Discrimination of Spatial Distribution of Aquatic Organisms in a Coastal Ecosystem Using eDNA
Abstract
Featured Application
Abstract
1. Introduction
2. Materials and Methods
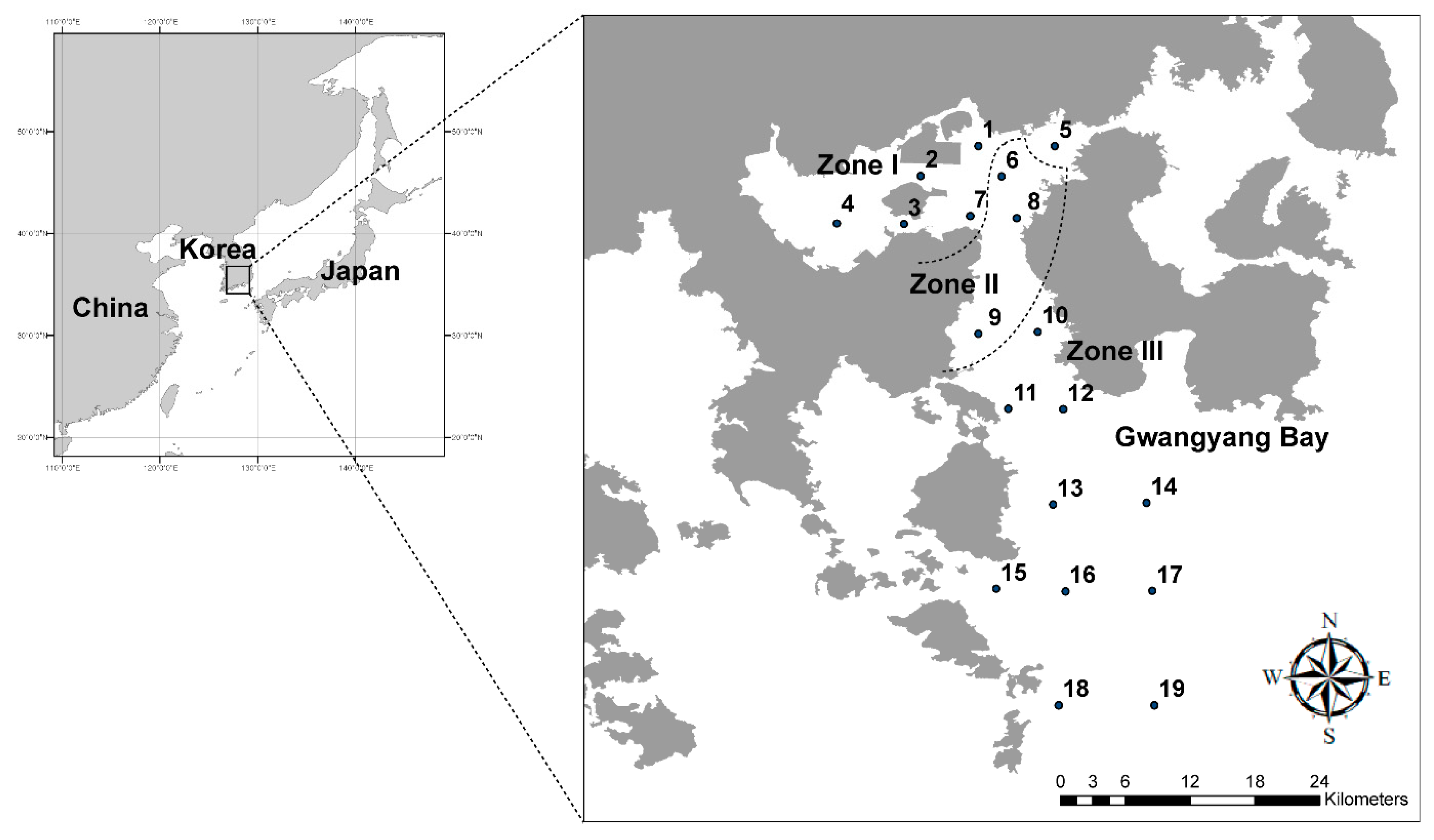
2.1. Study Area
2.2. Sampling, Data Collection and Primer Selection
2.3. DNA Extraction and Metagenomic Sequencing
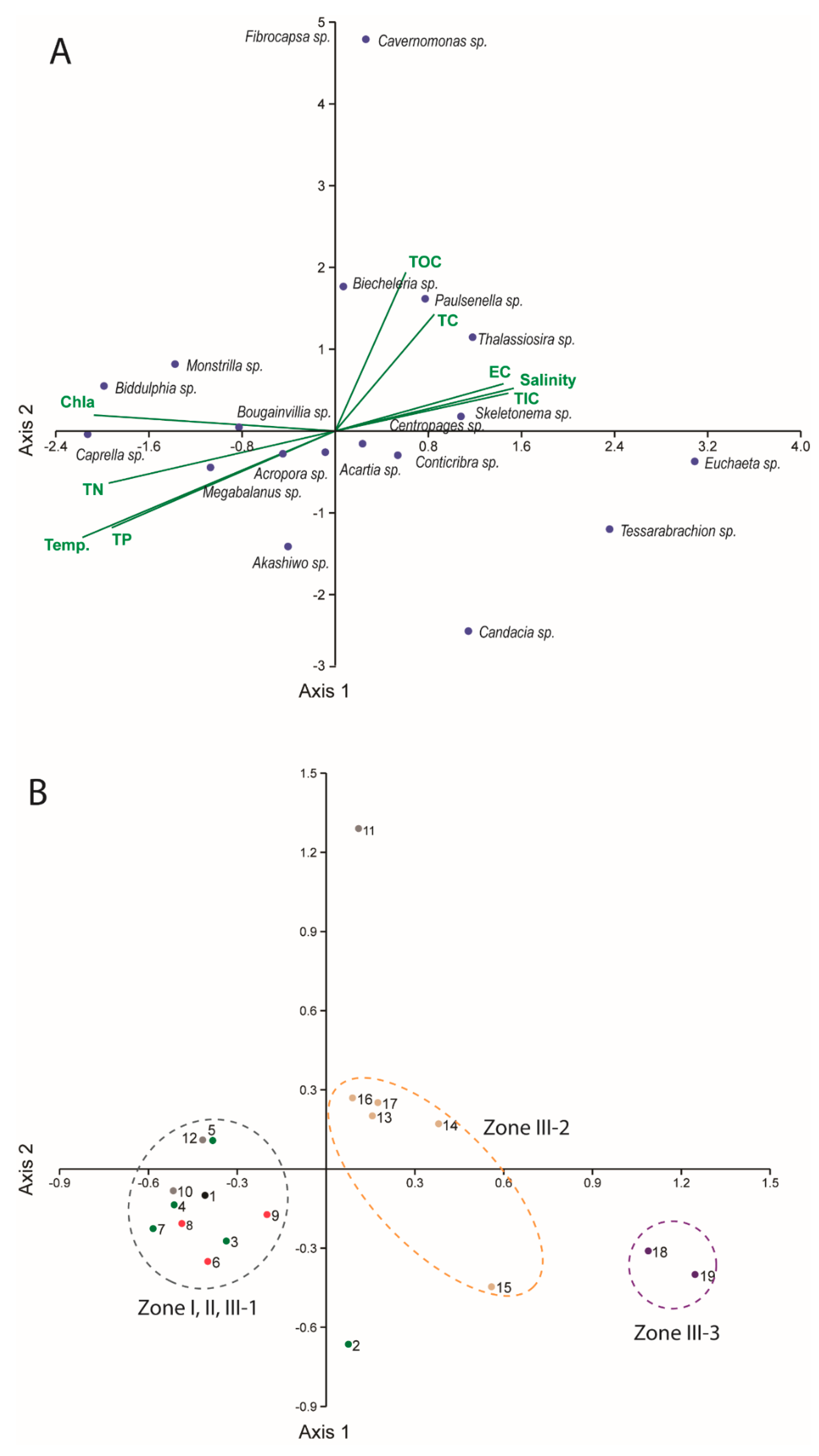
2.4. Data Analysis and Statistics
3. Results
3.1. Meta-Barcoding
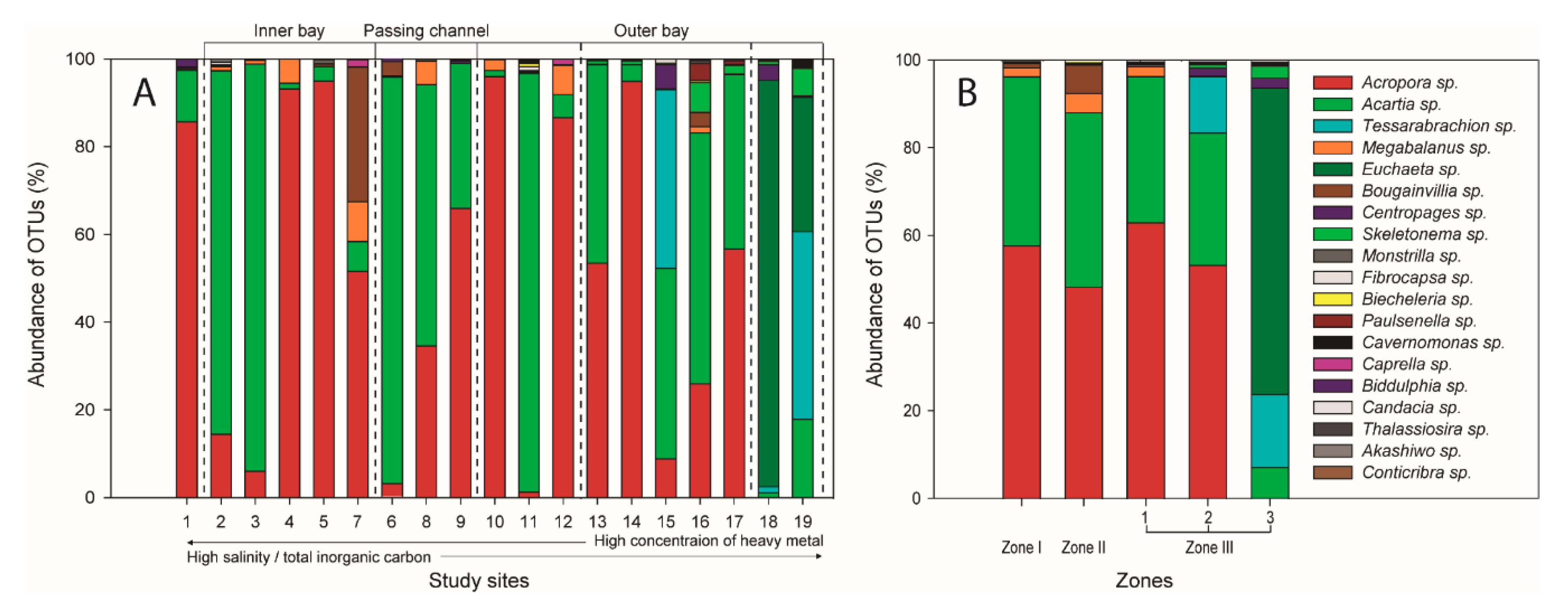
3.2. Spatial Distributions of Aquatic Organisms Based on Meta-Barcoding
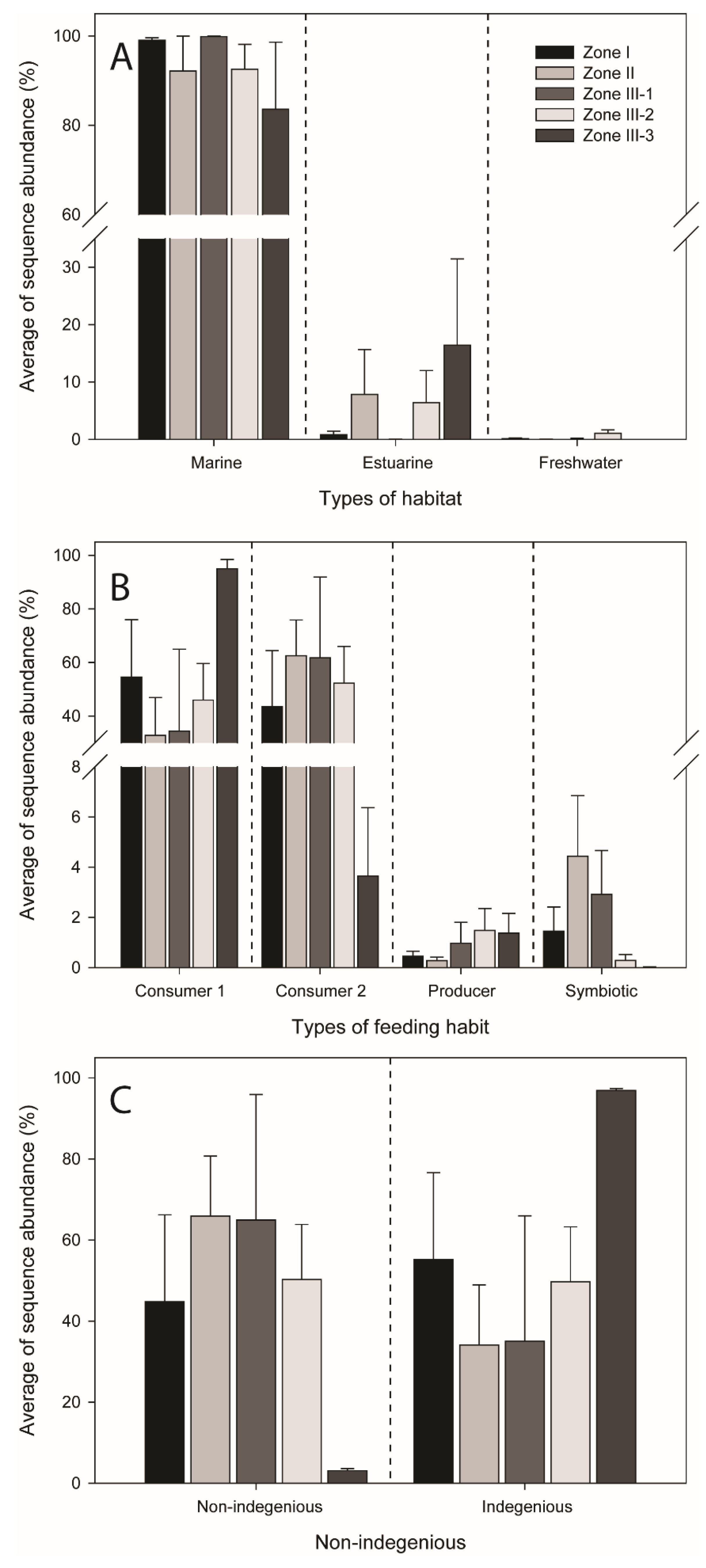
3.3. Functional Features and Non-Indigenous Species
4. Discussion
4.1. Effectiveness of eDNA Monitoring
4.2. Ecological Values of eDNA Monitoring
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Appendix A. Species Level Identification List Used in this Paper.
| Kingdom | Species |
| Eukaryota | Acropora granulosa |
| Eukaryota | Acartia omorii |
| Eukaryota | Tessarabrachion oculatum |
| Eukaryota | Megabalanus stultus |
| Eukaryota | Euchaeta indica |
| Eukaryota | Bougainvillia muscus |
| Eukaryota | Centropages typicus |
| Eukaryota | Skeletonema costatum |
| Eukaryota | Monstrilla sp. |
| Eukaryota | Fibrocapsa japonica |
| Eukaryota | Biecheleria brevisulcata |
| Eukaryota | Paulsenella vonstoschii |
| Eukaryota | Cavernomonas mira |
| Eukaryota | Caprella californica |
| Eukaryota | Biddulphia sp. |
| Eukaryota | Candacia bispinosa |
| Eukaryota | Thalassiosira mala |
| Eukaryota | Akashiwo sanguinea |
| Eukaryota | Conticribra weissflogiopsis |
References
- Jo, H.; Ventura, M.; Vidal, N.; Gim, J.S.; Buchaca, T.; Barmuta, L.A.; Erik, J.; Joo, G.J. Discovering hidden biodiversity: The use of complementary monitoring of fish diet based on DNA barcoding in freshwater ecosystems. Ecol. Evol. 2016, 6, 219–232. [Google Scholar] [CrossRef] [PubMed]
- Robertson, G.P.; Collins, S.L.; Foster, D.R.; Brokaw, N.; Ducklow, H.W.; Gragson, T.L.; Gries, C.; Hamilton, S.K.; McGuire, A.D.; Moore, J.C.; et al. Long-term ecological research in a human-dominated world. BioScience 2006, 62, 342–353. [Google Scholar] [CrossRef]
- Baird, D.J.; Hajibabaei, M. Biomonitoring 2.0: A new paradigm in ecosystem assessment made possible by next-generation DNA sequencing. Mol. Ecol. 2012, 21, 2039–2044. [Google Scholar] [CrossRef] [PubMed]
- Myers, R.A.; Worm, B. Rapid worldwide depletion of predatory fish communities. Nature 2003, 423, 280. [Google Scholar] [CrossRef] [PubMed]
- Pusceddu, A.; Bianchelli, S.; Martín, J.; Puig, P.; Palanques, A.; Masqué, P.; Danovaro, R. Chronic and intensive bottom trawling impairs deep-sea biodiversity and ecosystem functioning. Proc. Natl. Acad. Sci. USA 2014, 111, 8861–8866. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, G.M.; Fofonoff, P.W.; Carlton, J.T.; Wonham, M.J.; Hines, A.H. Invasion of coastal marine communities in North America: Apparent patterns, processes, and biases. Annu. Rev. Ecol. Syst. 2000, 31, 481–531. [Google Scholar] [CrossRef]
- Naylor, R.; Williams, S.L.; Strong, D.R. Aquaculture—A gateway for exotic species. Science 2001, 294, 1655–1656. [Google Scholar] [CrossRef]
- Padilla, D.K.; Williams, S.L. Beyond ballast water: Aquarium and ornamental trades as sources of invasive species in aquatic ecosystems. Front. Ecol. Environ. 2004, 2, 131–138. [Google Scholar] [CrossRef]
- Chapman, J.W.; Miller, T.W.; Coan, E.V. Live seafood species as recipes for invasion. Conserv. Biol. 2003, 17, 1386–1395. [Google Scholar] [CrossRef]
- Weigel, S.; Bester, K.; Hühnerfuss, H. Identification and quantification of pesticides, industrial chemicals, and organobromine compounds of medium to high polarity in the North Sea. Mar. Pollut. Bull. 2005, 50, 252–263. [Google Scholar] [CrossRef]
- Ardura, A.; Planes, S. Rapid assessment of non-indigenous species in the era of the eDNA barcoding: A Mediterranean case study. Estuar. Coast. Shelf Sci. 2017, 188, 81–87. [Google Scholar] [CrossRef]
- Carlton, J.T.; Geller, J.B. Ecological roulette: The global transport of nonindigenous marine organisms. Science 1993, 261, 78–82. [Google Scholar] [CrossRef]
- Williams, S.L.; Grosholz, E.D. The invasive species challenge in estuarine and coastal environments: Marrying management and science. Estuaries Coasts 2008, 31, 3–20. [Google Scholar] [CrossRef]
- Ogram, A.; Sayler, G.S.; Barkay, T. The extraction and purification of microbial DNA from sediments. J. Microbiol. Meth. 1987, 7, 57–66. [Google Scholar] [CrossRef]
- Taberlet, P.; Coissac, E.; Hajibabaei, M.; Rieseberg, L.H. Environmental DNA. Mol. Ecol. 2012, 21, 1789–1793. [Google Scholar] [CrossRef]
- Andersen, K.; Bird, K.L.; Rasmussen, M.; Haile, J.; Breuning-Medsen, H.; Kjaer, K.H.; Orlando, L.; Geilbert, M.T.P.; Willerslev, E. Meta-barcoding of ‘dirt’ DNA from soil reflects vertebrate biodiversity. Mol. Ecol. 2011, 21, 1966–1979. [Google Scholar] [CrossRef]
- Minamoto, T.; Yamanaka, H.; Takahara, T.; Honjo, M.N.; Kawabata, Z. Surveillance of fish species composition using environmental DNA. Limnology 2012, 13, 193–197. [Google Scholar] [CrossRef]
- Thomsen, P.; Kielgast, J.; Iversen, L.L.; Wiuf, C.; Rasmussen, M.; Gilbert, M.T.P.; Orlando, L.; Willerslev, E. Monitoring endangered freshwater biodiversity using environmental DNA. Mol. Ecol. 2012, 21, 2565–2573. [Google Scholar] [CrossRef]
- Thomsen, P.; Kielgast, J.; Iversen, L.L.; Møller, P.R.; Rasmussen, M.; Willerslev, E. Detection of a diverse marine fish fauna using environmental DNA from seawater samples. PLoS ONE 2012, 7, e41732. [Google Scholar] [CrossRef]
- Yamamoto, S.; Masuda, R.; Sato, Y.; Sado, T.; Araki, H.; Kondoh, M.; Toshifumi, M.; Miya, M. Environmental DNA metabarcoding reveals local fish communities in a species-rich coastal sea. Sci. Rep. 2017, 7, 40368. [Google Scholar] [CrossRef]
- Ficetola, G.F.; Miaud, C.; Pompanon, F.; Taberlet, P. Species detection using environmental DNA from water samples. Biol. Lett. 2008, 4, 423–425. [Google Scholar] [CrossRef]
- Goldberg, C.S.; Pilliod, D.S.; Arkle, R.S.; Waits, L.P. Molecular detection of vertebrates in stream water: A demonstration using Rocky Mountain tailed frogs and Idaho giant salamanders. PLoS ONE 2011, 6, e22746. [Google Scholar] [CrossRef]
- Hawkins, C.P.; Norris, R.H.; Hogue, J.N.; Feminella, J.W. Development and evaluation of predictive models for measuring the biological integrity of streams. Ecol. Appl. 2000, 10, 1456–1477. [Google Scholar] [CrossRef]
- Korean Statistical Information Service (KOSIS). Available online: http://kosis.kr (accessed on 9 June 2019).
- Kim, D.K.; Jo, H.; Han, I.; Kwak, I.S. Explicit Characterization of Spatial Heterogeneity Based on Water Quality, Sediment Contamination, and Ichthyofauna in a Riverine-to-Coastal Zone. Int. J. Environ. Res. Health 2019, 16, 409. [Google Scholar] [CrossRef]
- Kang, C.K.; Kim, J.B.; Lee, K.S.; Kim, J.B.; Lee, P.Y.; Hong, J.S. Trophic importance of benthic microalgae to macrozoobenthos in coastal bay systems in Korea: Dual stable C and N isotope analyses. Mar. Ecol. Prog. Ser. 2003, 259, 79–92. [Google Scholar] [CrossRef]
- Abad, D.; Albaina, A.; Aguirre, M.; Laza-Martínez, A.; Uriarte, I.; Iriarte, A.; Arantza, I.; Fernando, V.; Estonba, A. Is metabarcoding suitable for estuarine plankton monitoring? A comparative study with microscopy. Mar. Biol. 2016, 163, 149. [Google Scholar] [CrossRef]
- de Vargas, C.; Audic, S.; Henry, N.; Decelle, J.; Mahé, F.; Logares, R.; Lara, E.; Berney, C. Eukaryotic plankton diversity in the sunlit ocean. Science 2015, 348, 1261605. [Google Scholar] [CrossRef]
- Albaina, A.; Aguirre, M.; Abad, D.; Santos, M.; Estonba, A. 18S rRNA V9 metabarcoding for diet characterization: A critical evaluation with two sympatric zooplanktivorous fish species. Ecol. Evol. 2016, 6, 1809–1824. [Google Scholar] [CrossRef]
- Massana, R.; Gober, A.; Audic, S.; Bass, D.; Bittner, L.; Boutte, C.; Chambouvet, A.; Christen, R. Marine protist diversity in European coastal waters and sediments as revealed by high-throughput sequencing. Environ. Microbiol. 2015, 17, 4035–4049. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Huttley, G.A. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335. [Google Scholar] [CrossRef]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Hammer, Ø.; Harper, D.A.; Ryan, P.D. PAST: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 9. [Google Scholar]
- Addinsoft. XLSTAT Statistical and Data Analysis Solution; Addinsoft: New York, NY, USA, 2019. [Google Scholar]
- Thomsen, P.F.; Møller, P.R.; Sigsgaard, E.E.; Knudsen, S.W.; Jørgensen, O.A.; Willerslev, E. Environmental DNA from seawater samples correlate with trawl catches of subarctic, deepwater fishes. PLoS ONE 2016, 11, e0165252. [Google Scholar] [CrossRef]
- Bourlat, S.J.; Borja, A.; Gilbert, J.; Taylor, M.I.; Davies, N.; Weisberg, S.B.; Glöckner, F.O. Genomics in marine monitoring: New opportunities for assessing marine health status. Mar. Pollut. Bull. 2013, 74, 19–31. [Google Scholar] [CrossRef]
- Valentini, A.; Taberlet, P.; Miaud, C.; Civade, R.; Herder, J.; Thomsen, P.F.; Bellemain, E.; Besnard, A.; Coissac, E.; Boyer, F.; et al. Next-generation monitoring of aquatic biodiversity using environmental DNA barcoding. Mol. Ecol. 2016, 25, 929–942. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, J.L.; Kelly, R.P.; Shelton, A.O.; Samhouri, J.F.; Lowell, N.C.; Williams, G.D. Spatial distribution of environmental DNA in a nearshore marine habitat. Peer J. 2017, 5, e3044. [Google Scholar] [CrossRef] [PubMed]
- Djurhuus, A.; Pitz, K.; Sawaya, N.A.; Rojas-Márquez, J.; Michaud, B.; Montes, E.; Muller-Karger, F.; Breitbart, M. Evaluation of marine zooplankton community structure through environmental DNA metabarcoding. Limnol. Oceanogr. Methods 2018, 16, 209–221. [Google Scholar] [CrossRef]
- Hajibabaei, M.; Smith, M.A.; Janzen, D.H.; Rodriguez, J.J.; Whitfield, J.B.; Hebert, P.D. A minimalist barcode can identify a specimen whose DNA is degraded. Mol. Ecol. Notes 2006, 6, 959–964. [Google Scholar] [CrossRef]
- Darling, J.A.; Mahon, A.R. From molecules to management: Adopting DNA-based methods for monitoring biological invasions in aquatic environments. Environ. Res. 2011, 111, 978–988. [Google Scholar] [CrossRef]
- Victor, S.; Golbuu, Y.; Yukihira, H.; van Woesik, R. Acropora size-frequency distributions reflect spatially variable conditions on coral reefs of Palau. Bull. Mar. Sci. 2009, 85, 149–157. [Google Scholar]
- Wallace, C.C. Reproduction, recruitment and fragmentation in nine sympatric species of the coral genus Acropora. Mar. Biol. 1985, 88, 217–233. [Google Scholar] [CrossRef]
- Lewis, J.B. Recruitment, growth and mortality of a coral-inhabiting barnacle Megabalanus stultus (Darwin) upon the hydrocoral Millepora complanata Lamarck. J. Exp. Mar. Biol. Ecol. 1992, 162, 51–64. [Google Scholar] [CrossRef]
- Rothhaupt, K. Differences in particle size-dependent feeding efficiencies of closely related rotifer species. Limnol. Oceanogr. 1990, 35, 16–23. [Google Scholar] [CrossRef]
- Thouvenot, A.; Debroas, D.; Richardot, M.; Devaux, J. Impact of natural metazooplankton assemblage on planktonic microbial communities in a newly flooded reservoir. J. Plankton Res. 1999, 21, 179–199. [Google Scholar] [CrossRef][Green Version]
- Mohr, S.; Adrian, R. Reproductive success of the rotifer Brachionus calyciflorus feeding on ciliates and flagellates of different trophic modes. Freshwater Biol. 2002, 47, 1832–1839. [Google Scholar] [CrossRef]
- Devetter, M.; Sed’a, J. Rotifer fecundity in relation to components of microbial food web in a eutrophic reservoir. Hydrobiologia 2003, 504, 167–175. [Google Scholar] [CrossRef]
- Zhan, A.; Hulak, M.; Sylvester, F.; Huang, X.; Adebayo, A.A.; Abbott, C.L.; Adamowicz, S.J.; Heath, D.D. High sensitivity of 454 pyrosequencing for detection of rare species in aquatic communities. Methods Ecol. Evol. 2013, 4, 558–565. [Google Scholar] [CrossRef]
- Zaiko, A.; Samuiloviene, A.; Ardura, A.; Garcia-Vazquez, E. Metabarcoding approach for nonindigenous species surveillance in marine coastal waters. Mar. Pollut. Bull. 2015, 100, 53–59. [Google Scholar] [CrossRef]
- Comtet, T.; Sandionigi, A.; Viard, F.; Casiraghi, M. DNA (meta) barcoding of biological invasions: A powerful tool to elucidate invasion processes and help managing aliens. Biol. Invasions 2015, 17, 905–922. [Google Scholar] [CrossRef]
- Kelly, R.P.; Port, J.A.; Yamahara, K.M.; Martone, R.G.; Lowell, N.; Thomsen, P.F.; Mach, M.E.; Bennett, M. Environmental monitoring. Harnessing DNA to improve environmental management. Science 2014, 344, 1455–1456. [Google Scholar] [CrossRef]
| Variables | Temp | Salinity | EC | TP | TN | TOC | TIC | TC | Chla |
|---|---|---|---|---|---|---|---|---|---|
| Temp | 1 | −0.192 | −0.216 | 0.505 | 0.464 | −0.599 | −0.275 | −0.220 | 0.422 |
| Salinity | −0.192 | 1 | 0.996 | −0.586 | −0.532 | 0.301 | 0.435 | 0.486 | 0.006 |
| EC | −0.216 | 0.996 | 1 | −0.615 | −0.564 | 0.325 | 0.440 | 0.478 | −0.034 |
| TP | 0.505 | −0.586 | −0.615 | 1 | 0.904 | −0.505 | −0.634 | −0.715 | 0.192 |
| TN | 0.464 | −0.532 | −0.564 | 0.904 | 1 | −0.424 | −0.680 | −0.645 | 0.200 |
| TOC | −0.599 | 0.301 | 0.325 | −0.505 | −0.424 | 1 | 0.127 | 0.355 | −0.248 |
| TIC | −0.275 | 0.435 | 0.440 | −0.634 | −0.680 | 0.127 | 1 | 0.877 | 0.195 |
| TC | −0.220 | 0.486 | 0.478 | −0.715 | −0.645 | 0.355 | 0.877 | 1 | 0.263 |
| Chl−a | 0.422 | 0.006 | −0.034 | 0.192 | 0.200 | −0.248 | 0.195 | 0.263 | 1 |
| Marine | −0.197 | 0.879 | 0.878 | −0.343 | −0.195 | 0.314 | 0.051 | 0.151 | −0.082 |
| Fresh | 0.123 | −0.888 | −0.884 | 0.332 | 0.199 | −0.286 | −0.061 | −0.175 | 0.024 |
| Estuarine | 0.269 | −0.856 | −0.856 | 0.349 | 0.188 | −0.337 | −0.041 | −0.123 | 0.139 |
| First consumer | 0.195 | −0.283 | −0.271 | 0.164 | 0.120 | 0.152 | −0.329 | −0.269 | 0.159 |
| Second consumer | 0.011 | 0.387 | 0.373 | −0.329 | −0.070 | 0.159 | 0.239 | 0.346 | 0.026 |
| Producer | −0.054 | −0.319 | −0.307 | 0.288 | 0.040 | −0.194 | −0.162 | −0.281 | −0.064 |
| Symbiotic | 0.032 | −0.335 | −0.326 | 0.266 | 0.182 | 0.158 | −0.298 | −0.301 | 0.220 |
| Non-indigenous | −0.002 | −0.348 | −0.331 | 0.284 | 0.021 | −0.137 | −0.195 | −0.295 | −0.008 |
| Indigenous | 0.002 | 0.348 | 0.331 | −0.284 | −0.021 | 0.137 | 0.195 | 0.295 | 0.008 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jo, H.; Kim, D.-K.; Park, K.; Kwak, I.-S. Discrimination of Spatial Distribution of Aquatic Organisms in a Coastal Ecosystem Using eDNA. Appl. Sci. 2019, 9, 3450. https://doi.org/10.3390/app9173450
Jo H, Kim D-K, Park K, Kwak I-S. Discrimination of Spatial Distribution of Aquatic Organisms in a Coastal Ecosystem Using eDNA. Applied Sciences. 2019; 9(17):3450. https://doi.org/10.3390/app9173450
Chicago/Turabian StyleJo, Hyunbin, Dong-Kyun Kim, Kiyun Park, and Ihn-Sil Kwak. 2019. "Discrimination of Spatial Distribution of Aquatic Organisms in a Coastal Ecosystem Using eDNA" Applied Sciences 9, no. 17: 3450. https://doi.org/10.3390/app9173450
APA StyleJo, H., Kim, D.-K., Park, K., & Kwak, I.-S. (2019). Discrimination of Spatial Distribution of Aquatic Organisms in a Coastal Ecosystem Using eDNA. Applied Sciences, 9(17), 3450. https://doi.org/10.3390/app9173450







