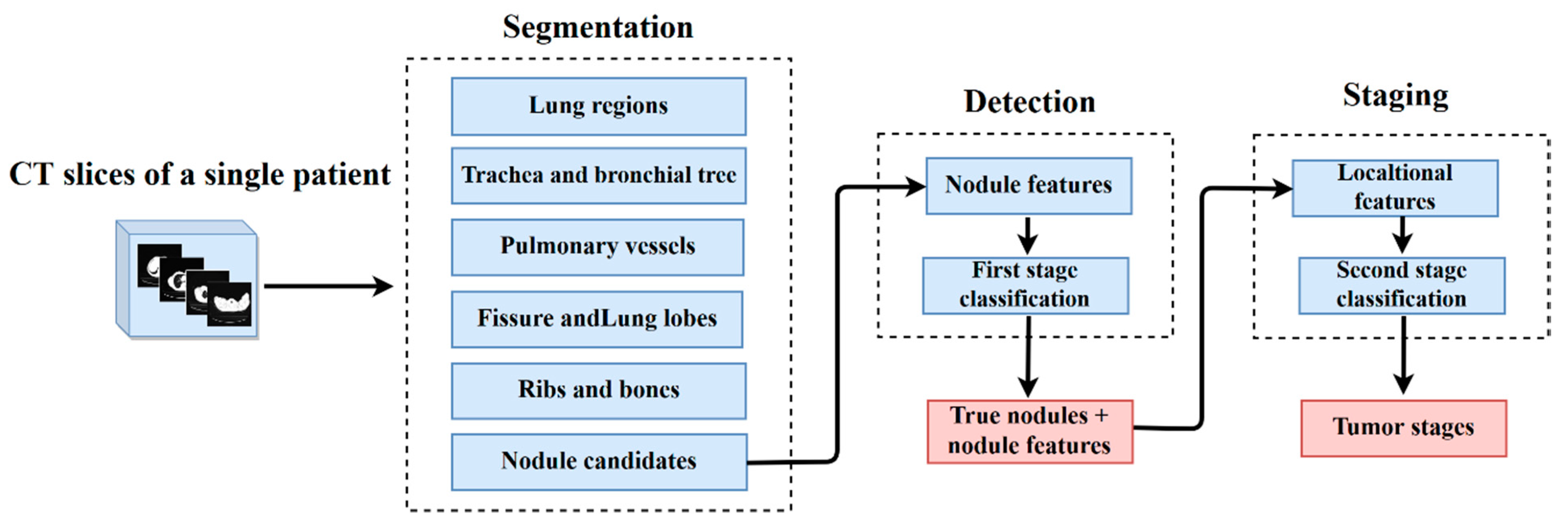
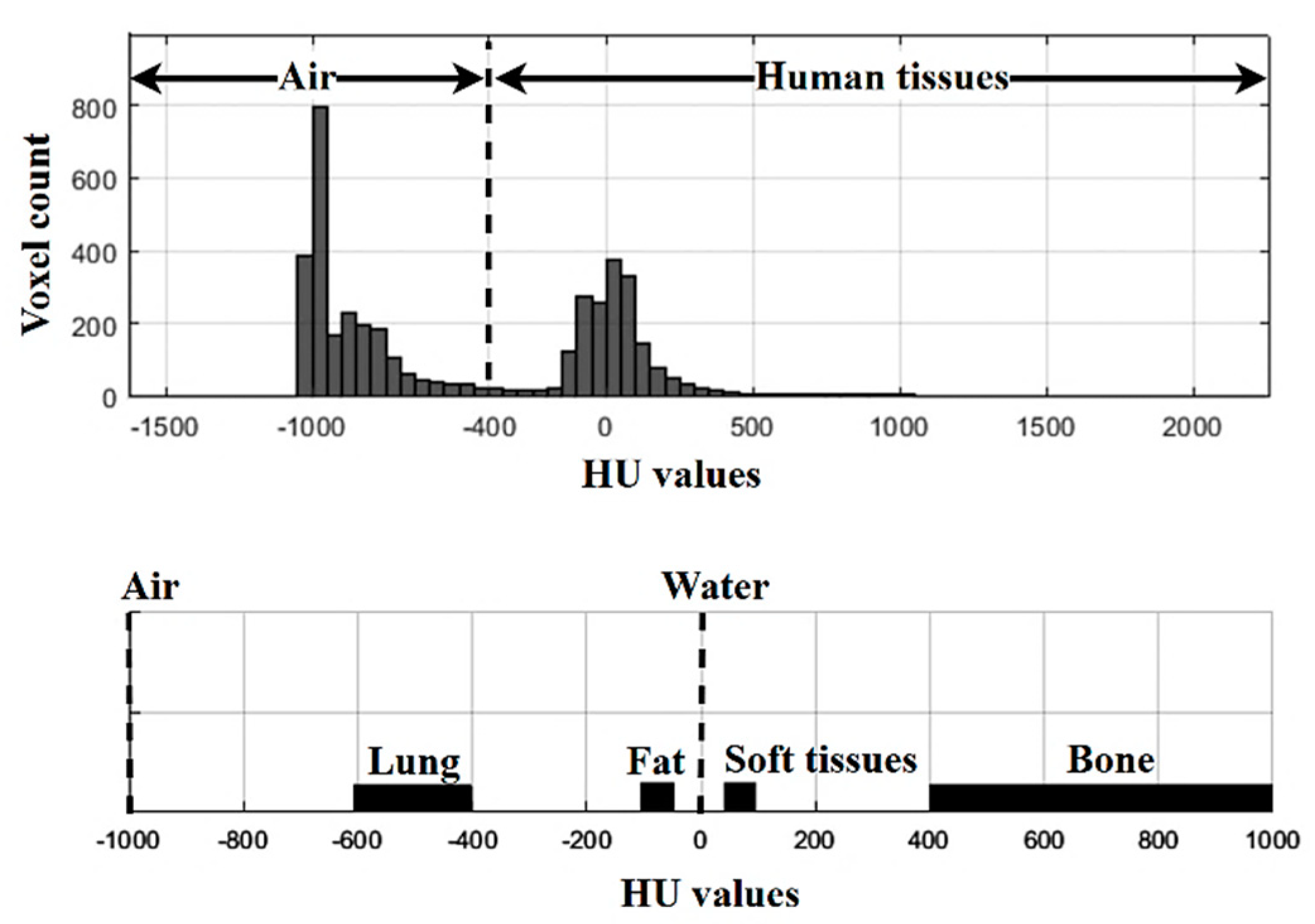
1. Introduction
Lung cancer is one of the most common diseases, and it has the highest morbidity and mortality rates of any cancer worldwide. As reported by global cancer statistics [
1], there were 2.1 million new cases and 1.8 million deaths from lung cancer in 2018. Approximately one in five cases (18.4%) of lung cancer leads to death. In oncology, medical imaging plays a vital role in reducing the mortality rate. Imaging is a non-invasive and painless procedure with the least harmful side effects for patients. It can provide detailed anatomical information about the disease by generating visual representations of tumors. Unlike other invasive methods such as surgeries and biopsies which extract and study a small portion of tumor tissue, imaging can give a more comprehensive view and analysis of the entire tumor region. Moreover, it is preferable for clinical routines that require an iterative analysis of the tumor during treatment [
2].
Computed tomography (CT) is a standard imaging modality for lung cancer diagnosis. The National Lung Screening Trial (NLST) reported that the lung cancer mortality rate of 15–20% could be reduced by performing low-dose CT screening [
3]. CTs display tumors on cross-sectional images of the body called “slices”. These slices can be stacked together to reconstruct a three-dimensional structure which makes possible more comprehensive analysis of tumors. However, the manual interpretation of the CT scans is prohibitively expensive in terms of time and effort. Also, it is subject to the skill and clinical practices of the interpreter; hence, the diagnosis results may suffer from inter- and intra-observer variation [
4]. Due to these inconveniences, any real-world application is necessary for automated interpretations of the CT scans. This fact has strongly motivated computer-aided diagnosis systems (CADs) to become an extensive research area in the field of biomedical engineering. Scientists have proposed a vast number of CADs for lung cancer diagnosis using modern image processing and machine learning techniques. Nonetheless, most of these CADs have focused only on the detection of pulmonary nodules. Nodules can evoke the possibility of lung cancer and appear as round or oval-shaped opacities on the CT scans [
5]. Nonetheless, the detection of nodules does not provide sufficient information about the severity of the disease to make proper treatment decisions. Generally, half of the patients who undergo CT screening present more than one nodule, but not all of these nodules are cancerous [
6]. Physicians usually use the terms “nodule” and “tumor” interchangeably when they are uncertain about the severity of the disease. Indeed, identifying the severity of a tumor nodule is a principal issue for lung cancer diagnosis.
In clinical practice, the severity of lung cancer can be expressed as different stages using the Tumor, Node, and Metastasis (TNM) system [
7]. As the name describes, this system assesses the severity of the disease based on three descriptors. The first descriptor (T) assesses the characteristics of the primary tumor such as its size, local invasion, and the presence of satellite tumor nodules. The CT modality performs T-staging well, because information about the tumor can be easily obtained from the CT scans. The second descriptor (N) assesses the involvement of the regional lymph nodes. On the CT scans, lymph nodes appear as small opacities with unclear silhouettes; thus, the CT modality performs weakly for N-staging [
8,
9]. The last descriptor (M) assesses the invasion of the tumor to the extra-thoracic organs, especially the brain, adrenal glands, liver and bones, and so forth [
9]. Staging of metastatic tumors usually requires additional examinations of the invaded body regions. For this reason, we focus only on the T-staging of tumor nodules in this study. T-stages can be further divided into sub-stages depending on the anatomic characteristics of the primary tumors.
Table 1 describes the detailed definitions of the T-stages, as determined by the eighth edition of the TNM system.
2. Related Works
The remaining problem of existing lung cancer diagnosis systems is that they are capable of detecting tumor nodules only. As there are numerous nodule-like lesions in the lungs, it is still challenging to detect real tumor nodules. Most current CADs thus try to reduce the number of false positives, which can hinder the detection of real nodules. This means that current CADs defer staging assessments. Even though there have been a lot of studies on the computerized detection of tumor nodules, we found a paucity of literature on staging. In 2014, Kulkarni and Panditrao [
10] proposed an automatic tumor staging system from chest CT scans using marker-controlled watershed and Support Vector Machine (SVM). As a pre-processing method, they smoothed the input CT images with a Gabor filter, and then possible tumor nodules from the smoothed CT slices were segmented using marker-controlled watershed. Next, three geometric features—area, perimeter and eccentricity—were extracted and fed into the Support Vector Machine (SVM) for classification. Their method produced diagnosis results in four stages (T1, T2, T3, and T4), and they evaluated 40 CT scans from the National Institute of Health/ National Cancer Institute (NIH/NCI) Lung Image Database Consortium (LIDC) dataset. However, the evaluation assessments were not described in detail.
Similarly, Ignatious et al. (2015) [
11] applied marker-controlled watershed for lung tumor detection and staging. Unlike [
10], they used the sharpening method to enhance the input CT slices and then five features—area, perimeter, eccentricity, convex area and mean intensity—were extracted. They selected the true tumors based on an empirical threshold value of area. After that, staging was conducted by four alternative classifiers: Support Vector Machine (SVM), Naive Bayes Multinomial classifier (NBM), Naive Bayes Tree (NB tree), and Random Tree. Their experiment was validated using 200 CT slices from Regional Cancer Centre Trivandrum, and an accuracy level of 94.4% was obtained by the Random Tree classifier. Nevertheless, tumor segmentation based on the marker-controlled watershed is hard to fully automate, and it has a low guarantee in detecting any shape and size of nodules, because creating internal and external markers for affine variant nodules is an arduous task.
Besides the aforementioned CADs, a multi-stage cancer detection method using watershed and multi-class SVM classifier was also proposed by Alam et al. (2018) [
12]. In their study, the median filter was used for the pre-processing of 500 lung CT images, and then thirteen different image features were extracted from the region of interest (ROI). In the first stage of classification, they try to detect any cancerous tumor nodules from the input CT exam. If there were cancerous tumors, they identified the clinical stages of those tumors in the second stage of classification. Otherwise, they calculated the probability of non-cancerous tumors becoming cancer in the third stage. For the first and second stages, they used the SVM classifier, and for the third stage, they calculated the area ratio (area of the nodule to the area of the entire lung). Their experiment reached accuracy levels of 97% for detection and 87% for staging, respectively. Our proposed CAD which applies two stages of classification for detection and staging is quite similar to that presented in [
12]. However, we differ from them by extracting the locational features of cancerous tumors and using alternative machine learning algorithms for classification. Moreover, we determine the clinical stages of tumors using the eighth edition of the TNM staging method, which is a global, up-to-date standard for cancer staging. In their approach, they classified the stages (initial, middle, and final) roughly; thus, it was not very beneficial for precise treatment.
Recently, deep learning models have made great breakthroughs for machine learning [
13]. Convolutional Neural Networks (CNNs) are the most frequently used deep learning model for the automated detection of lung tumors. However, for staging, CNN has been applied in very few studies. Kirienko et al. (2018) [
14] developed a CNN for T1–T2 or T3–T4 staging of lung tumors. Their staging referenced the seventh edition of the TNM system and tested with 472 FDG-PET/CT scans. Their proposed CNN was constructed using 2D patches that were cropped around the center of the tumor. Additionally, their CNN used two neural networks: (i) one as a feature extractor to extract the most relevant features from a single patch, and (ii) another as a classifier to perform classifications for all slices. They reported a high testing accuracy of 90%, but the outputs were presented as binary numbers (T1–T2 with label = 0 and T3–T4 with label = 1), that is, their method cannot give an accurate result for each specific stage. Moreover, their approach is not fully automated, because their CNN requires manually cropped patches as inputs.
Another tumor staging method using a double convolutional deep neural network (CDNN) was presented in [
15]. They used 73 CT exams from the Image and Data Archive of the University of South Carolina and the Laboratory of Neuro Imaging (LONI) dataset. They divided the input exams into two piles: pile 1 (with cancer) and pile 2 (without cancer). Then, all slices in both piles were further grouped depending on the angles using the K-means algorithm. Subsequently, they built a double CDNN containing double convolution layers and an additional max pooling layer. They reported a prediction accuracy of 99.62%. Nonetheless, their CDNN output only decimal values between 0 (non-cancer) and 1 (cancer); hence, a threshold value was necessary to determine the possibility of tumor stages.
The primary objective of this paper is to propose a CAD that is capable of not only the detection of tumor nodules, but also of their staging. We present two significant contributions in our work; the first is performing double-staged classification, and the second is the extraction of locational features. In the first contribution, we perform two stages of classifications: one for the detection task and another for the staging task. Performing double-staged classification helps to minimize false positives and makes the CAD more accurate and robust. In the second contribution, we extract the locational features to identify the stages of the lung tumor because the severity of lung cancer strongly depends on the sizes and locations of the tumor nodules. Moreover, knowing the exact locations of tumor nodules is implicitly beneficial for the surgical planning of higher staged tumors. The rest of this paper is organized as follow.
Section 3 describes the details of the methods applied in this study.
Section 4 presents the experimental results and performance evaluation of the proposed method. Finally,
Section 5 presents the discussion and conclusions of the paper.
4. Experimental Results
In this study, we propose a CAD that is capable of detecting and staging of the pulmonary tumors for lung cancer diagnosis. This CAD was developed and evaluated using four datasets, namely LIDC-IDRI [
29,
30], NSCLC-Radiomics-Genomics [
2,
31], NSCLC-Radiomics [
2,
32], and NSCLC Radiogenomics [
33,
34] (
Supplementary Materials) where NSCLC stands for non-small cell lung cancer. These datasets are downloadable from The Cancer Imaging Archive (TCIA) [
35] which is an open source of medical images for scientific and educational research. Expert radiologists confirmed all of the tumor nodules in these datasets, and the clinical diagnosis results were also provided. We applied these results as the gold standard to evaluate the performance of proposed CAD.
Table 4 presents detailed descriptions of each dataset used in this study.
As described earlier, our CAD has two primary purposes: detection and staging. The first purpose is more fundamental for the production of accurate diagnosis results because the correctly predicted tumor nodules from the first stage are further applied for staging. Among the datasets mentioned above, the LIDC-IDRI only provides XML files for the true nodule annotations. The clinical stages of the tumor nodules are not available in LIDC-IDRI. Therefore, we applied it only for the detection stage. Moreover, it is a huge dataset and had been employed by several researchers to develop CADs for nodule detection. There are 1010 exams in the LIDC-IDRI dataset, but only 888 exams were applied for our experiment because some exams contain missing and inconsistent slices. The application of LIDC-IDRI for the first purpose, detection, also acted as a separate validation for more robust detection.
Unlike LIDC-IDRI, the three NSCLC datasets can provide the data for clinical stages of tumor. Thus, we applied these three datasets for the second purpose. Indeed, the CT exams from these three datasets had to pass through the first stage classification to segregate the true tumor nodules. Subsequently, the true predicted tumors were categorized into different clinical stages by the second-stage classifier. Therefore, the first stage of classification was double validated by not only LIDC-IDRI, but also NSCLC.
However, the second stage of classification is a multi-classification problem that outputs seven classes. Class 1 represents T1a, class 2 represents T1b, class 3 represents T2a, class 4 represents T2b, class 5 represents T3, class 6 represents T5, and class 7 represents T0/Tis. The NSCLC datasets contain a different number of samples for each class, and the class distribution is profoundly different, as described in
Figure 12. In this figure, it is evident that there were few samples for class 6 (T5) and class 7 (T0/Tis). After removing the missing-class samples, the total number of samples from three NSCLC datasets became 672 and they were used for staging.
Before we started the experiment, we performed the preparation of the data for training and testing. As the data in the real world is fuzzy in nature, we validated our CAD using repeated random trials.
Figure 13 shows the preparation of data for repeated random trials in both first and second stage classifications. For first stage classification, the samples in the LIDC-IDRI dataset (888 samples) were randomly divided into five splits 50%-50%, 60%-40%, 70%-30%, 80%-20% and 90%-10% for training and testing. Similarly, for the second stage classification, samples in the NSCLC datasets (672 samples) were randomly divided into five trials. In each split, we repeatedly selected random samples
times as described in the figure. Since the samples were randomly selected, each iteration
generated different samples for training and testing. We trained and tested five different classifiers namely the Decision Tree (DT), K-nearest neighbor (KNN), Support Vector Machine (SVM), Ensemble Tree (ET), and the Back Propagation Neural Network (BPNN) on these random samples. In each iteration we calculated the performance measures of each classifier then calculated the mean and standard deviation of performance measures for each split.
The performance measurements which were calculated for each classifier are accuracy, sensitivity, specificity, precision, and the F-score. They can be computed by:
Nonetheless, the second stage of classification is a multi-classification problem unlike the first; it is therefore more complex to evaluate the performance. To solve this problem, we used the “one versus the rest method”, which calculates the performance of the classifier for an individual class and then calculates the overall performance. For example,
is an individual class of a multi-classification problem, and
is the total number of classes. From the counts of
, the assessments
,
,
and
were obtained. Then, the overall performance of the classifier for all classes was calculated by:
In addition to these measurements, we also take into account the area under the curve (AUC) and execution time of each classifier for performance evaluation.
Table 5 shows the mean and standard deviation of performance measurements obtained from each classifier in the first stage classification.
Similarly,
Table 6 describes the comparison of performance measurements generated by the five different classifiers for the second stage classification.
For graphical representation, box-plot distributions were applied to compare the performance of five alternative classifiers.
Figure 14 illustrates the box-plots comparison between mean performance measurements generated by each classifier in first stage classification. Similarly,
Figure 15 shows the box-plots comparison for second stage classification. Analyzing the distributions of box-plots, we can know that BPNN offers significantly higher performance than other classifiers in first stage classification. However, for the second stage classification, the performance of classifiers declined compared to the first stage because of class imbalance in the samples. The performance of DT, KNN and SVM decreased, and it indicates that they are not robust enough for random data with class imbalance. On the other hand, the ET and BPNN are robust for random data. The performance of ET seemed better than BPNN especially in specificity and precision because its box-plots had upper middle lines (higher median values) and small range of whiskers (condense data). But in other assessments such as accuracy, sensitivity, F1-score and AUC, BPNN showed higher values. Therefore, we decided to use BPNN for both stages of classification. As a result, BPNN reached an accuracy of 92.8%, a sensitivity of 93.6%, a specificity of 91.8%, a precision of 91.8%, an F1-score of 92.8% and an AUC of 96.8% for detection. For staging, it achieved an accuracy of 90.6%, a sensitivity of 77.4%, a specificity of 97.4%, a precision of 93.8%, an F1-score of 79.6% and an AUC of 84.6% respectively. Even though BPNN took more execution time, it can guarantee random and imbalanced samples.
As an example, the outputs of our proposed CAD are illustrated in
Figure 16. They demonstrate 3D visualizations of tumors of different stages.
Figure 16a represents a stage T1a tumor that is 1.83 cm in its greatest dimension and is located in the left lower lobe in an isolated manner.
Figure 16b demonstrate a stage T1b tumor which is also located in the left lower lob, but is bigger in size (2.14 cm) than the tumor in
Figure 16a.
Then,
Figure 16c,d represents stage T2a and T2b tumors, respectively. Both tumors are located in the right upper lobes. The tumor in
Figure 16c is 3.05 cm in its greatest dimension and is isolated, whereas the tumor in
Figure 16d is 2.10 cm and touches the visceral pleural. Hence, the tumor in
Figure 16d has a higher stage even though is smaller in size.
Figure 16e,f illustrates the stage T3 tumors. The tumor in
Figure 16e is located in the right upper lobe and invades the chest wall. Also,
Figure 16f demonstrates the presence of satellite nodules in the same lobe, the right upper lobe. Unlike
Figure 16f, the tumors in
Figure 16g contain satellite nodules in different lobes: one is in the right middle lobe, and another is in the left upper lobe. Thus, it is classified as stage T4. Finally, in
Figure 16h, we can see a T4-staged tumor nodule which invades the mediastinum.
Moreover, we compared the performance of our proposed CAD with methods presented in previous works, as stated in
Table 7. It was difficult to make a fair comparison because the CADs were developed using different datasets, different numbers of samples, and different methods. Thus, we could only make a quantitative comparison of the performance outcomes. Based on these quantitative comparative results, it is evident that our proposed CAD is validated using not only higher amount, but also random samples, and it can provide preferable performance in both detection and staging.
5. Discussion and Conclusions
This paper presents a CAD capable of the detection and staging of lung tumors from computed tomography (CT) scans. Our CAD has two major contributions. The first contribution is the extraction of locational features from tumor nodules for the T-staging of lung cancer. The second is the double use of the BPNN classifier for the detection and staging of the tumor nodules.
The CAD works in three pivotal phases called segmentation, detection, and staging. A single CT exam of a patient containing multiple 2D slices is an input of the proposed CAD. From each input exam, the lung anatomical structures, such as the lung regions, trachea, bronchial airway tree, pulmonary vessels, fissures, lobes, and nodule candidates, are initially segmented using gray-level thresholding and isotropic interpolation methods. Subsequently, 28 nodule features are extracted from the segmented nodule candidates to detect the true tumor nodules. These features include four geometric, five intensity, and nineteen texture features. Using these features, the first stage of classification is performed and the true tumor nodules are segregated.
Once the true tumor nodules have been achieved, the locational features are added for staging. With the help of the segmented lung anatomical structures from the first phase, the locational features can be defined by dividing the lungs into different zones. Moreover, the invasion and attachment of the tumor nodules to other pulmonary organs are also checked by using the 3D quick hull algorithm and logical array checking. As well as this, the number of true tumor nodules predicted in a single test and their lobe locations are also extracted in this stage. Using these features, the staging of tumor nodules is conducted by second-stage classification. In this study, we applied the BPNN for both classifications. Before selecting the BPNN, we tested and compared it with five alternative classifiers using repeated random trials. Based on the comparative results, we determined that the BPNN should be used due to its high performance and robustness.
Our proposed CAD was developed and tested using 1560 CT exams from four popular public datasets: LIDC-IDRI, NSCLC-Radiomics-Genomics, NSCLC-Radiomics and NSCLC Radiogenomics. In the performance evaluation, our proposed CAD achieved a detection accuracy of 92.8%, a sensitivity of 93.6%, a specificity of 91.8%, a precision of 91.8%, an F-score of 92.8%, and an AUC of 96.8%. For the staging, we achieved an accuracy of 90.6%, a sensitivity of 77.4%, a specificity of 97.4%, a precision of 93.8%, an F1-score of 79.6% and an AUC of 84.6% respectively. Compared with previous works, our CAD was evaluated with more samples and obtained auspicious performances.
Unlike most existing CADs for lung cancer diagnosis, our CAD makes possible not only the detection, but also the staging of lung tumor nodules. Our first contribution, the extraction of locational features, has advantages for the staging of tumor nodules, because the severity of lung cancer is strongly associated with the size and location of tumor nodules. Knowing the exact location of tumors reflects accurate staging. Patient survival rates can also be increased by providing precise treatment options depending on the tumor stage. Moreover, locational features produced by our CAD will be useful for the clinical surgery planning of seriously staged tumors. The next contribution, the double stage of classification using BPNN, makes the CDA more robust. As the first stage classification filters only the true tumor nodules, we do not need to extract the locational features from all nodule candidates; this saves both computational effort and time. Nonetheless, our study has some limitations in terms of the need for background knowledge for anatomical lung structures. Moreover, our CAD only focuses on the T-staging of lung cancer. Thus, there are some open issues which can be further upgraded for the N and M-staging of lung cancer.