Dual Aptamer-Functionalized 3D Plasmonic Metamolecule for Thrombin Sensing
Abstract
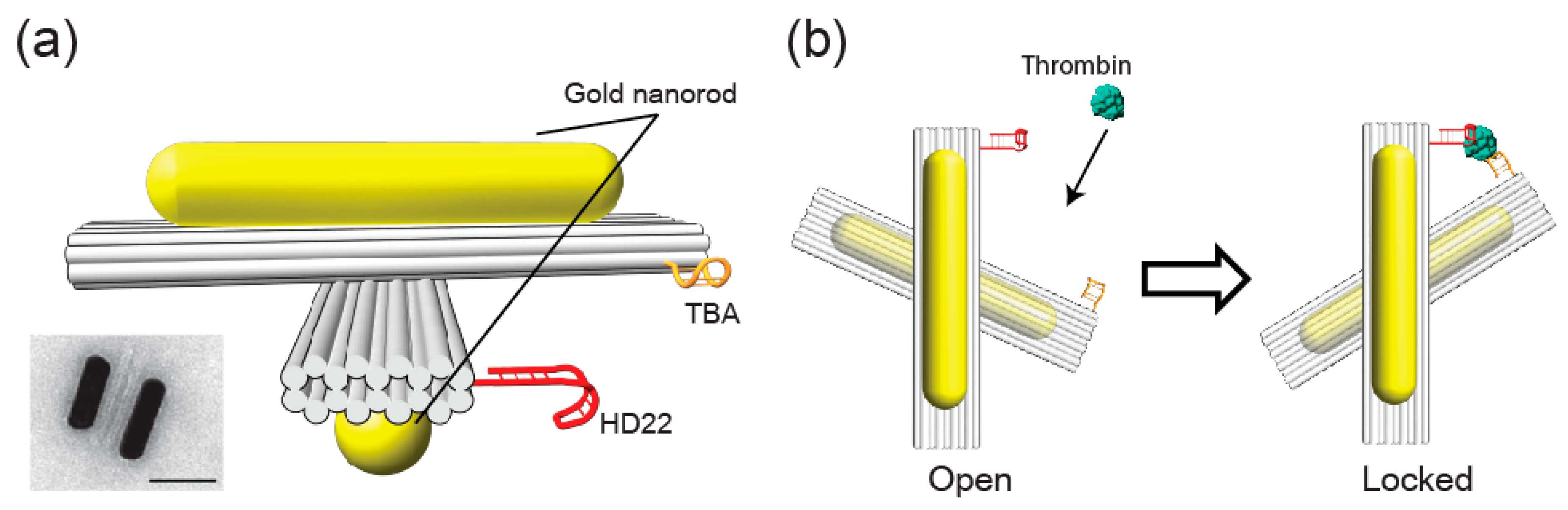
1. Introduction
2. Materials and Methods
2.1. Design, Folding, and Purification of DNA Nanostructures
2.2. Conjugation of Gold Nanorod
2.3. Preparing and Measuring 3D Plasmonic Metamolecule
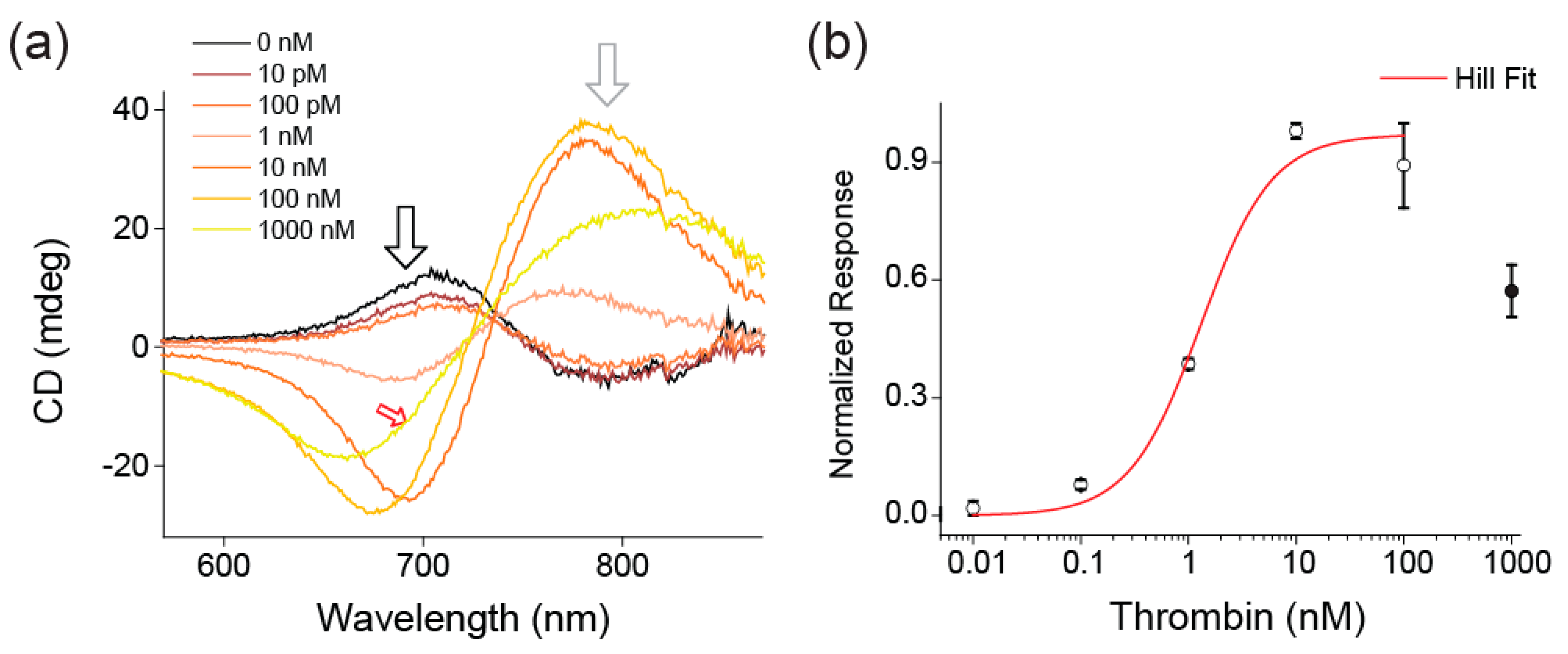
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Bock, L.C.; Griffin, L.C.; Latham, J.A.; Vermaas, E.H.; Toole, J.J. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature 1992, 355, 564–566. [Google Scholar] [CrossRef] [PubMed]
- Ellington, A.D.; Szostak, J.W. Selection in vitro of single-stranded DNA molecules that fold into specific ligand-binding structures. Nature 1992, 355, 850–852. [Google Scholar] [CrossRef] [PubMed]
- Pan, T.; Uhlenbeck, O.C. A small metalloribozyme with a two-step mechanism. Nature 1992, 358, 560–563. [Google Scholar] [CrossRef] [PubMed]
- Gold, L.; Polisky, B.; Uhlenbeck, O.; Yarus, M. Diversity of oligonucleotide functions. Annu. Rev. Biochem. 1995, 64, 763–797. [Google Scholar] [CrossRef] [PubMed]
- Wilson, D.S.; Szostak, J.W. In vitro selection of functional nucleic acids. Annu. Rev. Biochem. 1999, 68, 611–647. [Google Scholar] [CrossRef]
- Nimjee, S.M.; Rusconi, C.P.; Sullenger, B.A. Aptamers: An emerging class of therapeutics. Annu. Rev. Med. 2005, 56, 555–583. [Google Scholar] [CrossRef]
- Bunka, D.H.; Stockley, P.G. Aptamers come of age—At last. Nat. Rev. Microbiol. 2006, 4, 588–596. [Google Scholar] [CrossRef]
- Eyetech Study Group. Preclinical and phase 1A clinical evaluation of an anti-VEGF pegylated aptamer (EYE001) for the treatment of exudative age-related macular degeneration. Retina 2002, 22, 143–152. [Google Scholar] [CrossRef]
- Eyetech Study Group. Anti-vascular endothelial growth factor therapy for subfoveal choroidal neovascularization secondary to age-related macular degeneration: Phase II study results. Ophthalmology 2003, 110, 979–986. [Google Scholar] [CrossRef]
- Liu, J.; Lu, Y. A colorimetric lead biosensor using DNAzyme-directed assembly of gold nanoparticles. J. Am. Chem. Soc. 2003, 125, 6642–6643. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Lu, Y. Fast colorimetric sensing of adenosine and cocaine based on a general sensor design involving aptamers and nanoparticles. Angew. Chem. Int. Ed. Engl. 2005, 45, 90–94. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Brown, A.K.; Meng, X.; Cropek, D.M.; Istok, J.D.; Watson, D.B.; Lu, Y. A catalytic beacon sensor for uranium with parts-per-trillion sensitivity and millionfold selectivity. Proc. Natl. Acad. Sci. USA 2007, 104, 2056–2061. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.; Baker, B.R.; Wachsmann-Hogiu, S.; Pagba, C.V.; Laurence, T.A.; Lane, S.M.; Lee, L.P.; Tok, J.B. Aptamer-based SERRS sensor for thrombin detection. Nano Lett. 2008, 8, 4386–4390. [Google Scholar] [CrossRef] [PubMed]
- Pavlov, V.; Xiao, Y.; Shlyahovsky, B.; Willner, I. Aptamer-functionalized Au nanoparticles for the amplified optical detection of thrombin. J. Am. Chem. Soc. 2004, 126, 11768–11769. [Google Scholar] [CrossRef] [PubMed]
- Tombelli, S.; Minunni, M.; Luzi, E.; Mascini, M. Aptamer-based biosensors for the detection of HIV-1 Tat protein. Bioelectrochemistry 2005, 67, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Davis, J.H.; Szostak, J.W. Isolation of high-affinity GTP aptamers from partially structured RNA libraries. Proc. Natl. Acad. Sci. USA 2002, 99, 11616–11621. [Google Scholar] [CrossRef]
- Hasegawa, H.; Savory, N.; Abe, K.; Ikebukuro, K. Methods for Improving Aptamer Binding Affinity. Molecules 2016, 21, 421. [Google Scholar] [CrossRef]
- Rinker, S.; Ke, Y.; Liu, Y.; Chhabra, R.; Yan, H. Self-assembled DNA nanostructures for distance-dependent multivalent ligand-protein binding. Nat. Nanotechnol. 2008, 3, 418–422. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, H.; Taira, K.I.; Sode, K.; Ikebukuro, K. Improvement of Aptamer Affinity by Dimerization. Sensors (Basel) 2008, 8, 1090–1098. [Google Scholar] [CrossRef]
- Rothemund, P.W. Folding DNA to create nanoscale shapes and patterns. Nature 2006, 440, 297–302. [Google Scholar] [CrossRef] [PubMed]
- Dietz, H.; Douglas, S.M.; Shih, W.M. Folding DNA into twisted and curved nanoscale shapes. Science 2009, 325, 725–730. [Google Scholar] [CrossRef] [PubMed]
- Castro, C.E.; Kilchherr, F.; Kim, D.N.; Shiao, E.L.; Wauer, T.; Wortmann, P.; Bathe, M.; Dietz, H. A primer to scaffolded DNA origami. Nat. Methods 2011, 8, 221–229. [Google Scholar] [CrossRef] [PubMed]
- Han, D.; Pal, S.; Nangreave, J.; Deng, Z.; Liu, Y.; Yan, H. DNA origami with complex curvatures in three-dimensional space. Science 2011, 332, 342–346. [Google Scholar] [CrossRef] [PubMed]
- Kuzyk, A.; Schreiber, R.; Zhang, H.; Govorov, A.O.; Liedl, T.; Liu, N. Reconfigurable 3D plasmonic metamolecules. Nat. Mater. 2014, 13, 862–866. [Google Scholar] [CrossRef] [PubMed]
- Nickels, P.C.; Wunsch, B.; Holzmeister, P.; Bae, W.; Kneer, L.M.; Grohmann, D.; Tinnefeld, P.; Liedl, T. Molecular force spectroscopy with a DNA origami-based nanoscopic force clamp. Science 2016, 354, 305–307. [Google Scholar] [CrossRef]
- Ijäs, H.; Nummelin, S.; Shen, B.; Kostiainen, M.A.; Linko, V. Dynamic DNA Origami Devices: From Strand-Displacement Reactions to External-Stimuli Responsive Systems. Int. J. Mol. Sci. 2018, 19, 2114. [Google Scholar] [CrossRef] [PubMed]
- Endo, M.; Sugiyama, H. DNA Origami Nanomachines. Molecules 2018, 23, 1766. [Google Scholar] [CrossRef]
- Bae, W.; Kocabey, S.; Liedl, T. DNA nanostructures in vitro, in vivo and on membranes. Nano Today 2019, 26, 98–107. [Google Scholar] [CrossRef]
- Kuzyk, A.; Jungmann, R.; Acuna, G.P.; Liu, N. DNA Origami Route for Nanophotonics. ACS Photonics 2018, 5, 1151–1163. [Google Scholar] [CrossRef]
- Govorov, A.O.; Fan, Z.; Hernandez, P.; Slocik, J.M.; Naik, R.R. Theory of circular dichroism of nanomaterials comprising chiral molecules and nanocrystals: Plasmon enhancement, dipole interactions, and dielectric effects. Nano Lett. 2010, 10, 1374–1382. [Google Scholar] [CrossRef] [PubMed]
- Greenfield, N.J. Using circular dichroism spectra to estimate protein secondary structure. Nat. Protoc. 2006, 1, 2876–2890. [Google Scholar] [CrossRef] [PubMed]
- Kypr, J.; Kejnovska, I.; Renciuk, D.; Vorlickova, M. Circular dichroism and conformational polymorphism of DNA. Nucleic Acids Res. 2009, 37, 1713–1725. [Google Scholar] [CrossRef] [PubMed]
- Martin, S.R.; Schilstra, M.J. Circular dichroism and its application to the study of biomolecules. Methods Cell Biol. 2008, 84, 263–293. [Google Scholar] [CrossRef] [PubMed]
- Funck, T.; Nicoli, F.; Kuzyk, A.; Liedl, T. Sensing Picomolar Concentrations of RNA Using Switchable Plasmonic Chirality. Angew. Chem. Int. Ed. Engl. 2018, 57, 13495–13498. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Xin, L.; Duan, X.; Urban, M.J.; Liu, N. Dynamic Plasmonic System That Responds to Thermal and Aptamer-Target Regulations. Nano Lett. 2018, 18, 7395–7399. [Google Scholar] [CrossRef] [PubMed]
- Tasset, D.M.; Kubik, M.F.; Steiner, W. Oligonucleotide inhibitors of human thrombin that bind distinct epitopes. J. Mol. Biol. 1997, 272, 688–698. [Google Scholar] [CrossRef] [PubMed]
- Vagner, J.; Handl, H.L.; Gillies, R.J.; Hruby, V.J. Novel targeting strategy based on multimeric ligands for drug delivery and molecular imaging: Homooligomers of alpha-MSH. Bioorg. Med. Chem. Lett. 2004, 14, 211–215. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Wang, E.; Dong, S. G-quadruplex-based DNAzyme for facile colorimetric detection of thrombin. Chem. Commun. (Camb.) 2008, 31, 3654–3656. [Google Scholar] [CrossRef]
- Yang, R.; Tang, Z.; Yan, J.; Kang, H.; Kim, Y.; Zhu, Z.; Tan, W. Noncovalent assembly of carbon nanotubes and single-stranded DNA: An effective sensing platform for probing biomolecular interactions. Anal. Chem. 2008, 80, 7408–7413. [Google Scholar] [CrossRef]
- Zhao, Q.; Li, X.F.; Le, X.C. Aptamer-modified monolithic capillary chromatography for protein separation and detection. Anal. Chem. 2008, 80, 3915–3920. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Fu, H.E.; Wu, L.J.; Zheng, A.X.; Chen, G.N.; Yang, H.H. General colorimetric detection of proteins and small molecules based on cyclic enzymatic signal amplification and hairpin aptamer probe. Anal. Chem. 2012, 84, 5309–5315. [Google Scholar] [CrossRef] [PubMed]
- Heyduk, E.; Heyduk, T. Nucleic acid-based fluorescence sensors for detecting proteins. Anal. Chem. 2005, 77, 1147–1156. [Google Scholar] [CrossRef] [PubMed]
- Di Giusto, D.A.; Wlassoff, W.A.; Gooding, J.J.; Messerle, B.A.; King, G.C. Proximity extension of circular DNA aptamers with real-time protein detection. Nucleic Acids Res. 2005, 33, 64. [Google Scholar] [CrossRef] [PubMed]
- Kielar, C.; Xin, Y.; Shen, B.; Kostiainen, M.A.; Grundmeier, G.; Linko, V.; Keller, A. On the Stability of DNA Origami Nanostructures in Low-Magnesium Buffers. Angew. Chem. Int. Ed. Engl. 2018, 57, 9470–9474. [Google Scholar] [CrossRef] [PubMed]
| Staple Strand | Sequence |
|---|---|
| Arm a TBA | CAA ATT AAC TGA ACA CAA GAA TTG AGT TAA GCC TTA CAG AAT ttG G |
| Arm b2 HH22 | CAT CAC TAA GGG AAG AAA GCG CCC CGC TGC GGG AGt tAG TCC GTG GTA GGG CAG GTT GGG GTG ACT |
| Arm b blank | AAG CCT TTA TTT CAT CCC GCC AAA ATA AAA AGG AGT TGA TTA AAG A |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Funck, T.; Liedl, T.; Bae, W. Dual Aptamer-Functionalized 3D Plasmonic Metamolecule for Thrombin Sensing. Appl. Sci. 2019, 9, 3006. https://doi.org/10.3390/app9153006
Funck T, Liedl T, Bae W. Dual Aptamer-Functionalized 3D Plasmonic Metamolecule for Thrombin Sensing. Applied Sciences. 2019; 9(15):3006. https://doi.org/10.3390/app9153006
Chicago/Turabian StyleFunck, Timon, Tim Liedl, and Wooli Bae. 2019. "Dual Aptamer-Functionalized 3D Plasmonic Metamolecule for Thrombin Sensing" Applied Sciences 9, no. 15: 3006. https://doi.org/10.3390/app9153006
APA StyleFunck, T., Liedl, T., & Bae, W. (2019). Dual Aptamer-Functionalized 3D Plasmonic Metamolecule for Thrombin Sensing. Applied Sciences, 9(15), 3006. https://doi.org/10.3390/app9153006







