance Test
We use three machines (Name node: the machine with Intel® Core™ i5-2300 CPU @ 2.80 GHz × 4 and 16 G RAM, Data nodes: two machines with Intel® Core™ i5-4590 CPU @ 3.30 GHz × 4 and 7.9 GB RAM) to setup the Spark-Hadoop cluster. Each machine has 4 cores.
By comparing the computational performance of Spark, MapReduce and serial programming, the computation time of Spark is significantly less than MapReduce and serial programming. The results are as follows:
As shown in
Figure 15, when the dataset size increases, the efficiency of Spark has a great advantage than MapReduce and serial programming. The computation time of Spark is less than that of MapReduce and serial program in the data size of 550M, 826M and 1034M.
For the different sizes of datasets, we controlled the degree of parallelism by the number of cores and test the performance of Spark. As shown in
Figure 16, when the size of datasets is small, we can see the increase of the number of cores couldn’t improve the performance, even make the performance down slightly, such as the datasets of Dataset_168M and Dataset_550M. When the size of datasets becomes bigger, the increase of the number of cores would get remarkable improvement in the performance, such as the dataset of Dataset-826M and Dataset-1034M. Spark is more suitable for the online analysis and computing of the large dataset of plant electrical signals [
50].
4.6. Discussion
PlantES, a plant electrophysiological multi-source data sharing prototype system, realized the visualization for many plant electrophysiological data (extracellular recording data, plant fluorescence image series data, MEA data with typical size of several GB) and integrated various data analysis algorithms for the online analysis and visualization.
Compared with other electrophysiological platforms for medical application, our system integrated with many signal analysis algorithms, such as signal classification, feature extraction, and fluorescence image electrical signals extraction, which are convenient for researchers’ online analysis and further reduce the user threshold. In addition, we added the interactivity to the data visualization. Although plant electrical signals are similar to animal electrophysiology, the distinct difference of plant cell and animal cell makes researchers usually have to use the customization system. In particular, the difference of variety, measuring parts, growth phases, and size leads to the diversity of the plant electrophysiological data formats. To solve the problem, we adopted different storage schemas for different types of data by using HBase and Hadoop to achieve effective data management, storage, and retrieval. For the problem of latency for large-scale data analysis, Spark was used to process the parallel computing tasks. The experiment results demonstrated that Spark was more effective than MapReduce and the improvement of Spark in large-scale data analysis was more pronounced than that in small-scale data analysis. It provided a flexible and scalable solution for the plant electrophysiological data sharing, management and analysis.
We developed the plant electrophysiological data sharing and analysis platform. To adapt to large-scale data analysis, we used the technology stack of Hadoop to read, write and compute the datasets. The parallel computing on Spark cluster enabled online analysis to be efficiently executed. Based on the web platform, the efficient distributed computing and storage architecture were designed to realize the real-time or near real-time online calculation of plant electrophysiological data, which can greatly improve the efficiency of data processing. It also integrated various data analysis algorithms for the online analysis and visualization. Researchers can comfortably visualize, analyze and share plant electrophysiological datasets online by browser.
At the present stage, our research is only an initial trial. In the design process, the existing EEG-ECG electrophysiological platforms have given us a lot of inspiration [
30,
31,
32,
33,
34,
35,
36,
37,
38,
40]. Although the functions of our system are similar to those platforms for medical applications, our system is used to deal with plant electrophysiological data. In future, we will develop a more effective storage format for plant electrophysiological data. For multi-source scientific data, e.g., MEA data, extracellular recordings data, fluorescent images and other data, standardized file format needs to be designed reasonably, and then metadata, data, and annotation data information need to be organized much more properly. As a standardized scientific data storage format, HDF5 is the potential scheme of standardized storage for the plant electrical signals data [
51]. Next, the coupling between the standardized data storage format and the storage platform needs to be implemented through providing a uniform data operation interface for the upper application, designing fast retrieval algorithm of plant electrical signals, constructing new template index to reduce the time of template matching, and building standardized plant electrical signals data storage. In addition, web service interfaces provide external applications to download experimental data and invoke computational programs for data analysis and obtaining analysis results. The scientific workflow of plant electrical signals analysis also should be integrated.
5. Conclusions and Future Works
In this paper, we developed the platform for the online analysis and sharing of the plant electrophysiological multi-source data. It integrated data storage, management, visualization, analysis, and sharing. In summary, the system prototype is as follows.
- (1)
By integrating plant electrical signals extraction, classification algorithm and other methods, our system provided a simple and user-friendly interface for data analysis.
- (2)
The web-based visualization and annotation of plant electrical signals allow users to obtain the intuitive of data, which can communicate information clearly and effectively.
- (3)
We designed a suitable storage schema to adapt multi-source plant electrophysiological big data. HBase and HDFS were integrated to storage the different types of files in plant electrophysiological data respectively.
- (4)
For different online computing tasks in the analysis of plant electrical signals, by using Spark, complex tasks can be parallelized to improve the computing time.
In brief, our proposed system prototype is efficient for sharing and analysis of plant electrical signals online. In the future, more efficient feature selection, retrieval and classification algorithms will be paralleled based Spark. The web service interfaces will provide external applications to download experimental data and call computational programs for data analysis and obtaining analysis results. The scientific workflow of plant electrical signals analysis also will be integrated in the system.