Exoproduction and Molecular Characterization of Peroxidase from Ensifer adhaerens
Abstract
Featured Application
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation of Ligninolytic and Peroxidase-Producing Bacteria
2.2. Exoperoxidase Production under Submerged Fermentation (SMF)
2.3. Determination of Peroxidase Activity
2.4. Determination of Process Parameters for Optimal Exoperoxidase Production
2.5. Exoperoxidase Production Over a Time Course
2.6. Utilization of Agricultural Residues for Exoperoxidase Production
2.7. Polymerase Chain Reaction (PCR) Detection of Peroxidase Gene
2.8. Bioinformatics Analysis
2.9. Data Analysis
2.10. Accession Number
3. Results and Discussion
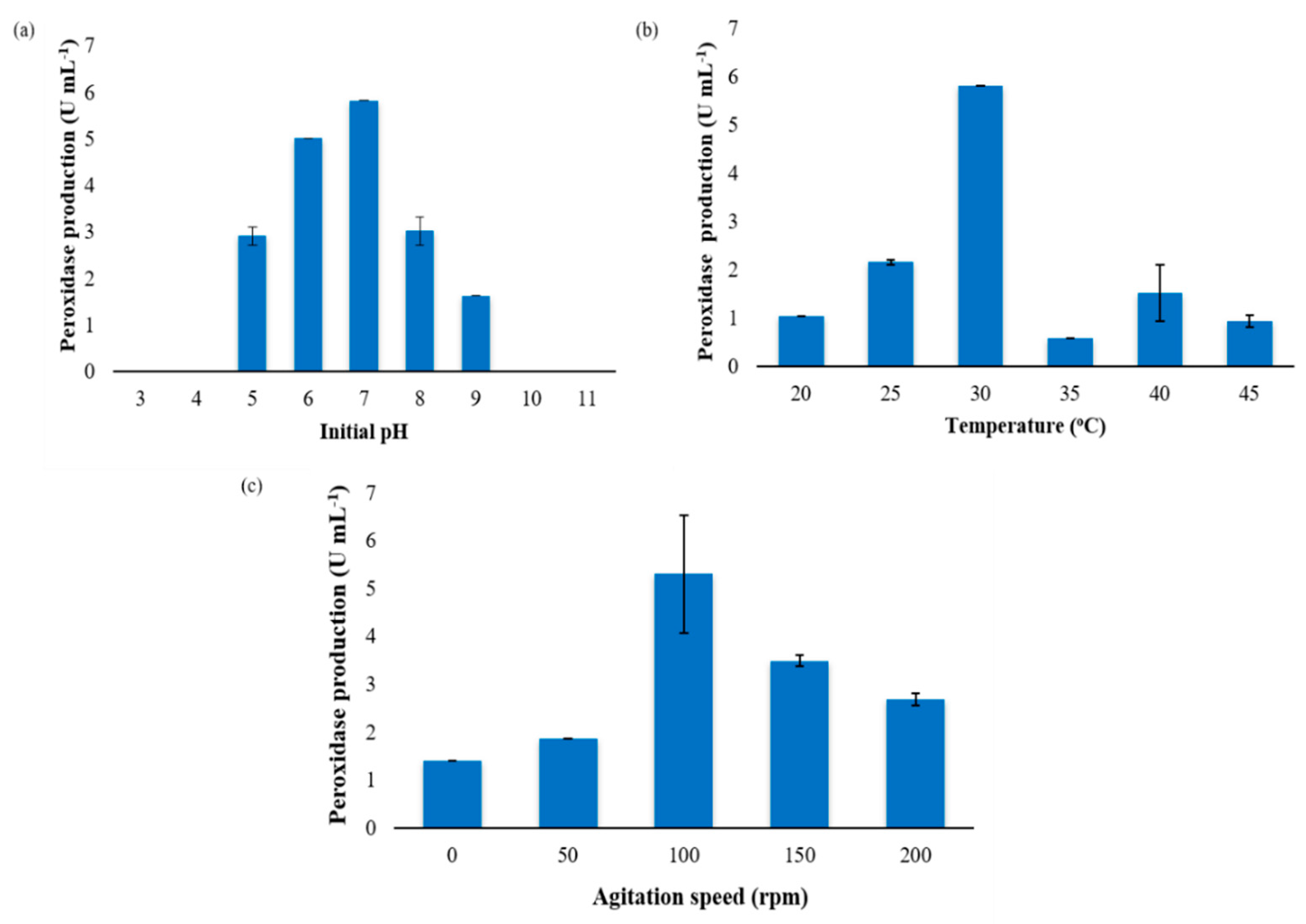
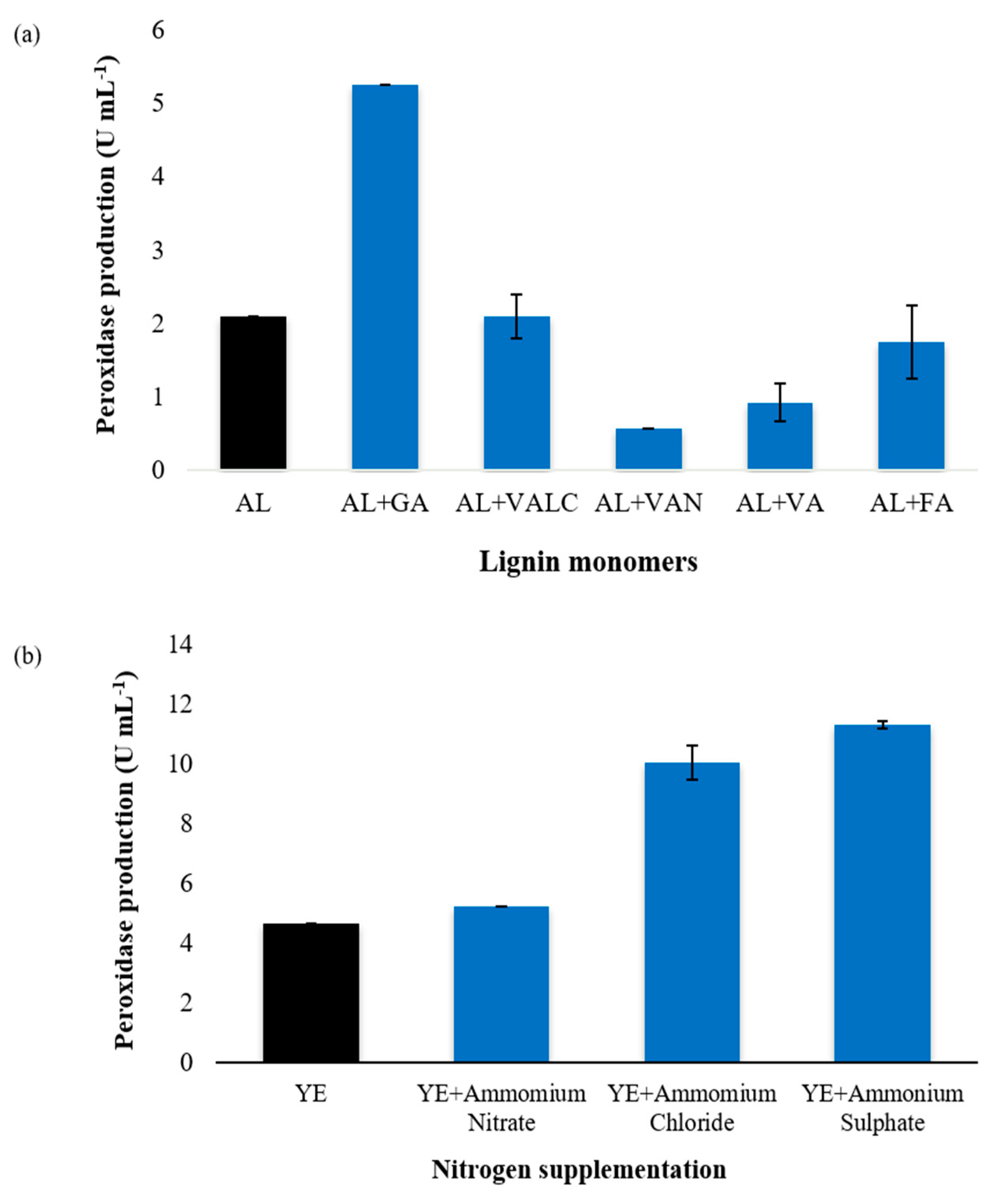
3.1. Determination of Process Parameters for Optimal Exoperoxidase Production
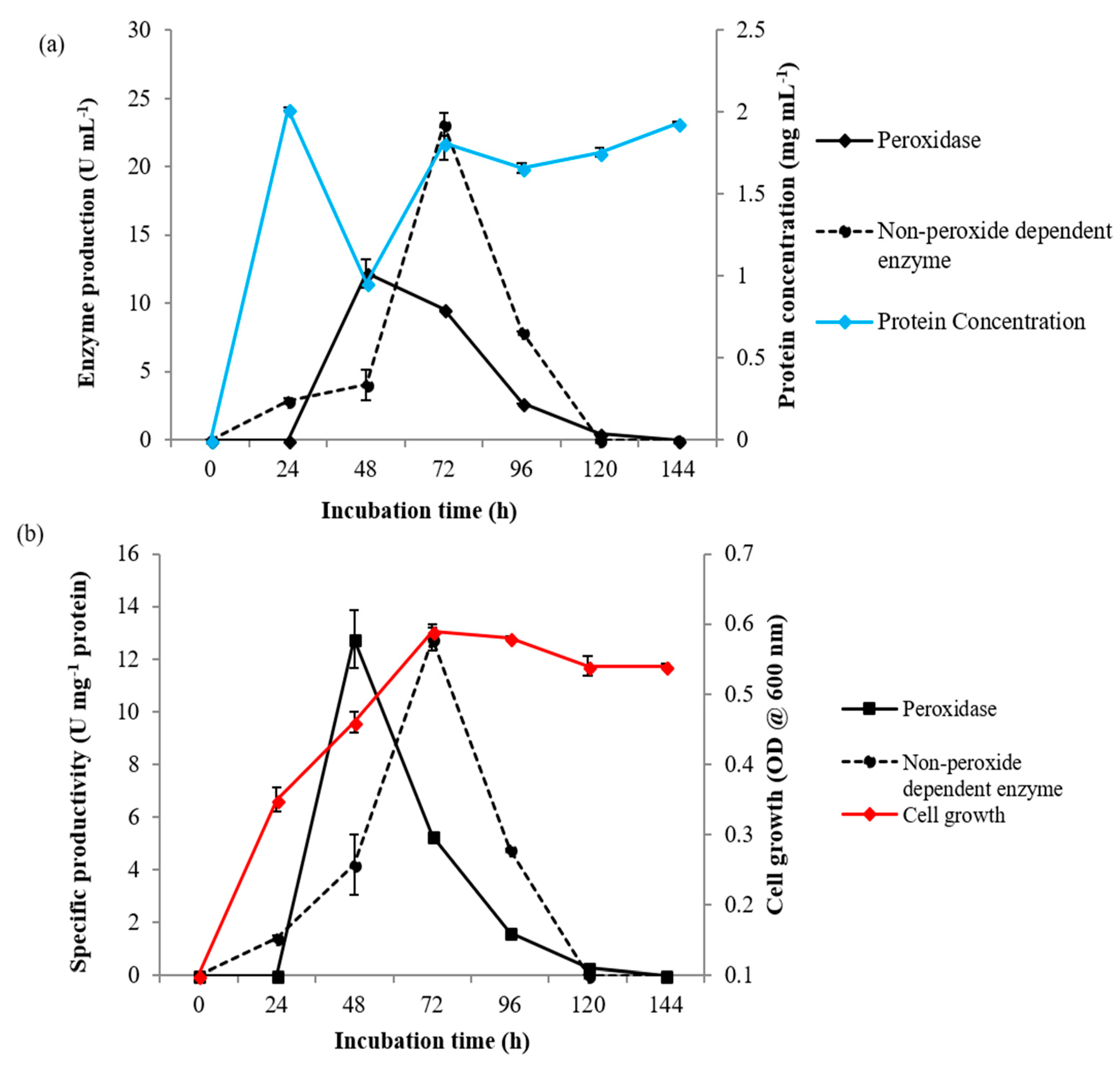
3.2. Exoperoxidase Production Over a Time Course
3.3. Assessment of Agricultural Residues for Exoperoxidase Production
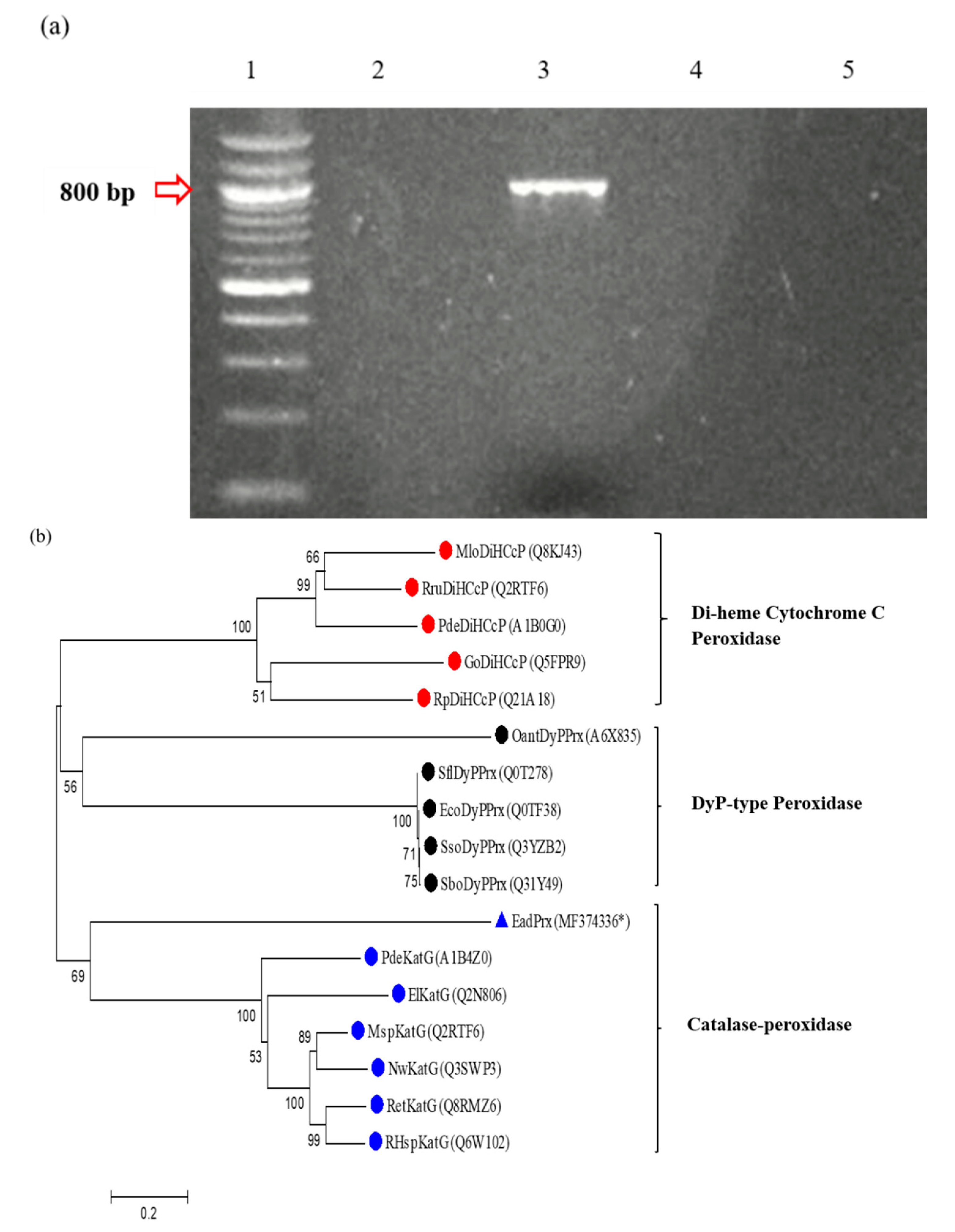
3.4. PCR Detection of Peroxidase Gene
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AL | alkali lignin |
| ALM | alkali lignin medium |
| ANOVA | analysis of variance |
| BLAST | basic local alignment search tool |
| FA | ferulic acid |
| GA | guaiacol |
| LMEs | lignin modifying enzymes |
| MEGA | molecular evolutionary genetics analysis |
| NAD | not detected |
| NCBI | National Center for Biotechnology Information |
| OD | optical density |
| SMF | submerged fermentation |
| SSF | solid state fermentation |
| UV | ultraviolet |
| VALC | veratryl alcohol |
| VAN | vanillin |
| VA | vanillic acid |
References
- Draelos, Z.D. A split-face evaluation of a novel pigment-lightening agent compared with no treatment and hydroquinone. J. Am. Acad. Dermatol. 2015, 72, 105–107. [Google Scholar] [CrossRef] [PubMed]
- Taboada-Puig, R.; Lu-Chau, T.A.; Eibes, G.; Feijoo, G.; Moreira, M.T.; Lema, J.; Taboada-Puig, R.; Lú-Chau, T.A. Continuous removal of endocrine disruptors by versatile peroxidase using a two-stage system. Biotechnol. Proc. 2015, 31, 908–916. [Google Scholar] [CrossRef] [PubMed]
- Ferrer, I.; Dezotti, M.; Duran, N. Decolourisation of kraft effluent by free and immobilised lignin peroxidases and horseradish peroxidase. Biotechnol. Lett. 1991, 13, 577–582. [Google Scholar] [CrossRef]
- Torres, E.; Bustos-Jaimes, I.; Le Borgne, S. Potential use of oxidative enzymes for the detoxification of organic pollutants. Appl. Catal. B Environ. 2003, 46, 1–15. [Google Scholar] [CrossRef]
- Valderrama, B.; Ayala, M.; Vasquez-Duhalt, R. Suicide inactivation of peroxidases and the challenge of engineering more robust enzymes. Chem. Biol. 2002, 9, 555–565. [Google Scholar] [CrossRef]
- Ikehata, K.; Pickard, M.A.; Buchanan, I.D.; Smith, D.W. Optimization of extracellular fungal peroxidase production by 2 Coprinus species. Can. J. Microbiol. 2004, 50, 1033–1040. [Google Scholar] [CrossRef]
- Urek, R.O.; Pazarlioglu, N.K. Enhanced production of manganese peroxidase by Phanerochaete chrysosporium. Braz. Arch. Biol. Technol. 2007, 50, 913–920. [Google Scholar] [CrossRef]
- Hariharan, S.; Nambisan, P. Optimization of lignin peroxidase, manganese peroxidase and lac production from Ganoderma lucidum under solid state fermentation of pineapple leaf. BioResources 2013, 8, 250–271. [Google Scholar] [CrossRef]
- Mercer, D.K.; Iqbal, M.; Miller, P.G.G.; McCarthy, A.J. Screening actinomycetes for extracellular peroxidase activity. Appl. Environ. Microbiol. 1996, 62, 2186–2190. [Google Scholar]
- Tuncer, M.; Kuru, A.; Isikli, M.; Sahin, N.; Çelenk, F. Optimization of extracellular endoxylanase, endoglucanase, and peroxidase production by Streptomyces sp. F2621 isolated in Turkey. J. Appl. Microbiol. 2004, 97, 783–791. [Google Scholar] [CrossRef]
- Tuncer, M.; Kuru, A.; Sahin, N.; Isikli, M. Production and partial characterisation of extracellular peroxidase produced by Streptomyces sp. F6616 isolated in Turkey. Ann. Microbiol. 2009, 59, 323–334. [Google Scholar] [CrossRef]
- Musengi, A.; Khan, N.; Le Roes-Hill, M.; Pletschke, B.I. Increasing the scale of peroxidase production by Streptomyces sp. strain BSII#1. J. Appl. Microbiol. 2014, 116, 554–562. [Google Scholar] [PubMed]
- Anbu, P.; Gopinath, S.C.B.; Cihan, A.C.; Chaulagain, B.P. Microbial enzymes and their applications in industries and medicine. BioMed. Res. Int. 2017. [Google Scholar] [CrossRef] [PubMed]
- Falade, A.O.; Eyisi, O.A.L.; Mabinya, L.V.; Nwodo, U.U.; Okoh, A.I. Peroxidase production and ligninolytic potentials of freshwater bacteria Raoutella ornithinolytica and Ensifer adhaerens. Biotechnol. Rep. 2017, 16, 12–17. [Google Scholar] [CrossRef]
- Bugg, T.D.H.; Ahmad, M.; Hardiman, E.M.; Singh, R. The emerging role for bacteria in lignin degradation and bio-product formation. Curr. Opin. Biotechnol. 2011, 22, 394–400. [Google Scholar] [CrossRef] [PubMed]
- Taylor, C.; Hardiman, E.; Ahmad, M.; Sainsbury, P.; Norris, P.; Bugg, T. Isolation of bacterial strains able to metabolize lignin from screening of environmental samples. J. Appl. Microbiol. 2012, 113, 521–530. [Google Scholar] [CrossRef] [PubMed]
- Prabhakaran, M.; Couger, M.B.; Jackson, C.A.; Weirick, T.; Fathepure, B.Z.; Lopez-Alonso, V.; Ortiz, S.; Martínez-Suárez, J.V. Genome sequences of the lignin-degrading Pseudomonas sp. strain YS-1p and Rhizobium sp. strain YS-1r isolated from decaying wood. Genome Announc. 2015, 3, e00019-15. [Google Scholar] [CrossRef]
- Woo, H.L.; Utturkar, S.; Klingeman, D.; Simmons, B.A.; DeAngelis, K.M.; Brown, S.D.; Hazen, T.C. Draft genome sequence of the lignin-degrading Burkholderia sp. strain LIG30 isolated from wet tropical forest soil. Genome Announc. 2014, 2, e00637-14. [Google Scholar] [CrossRef]
- Kalyani, D.C.; Phugare, S.S.; Shedbalkar, U.U.; Adhar, J.P. Purification and characterization of a bacterial peroxidase from the isolated strain Pseudomonas sp. SUK1 and its application for textile dye decolourization. Ann. Microbiol. 2011, 61, 483–491. [Google Scholar] [CrossRef]
- Sasikumar, V.; Priya, V.; Shiv, S.C.; Sathish, S.D. Isolation and preliminary screening of lignin degrading microbes. J. Acad. Indus. Res. 2014, 3, 291–294. [Google Scholar]
- Chance, B.; Maehly, A.C. Assay of catalases and peroxidases. Methods Enzymol. 1955, 2, 773–775. [Google Scholar]
- Fatokun, E.N.; Nwodo, U.U.; Okoh, A.I. Classical optimization of cellulase and xylanase production by a marine Streptomyces species. Appl. Sci. 2016, 6, 286. [Google Scholar] [CrossRef]
- Tuncer, M.; Rob, A.; Ball, A.S.; Wilson, M.T. Optimisation of extracellular lignocellulolytic enzyme production by a thermophilic actinomycete Thermomonospora fusca BD25. Enzym. Microb. Technol. 1999, 25, 38–47. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Falade, A.O.; Mabinya, L.V.; Okoh, A.I.; Nwodo, U.U. Agrowastes utilization by Raoultella ornithinolytica for optimal extracellular peroxidase activity. Biotechnol. Appl. Biochem. 2019, 66, 60–67. [Google Scholar] [CrossRef]
- Neifar, M.; Chouchane, H.; Mahjoubi, M.; Jaouani, A.; Cherif, A. Pseudomonas extremorientalis BU118: A new salt-tolerant laccase-secreting bacterium with biotechnological potential in textile azo dye decolourization. 3 Biotech 2016, 6, 107–115. [Google Scholar] [CrossRef] [PubMed]
- Maugeri, T.L.; Carbone, M.; Tera, M.T.; Gugliandolo, C. Detection and differentiation of Vibrio vulnificus in seawater and plankton of a coastal zone of the Mediterranean Sea. Res. Microbiol. 2006, 157, 194–200. [Google Scholar] [CrossRef]
- Falade, A.O.; Mabinya, L.V.; Okoh, A.I.; Nwodo, U.U. Biochemical and molecular characterization of a novel dye-decolourizing peroxidase from Raoultella ornithinolytica OKOH-1. Int. J. Biol. Macromol. 2019, 121, 454–462. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- McCarthy, A.J. Lignocellulose degrading actinomycetes. FEMS Microbiol. Rev. 1987, 46, 145–163. [Google Scholar] [CrossRef]
- Niladevi, K.N.; Prema, P. Effect of inducers and process parameters on laccase production by Streptomyces psammoticus and its application in dye decolourization. Bioresour. Technol. 2008, 99, 4583–4589. [Google Scholar] [CrossRef] [PubMed]
- Salehizadeh, H.; Shojaosadati, S.A. Extracellular biopolymeric flocculants: Recent trends and biotechnological importance. Biotechnol. Adv. 2001, 19, 371–385. [Google Scholar] [CrossRef]
- Makapela, B.; Okaiyeto, K.; Ntozonke, N.; Nwodo, U.U.; Green, E.; Mabinya, L.; Okoh, A. Assessment of Bacillus pumilus isolated from freshwater milieu for bioffloculant production. Appl. Sci. 2016, 6, 211. [Google Scholar] [CrossRef]
- Rob, A.; Hernandez, M.; Ball, A.S.; Tuncer, M.; Arias, M.E.; Wilson, M.T. Production and partial characterization of extracellular peroxidase produced by Streptomyces avermitilis UAH30. Appl. Biochem. Biotechnol. 1997, 62, 159–174. [Google Scholar] [CrossRef]
- Nour El-Dein, M.M.; Shereif, A.E.A.; Mansour, F.A.; Abou-Dobara, M.I.; Ball, A.S. Optimization of xylanase and peroxidase production from Streptomyces sp. K37. J. BioSci. Biotechnol. 2014, 3, 29–42. [Google Scholar]
- Xia, S.Q.; Zhang, Z.Q.; Wang, X.J.; Yang, A.M.; Chen, L.; Zhao, J.; Leonard, D.; Jaffrezic-Renault, N. Production and characterization of a bioffloculant by Proteus mirabilis TJ-1. Bioresour. Technol. 2008, 99, 6520–6527. [Google Scholar] [CrossRef]
- Rajkumar, R.; Yaakob, Z.; Takriff, M.S.; Kamarudin, K.F. Optimization of medium composition for production of peroxidase by Bacillus sp. Der Pharma Chem. 2013, 5, 167–174. [Google Scholar]
- Rao, P.R.; Kavya, P. Production, isolation and purification of peroxidase using Bacillus subtilis. 2014 1st International Congress on Environmental, Biotechnology, and Chemistry Engineering. Int. Cong. Environ. Biotechnol. Chem. Eng. 2014, 64, 21–27. [Google Scholar]
- Ray, A.K.; Bairagi, A.; Ghosh, K.S.; Sen, S.K. Optimization of fermentation conditions for cellulase production by Bacillus subtilis CY5 and Bacillus circulans TP3 isolated from fish gut. Acta Ichthyol. Piscat. 2007, 3, 47–53. [Google Scholar] [CrossRef]
- Giavasis, I.; Harvey, L.M.; McNeil, B. The effect of agitation and aeration on the synthesis and molecular weight of gellan in batch cultures of Sphingomonas paucimobilis. Enzym. Microb. Technol. 2006, 38, 101–108. [Google Scholar] [CrossRef]
- Sepahy, A.A.; Ghazi, S.; Sepahy, M.A. Cost-effective production and optimization of alkaline xylanase by indigenous Bacillus mojavensis AG137 fermented on waste. Enzym. Res. 2011, 593624. [Google Scholar] [CrossRef]
- Patil, S.R. Production and purification of lignin peroxidase from Bacillus megaterium and its application in bioremediation. CIB Tech J. Microbiol. 2014, 3, 22–28. [Google Scholar]
- Niku-Paavola, M.-L.; Karhunen, E.; Kantelinen, A.; Viikari, L.; Lundell, T.; Hatakka, A. The effect of culture conditions on the production of lignin modifying enzymes by the white rot fungus Phlebia radiata. J. Biotechnol. 1990, 13, 211–221. [Google Scholar] [CrossRef]
- Mester, T.; de Jong, E.; Field, J.A. Manganese regulation of veratryl alcohol in white rot fungi and its indirect effect of lignin peroxidase. Appl. Environ. Microbiol. 1995, 61, 1881–1887. [Google Scholar] [PubMed]
- Couto, S.R.; Longo, M.A.; Cameselle, C.; Sanromán, A. Ligninolytic enzymes from corncob cultures of Phanerochaete chrysosporium under semi-solid-state conditions. Acta Biotechnol. 1999, 19, 17–25. [Google Scholar] [CrossRef]
- Kachlishvili, E.; Penninck, M.J.; Tsiklauri, N.; Elisashvili, V. Effect of nitrogen source on lignocellulolytic enzyme production by white-rot basidiomycetes under solid state cultivation. World J. Microbiol. Biotechnol. 2005, 22, 391–397. [Google Scholar] [CrossRef]
- Mikiashvili, N.; Wasser, S.P.; Nevo, E.; Elisashvili, V. Effects of carbon and nitrogen sources on Pleurotus ostreatus ligninolytic enzyme activity. World J. Microbiol. Biotechnol. 2006, 22, 999–1002. [Google Scholar] [CrossRef]
- Stajic, M.; Persky, L.; Friesem, D.; Hadar, Y.; Wasser, S.P.; Nevo, E.; Vukojević, J. Effect of different carbon and nitrogen sources on laccase and peroxidases production by selected Pleurotus species. Enzym. Microb. Technol. 2006, 38, 65–73. [Google Scholar] [CrossRef]
- Kaal, E.E.J.; Field, J.A.; Joyce, T.W. Increasing ligninolytic enzyme activities in several white-rot basidiomycetes by nitrogen sufficient media. Bioresour. Technol. 1995, 53, 133–139. [Google Scholar] [CrossRef]
- Mester, T.A.; Field, A.J. Optimization of manganese peroxidase production by the white-rot fungus Bjerkandera sp. strain BOS55. FEMS Microbiol. Lett. 1997, 155, 161–168. [Google Scholar] [CrossRef]
- Gianfreda, L.; Xu, F.; Bollag, J. Laccases: A useful group of oxidoreductive enzymes. Bioremediat. J. 1999, 3, 1–25. [Google Scholar] [CrossRef]
- Galhaup, C.; Wagner, H.; Hinterstoisser, B.; Haltrich, D. Increased production of laccase by the wood-degrading basidiomycete Trametes pubescens. Enzym. Microb. Technol. 2002, 30, 529–536. [Google Scholar] [CrossRef]
- Buswell, J.A. Fungal degradation of lignin. In Handbook of Applied Mycology; Arora, D.K., Rai, B., Mukerji, K.G., Kundsen, G., Eds.; Marcel Dekker: New York, NY, USA, 1992; pp. 425–480. [Google Scholar]
- Papagianni, M.; Moo-Young, M. Protease secretion in glucoamylase producer Aspergillus niger cultures: Fungal morphology and inoculum effects. Process Biochem. 2002, 37, 1271–1278. [Google Scholar] [CrossRef]
- Falade, A.O.; Nwodo, U.U.; Iweriebor, B.C.; Green, E.; Mabinya, L.V.; Okoh, A.I. Lignin peroxidase functionalities and prospective applications. Microbiologyopen 2017, 6, e00394. [Google Scholar] [CrossRef]
- Knezevic, A.; Milovanovic, I.; Stajic, M.; Vakojevic, J. Trametes suaveolens as ligninolytic enzyme producer. J. Nat. Sci. 2013, 124, 437–444. [Google Scholar] [CrossRef]
- Saratale, G.D.; Kshirsagar, S.D.; Sampange, V.T.; Saratale, R.G.; Oh, S.-E.; Govindwar, S.P.; Oh, M.-K. Cellulolytic enzymes production by utilizing agricultural wastes under solid state fermentation and its application for biohydrogen production. Appl. Biochem. Biotechnol. 2014, 174, 2801–2817. [Google Scholar] [CrossRef]
- Sharma, R.; Rawat, R.; Bhogal, R.S.; Oberoi, H.S. Multi-component thermostable cellulolytic enzyme production by Aspergillus niger HN-1 using pea pod waste: Appraisal of hydrolytic potential with lignocellulosic biomass. Process Biochem. 2015, 50, 696–704. [Google Scholar] [CrossRef]
- Olajuyigbe, F.M.; Ogunyewo, O.A. Enhanced production and physicochemical properties of thermostable crude cellulose from Sporothrix carnis grown on corn cobs. Biocatal. Agric. Biotechnol. 2016, 7, 110–117. [Google Scholar] [CrossRef]
- Muthukumarasamy, N.; Murugan, S. Production, purification and application of bacterial laccase: A review. Biotechnol. Adv. 2014, 13, 196–205. [Google Scholar]
- Niladevi, K.N.; Sukumaran, R.K.; Prema, P. Utilization of rice straw for laccase production by Streptomyces psammoticus in solid state fermentation. J. Ind. Microbiol. Biotechnol. 2007, 34, 665–674. [Google Scholar] [CrossRef] [PubMed]
- Kamsani, N.; Salleh, M.M.; Yahya, A.; Chong, C.S. Production of lignocellulolytic enzymes by microorganisms isolated from Bulbitermes sp. termite gut in solid-state fermentation. Waste Biomass Valor. 2016, 7, 357. [Google Scholar] [CrossRef]
- Unuofin, J.O.; Okoh, A.I.; Nwodo, U.U. Utilization of agricultural wastes for the production of laccase by Achromobacter xylosoxidans HWN16 and Bordetella bronchiseptica HSO16. J. Environ. Manag. 2019, 231, 222–231. [Google Scholar] [CrossRef] [PubMed]
- Unuofin, J.O.; Okoh, A.I.; Nwodo, U.U. Maize stover as a feedstock for enhanced laccase production by two gamma proteobacteria: A solution to agroindustrial waste stockpiling. Ind. Crop Prod. 2019, 129, 611–623. [Google Scholar] [CrossRef]
- Zamocky, M.; Furtmuller, P.G.; Obinger, C. Evolution of catalases from bacteria to humans. Antioxid. Redox Signal. 2008, 10, 1527–1547. [Google Scholar] [CrossRef] [PubMed]
- Zamocky, M.; Obinger, C. Molecular phylogeny of heme peroxidases. In Biocatalysis Based on Heme Peroxidases; Torres, E., Ayala, M., Eds.; Springer: Heidelberg, Germany, 2010; pp. 7–35. [Google Scholar]
- Brown, M.E.; Walker, M.C.; Nakashige, T.G.; Iavarone, A.T.; Chang, M.C.Y. Discovery and characterization of heme enzymes from unsequenced bacteria: Application to microbial lignin degradation. J. Am. Chem. Soc. 2011, 133, 18006–18009. [Google Scholar] [CrossRef] [PubMed]
- De Gonzalo, G.; Colpa, D.I.; Habib, M.H.M.; Fraaije, M.W. Bacterial enzymes involved in lignin degradation. J. Biotechnol. 2016, 236, 110–119. [Google Scholar] [CrossRef]
| Agricultural Residue | Protein Concentration (mg mL−1) | Enzyme Assay (With H2O2) | Enzyme Assay (Without H2O2) | ||
|---|---|---|---|---|---|
| Peroxidase Production (U mL−1) | Specific Productivity (U mg−1) | Probable Laccase Production (U mL−1) | Specific Productivity (U mg−1) | ||
| Sawdust | 0.028 ± 0.0 a | 1.05 ± 0.00 a | 37.50 ± 0.00 a | NAD * | NAD * |
| Wheat Straw | 1.023 ± 0.116 b | 5.37 ± 0.00 b | 5.25 ± 0.00 b | NAD * | NAD * |
| Corn Stover | 1.366 ± 0.021 b | 5.13 ± 0.00 b | 3.76 ± 0.00 c | 1.93 ± 0.53 | 1.23 ± 0.21 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Falade, A.; Jaouani, A.; Mabinya, L.; Okoh, A.; Nwodo, U. Exoproduction and Molecular Characterization of Peroxidase from Ensifer adhaerens. Appl. Sci. 2019, 9, 3121. https://doi.org/10.3390/app9153121
Falade A, Jaouani A, Mabinya L, Okoh A, Nwodo U. Exoproduction and Molecular Characterization of Peroxidase from Ensifer adhaerens. Applied Sciences. 2019; 9(15):3121. https://doi.org/10.3390/app9153121
Chicago/Turabian StyleFalade, Ayodeji, Atef Jaouani, Leonard Mabinya, Anthony Okoh, and Uchechukwu Nwodo. 2019. "Exoproduction and Molecular Characterization of Peroxidase from Ensifer adhaerens" Applied Sciences 9, no. 15: 3121. https://doi.org/10.3390/app9153121
APA StyleFalade, A., Jaouani, A., Mabinya, L., Okoh, A., & Nwodo, U. (2019). Exoproduction and Molecular Characterization of Peroxidase from Ensifer adhaerens. Applied Sciences, 9(15), 3121. https://doi.org/10.3390/app9153121






