A Dynamic Programmable Network for Large-Scale Scientific Data Transfer Using AmoebaNet
Abstract
1. Introduction
2. Proposed Solution
2.1. Overview of AmoebaNet Solution
- To provide a dynamic programmability feature for underlying network resources, AMQP interface [14] was provided. The JSON-RPC style based communication system of AMQP will allow an application to interact with the underlying networks in a flexible manner.
- QoS-guaranteed path computation service along with route selection.
- Provide “Application-aware” networking services.
- Support differentiated service to ensure the required QoS services for priority traffic.
- Provision the QoS-guaranteed network path between two ends with a fastest possible time.
- REST-based network initialization and configuration.
- Path resiliency.
- Advanced reservation system.
- Service locking/unlocking.
- By realizing network as a service, AmoebaNet supports various extreme-scale scientific applications and provides the rich and powerful set of dynamic programming primitives. These dynamic programming primitives make it possible for scientific applications to program the networks in more flexible ways. AmoebaNet also provides run-time programming capability for bulk data transfer, and utilizes the underlying network resources in an efficient and optimum way. These supported features of AmoebaNet address the agility, automation, programmability challenges.
- To address the last mile challenges, AmoebaNet provides a newly QoS-aware end-to-end bandwidth reservation feature. It is a new rate enforcement mechanism by combining the meters and queuing based techniques and characterizes the traffic into two classes of services: best effort and priority. AmoebaNet also has a feature of a new path computation service by using the largest bottleneck bandwidth algorithm [12]. In this mechanism the bandwidth can be considered as constraints and the best available paths between two end-points can be calculated. To solve the scalability challenges of science networks, AmoebaNet uses the spoke-hub distribution model. AmoebaNet at each site supports a hub-spoke distribution model for LAN or campus network. It allows seamless integration of local networks and wide area networks. When two systems are trying to send/receive the traffic to/from two physically isolated sites, then the service of AmoebaNet carefully identifies the corresponding p2p layer-2 circuits between sites and properly multiplexed and/or de-multiplexed the traffics accordingly.
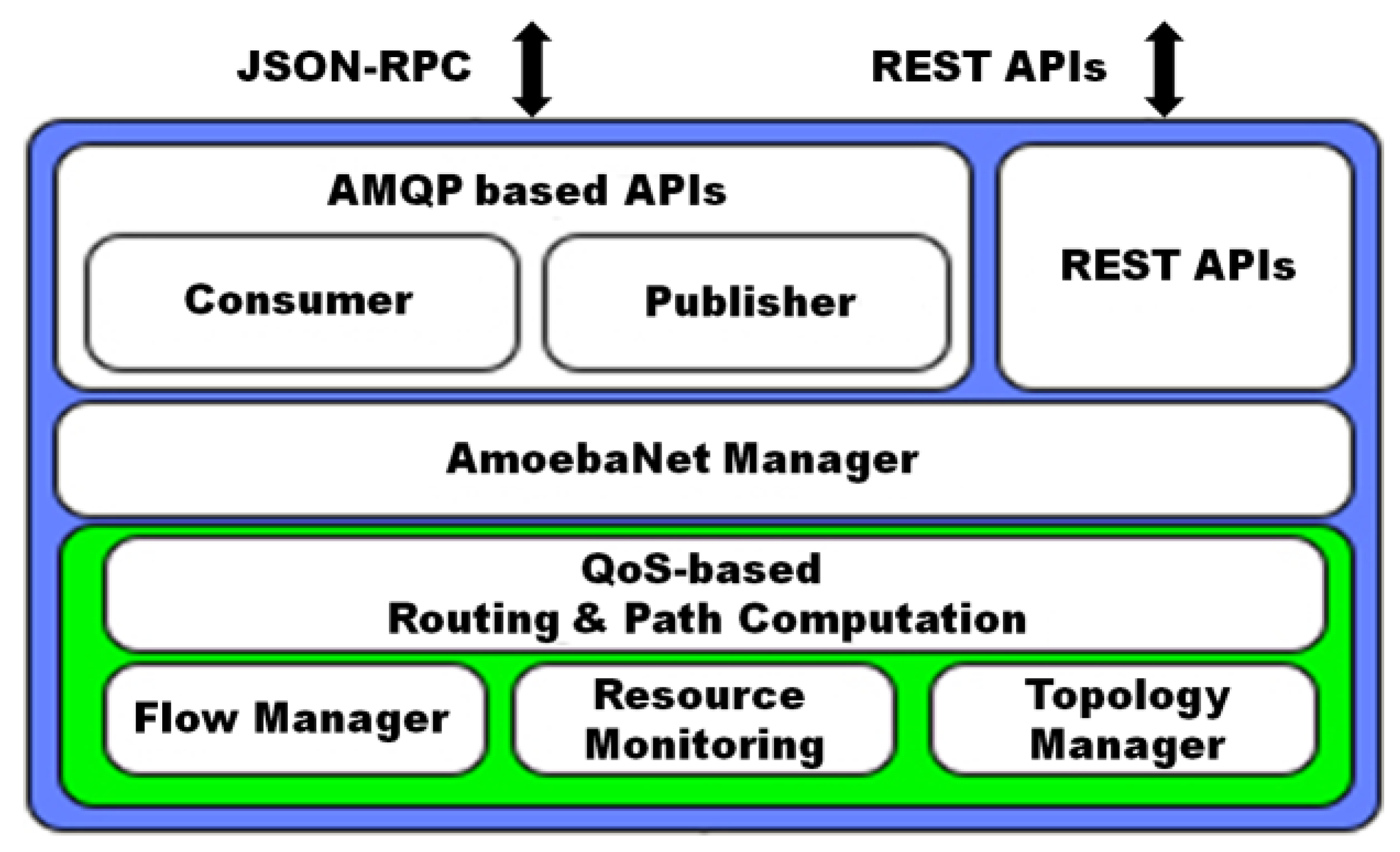
- AmoebaNet is able to provide a novel control plane logic, which consists of different sub-modules. Each sub-module of control plane architecture is dedicated for a specific task and interacts with each other. The entire control plane architecture is implemented inside ONOS SDN controller as a service and it uses north-bound and south-bound APIs to interact with applications and data plane, respectively.
- To avoid the unsynchronized access and provides advanced reservation capability to network resources, we introduced the new locking/unlocking and scheduler mechanisms in AmoebaNet’s solution.
2.2. AmoebaNet’s Programming Primitives
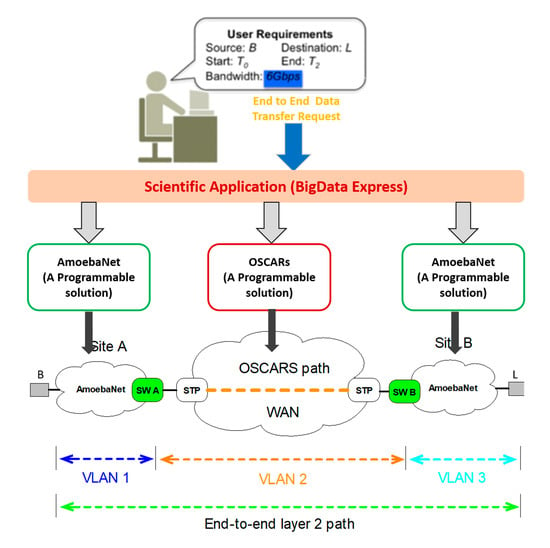
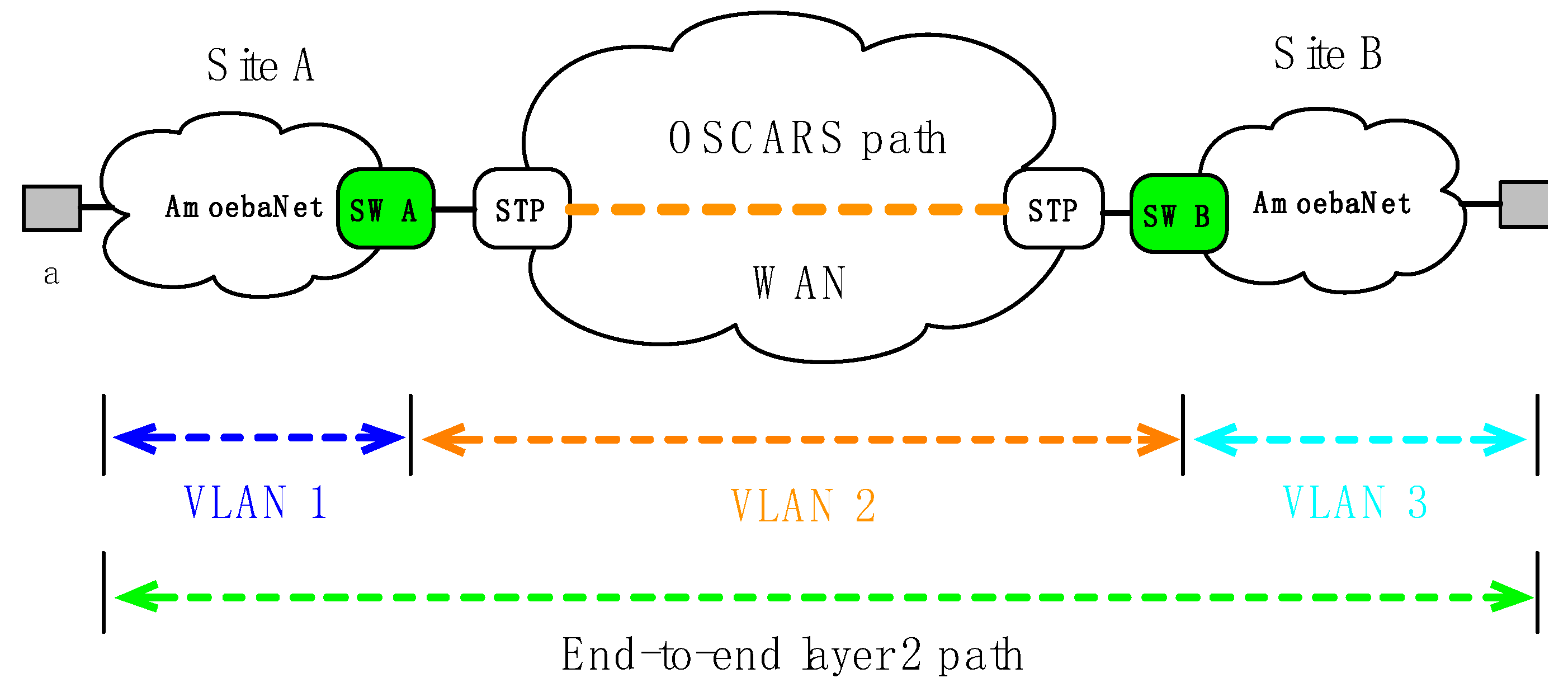
- initial_config(): By using initial configuration programming primitive, an application can configure the gateway switches’ interfaces/ports. The gateway switches are connecting the internal campus networks with the external networks. To provision the layer 2 end-to-end network paths required switching of different path segments at gateways. The initial configuration will help to setup VLANs and perform operations such as push, pop, and swap VLAN tags from internal to external network and vice versa. An example of the layer 2 end-to-end network path is shown in Figure 2, gateway switches (A and B) are configured for two sites (A and B), respectively. In fact, the path segments is made of different VLANs (VLAN1 ↔ VLAN2 and VLAN2 ↔ VLAN3).
- lock()/unlock(): It is a new feature introduced in our proposed solution, which is called lock/unlock. As per best of our knowledge, no API of ONOS is found, which is providing the same functional features to applications. AmoebaNet provides the lock/unlock mechanism to prevent the resource contention and unsynchronized access of resources among the applications. When an application sends the query request for available resources, then the entire session will be locked for that request. It keeps the lock until reservation requests are committed or the default session time expires (i.e., five seconds). Some applications (like BigData Express) require to hold the resources for a while for coordination purpose and others have not such type of requirements. Therefore, it can be claimed that AmoebaNet supports wide spectrum of applications; it provides the flexible usage of lock/unlock mechanism as per the requirements of applications. Simply, an application can enable or disable the lock/unlock during the query request. At this moment, AmoebaNet supports a very high-level coarse-grain lock/unlock, but our target is to provide the fine-grain for lock/unlock in future.
- query_virtual_netslice(): The current intent framework of ONOS can provide a functionality to install multi-to-single host or single-to-multi host paths. However, this intent framework is not providing the functionality of virtual slicing based on VLANs along with QoS-guaranteed features. In our proposed AmoebaNet solution, we added this feature by using virtual network slicing programming primitives.
- query_path(): By using the query path programming primitive, an application can view the current setting of a particular path;
- query_host_to_host(): This programming primitive helps to view the available maximum bandwidth between two DTNs in a network segment;
- query_host_to_gateway(): To check the available bandwidth from a host to a gateway switch;
- create_path(): Using this programming primitive an application can dynamically establish a layer 2/3 path between two end points. It is also necessary to specify the type of traffic and required amount of bandwidth for a specific time period;
- create_virtual_netslice(): Given a list of IPs, it creates or reserves a virtual network slice among them with a specified bandwidth and at a specified time slot;
- update_path(): Used to change the settings of installed end-to-end path; this programming primitive will seamlessly update or modify the paths information;
- update_virtual_netslice(): Used to change the settings of installed virtual slice path; it seamlessly updates or modifies the slice paths information;
- release_path(): Torn down and deleted the entries of a particular path;
- release_virtual_net_slice(): Torn down and deleted the entries of a particular virtual network slice.
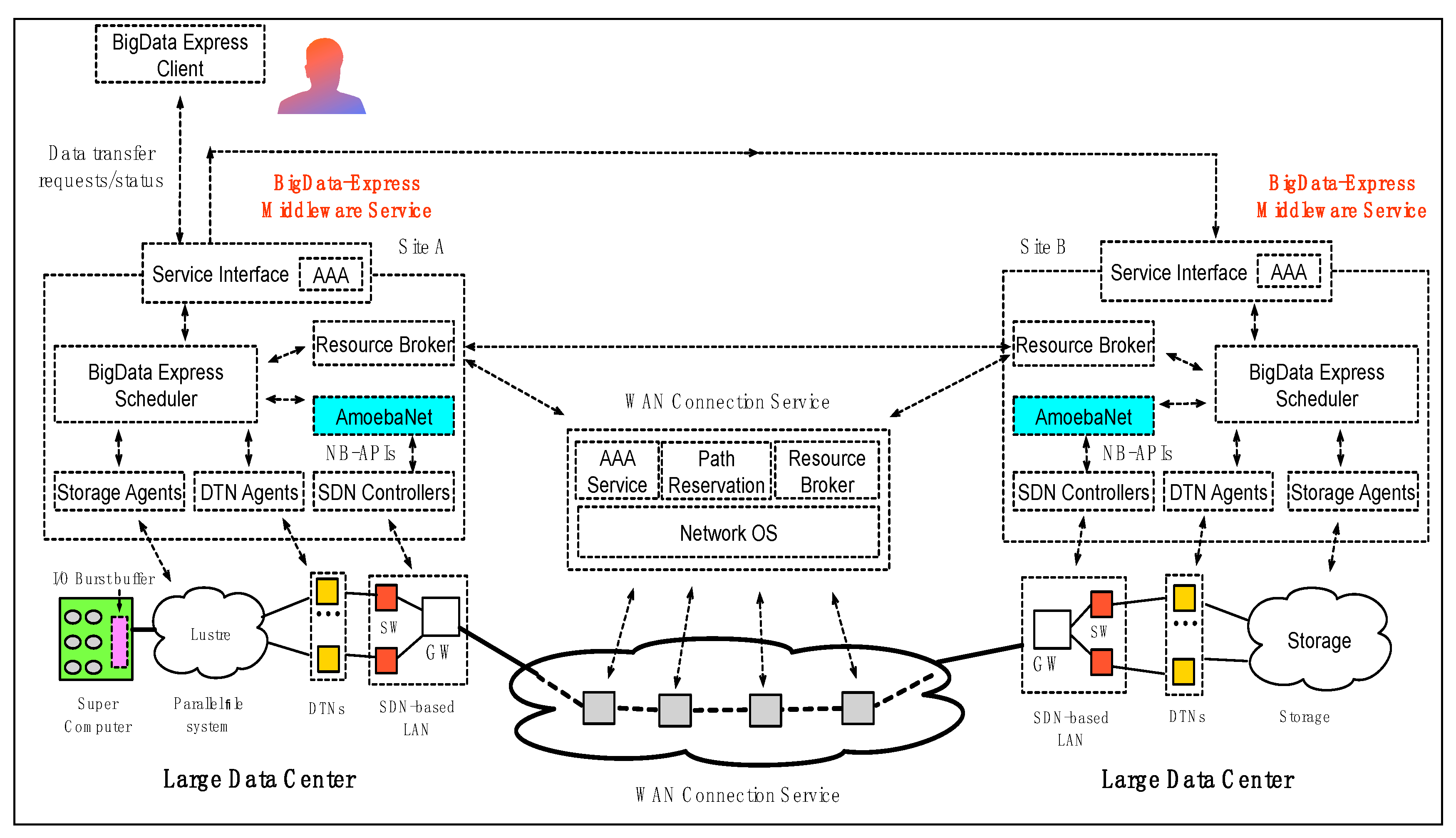
2.3. AmoebaNet Solution for the BigData Express (BDE) Project
3. Related Work
4. Evaluation and Results
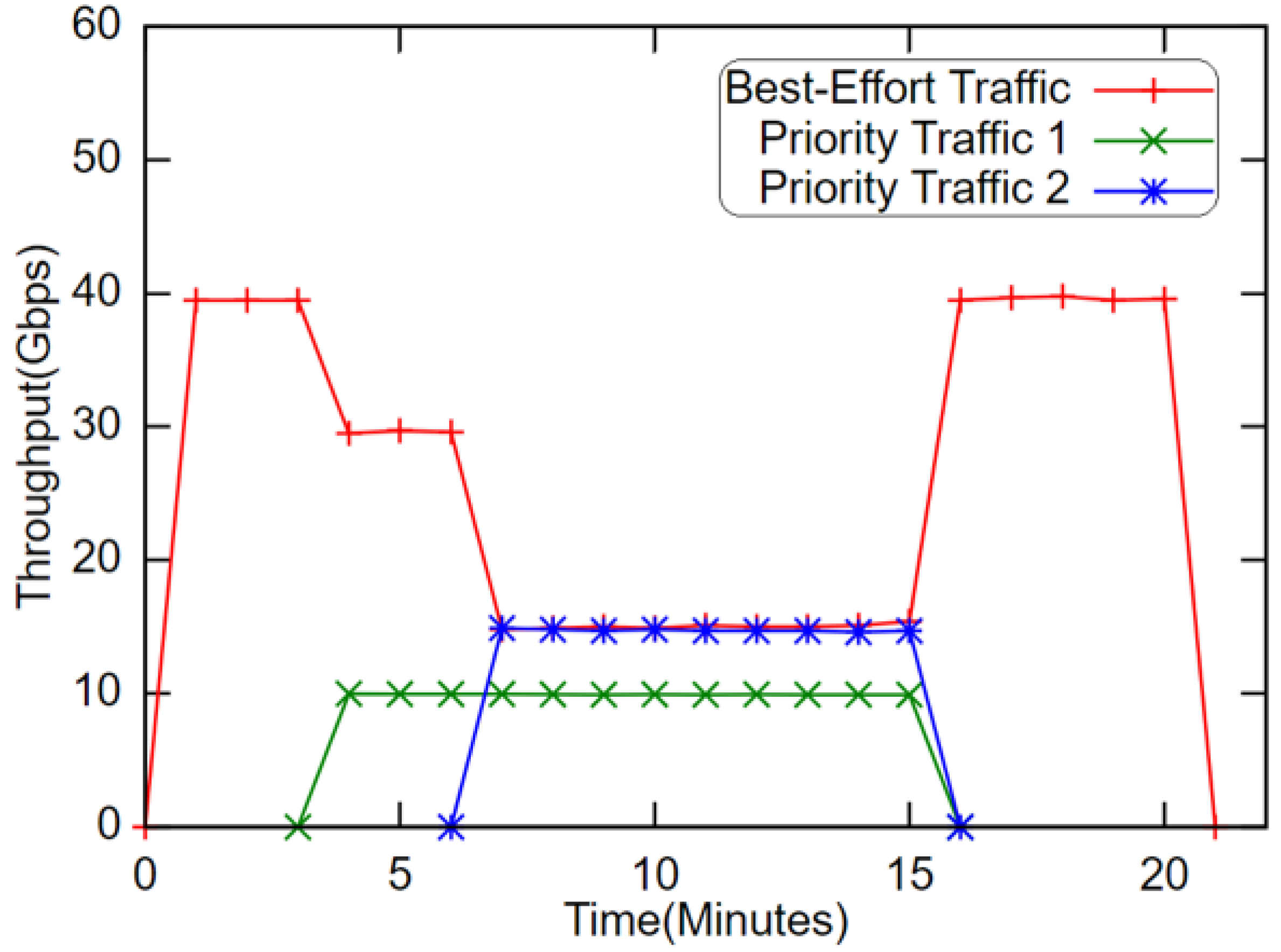
4.1. Test Case I: Extreme-Scale Data Transfer in a Single Domain
- Priority traffic 1, which is sent from 192.3.3.2 (vlan3 nic@DTN2) to 192.3.3.4 (vlan3 nic@DTN4). The data transfer starts at 4th min, and ends at 15th min, wherein the maximum bandwidth is capped at 10 Gbps.
- Priority traffic 2, which sent from 192.4.4.3 (vlan4 nic@DTN3) to 192.4.4.4 (vlan4 @DTN4). The data transfer starts at the 7th min and ends at the 18th min, wherein the maximum bandwidth is capped at 15 Gbps.
- Best-effort traffic, which is sent from 192.2.2.1 (vlan2 nic@DTN1) to 192.2.2.4 (vlan2 nic@DTN4). The data transfer starts at the 1st min and ends at the 20th min.
- (1)
- Calculate and route local LAN paths.
- (2)
- Install flow rules to set up local LAN paths.
- (3)
- Install meters and queues to enforce rate control and QoS guarantee for priority traffic.
{
“cmd”:“create_path”,
“hosts”:
[{
“startTime”: “immediate”,
“endTime”: “2019-09-07 13:00”,
“srcIp”:“192.3.3.2”,
“rate”:“10000”, // in Mbps
“dstIp”:“192.3.3.4”,
“trafficType”:“priority traffic”,
“vlanId”:“3”,
“routeType”: “host-to-host”
}]
}
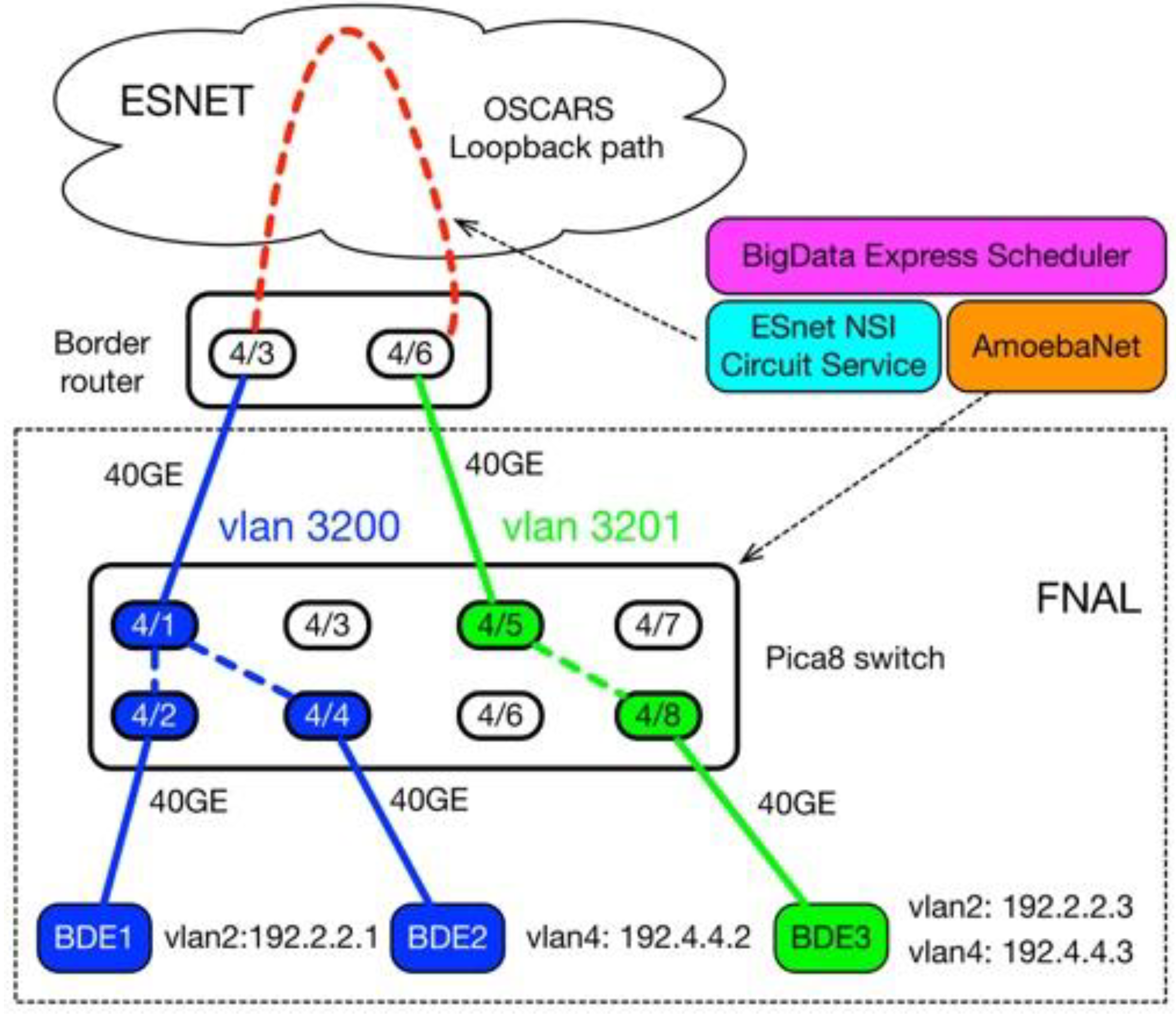
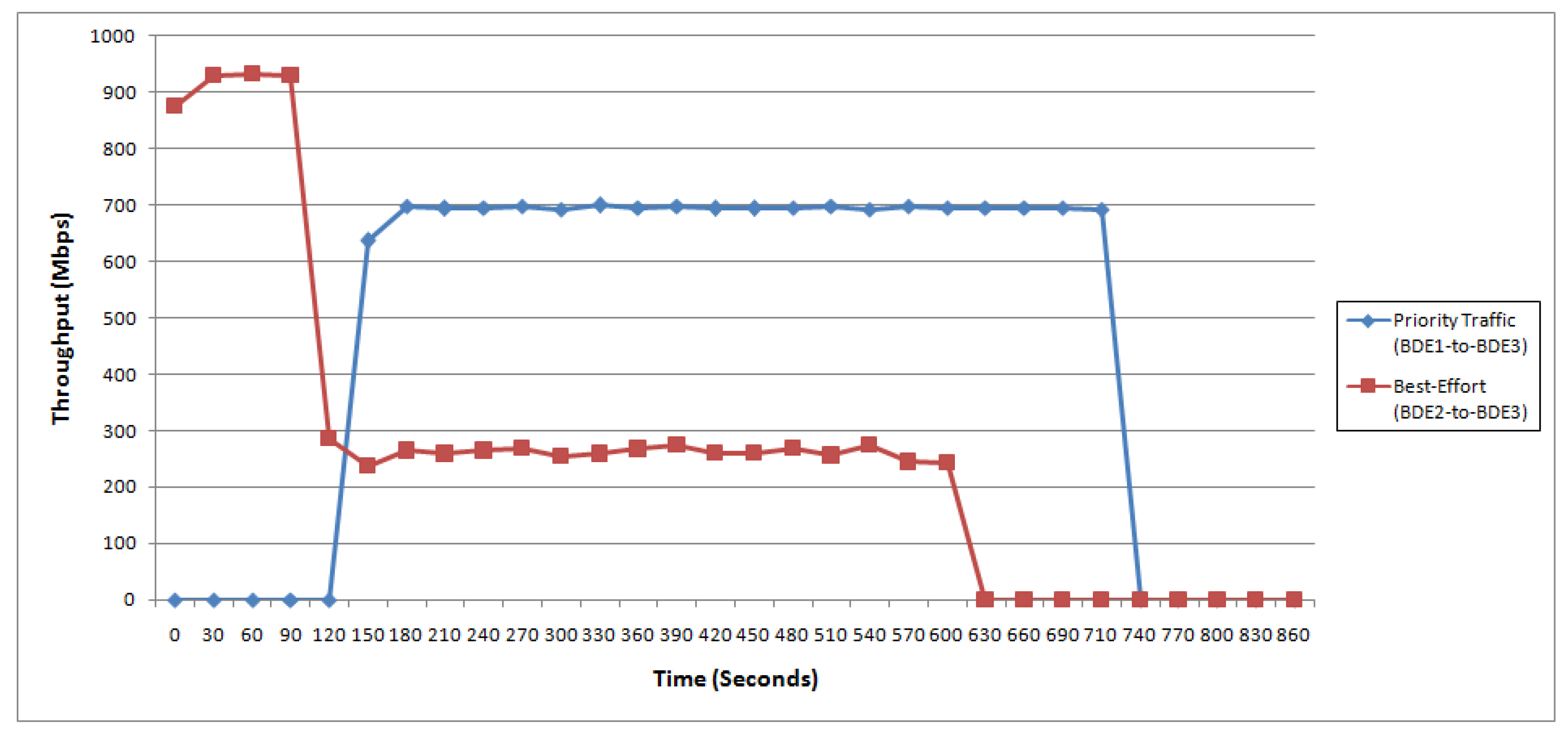
4.2. Test Case II: Cross Domain Data Transfer
- Priority traffic: In this case, we sent the data from BDE1 data transfer node (192.2.2.1 with vlan2) to BDE3 data transfer node (192.2.2.3 with vlan2). The traffic started at 120 s, and ended at 720 s, wherein the maximum bandwidth was capped at 700 Mbps.
- Best-effort traffic: In this case, data was sent from BDE2 data transfer node (192.4.4.2 with vlan4) to BDE3 data transfer node (192.4.4.3 with vlan4). The traffic started at 0 s, and ended at 600 s.
4.3. Test Case III: Cross Pacific QoS-Guaranteed Data Transfer
- FNAL sites:
- ○
- Site 1: Fermi National Accelerator Laboratory (FNAL):
- ▪
- DTNs: DTN1 (BDE1), DTN2 (BDE2), and DTN3 (BDE3).
- ▪
- Data transfer nodes equipped with: a 40GE Mellanox NIC and an Intel NVMe storage drive.
- ▪
- Switches: one SDN enabled switch (Pica8 P5101 running PicaOS).
- ▪
- ONOS SDN controller along with AmoebaNet solution.
- ○
- Site 2: Fermi National Accelerator Laboratory (FNAL S2):
- ▪
- DTNs: DTN4 (BDE4), and DTN5 (BDE-HP5).
- ▪
- Data transfer nodes equipped with: a 40GE Mellanox NIC and an Intel NVMe storage drive.
- ▪
- Switches: one SDN enabled switch (Pica8 P3930 running PicaOS).
- ▪
- ONOS SDN controller along with AmoebaNet solution.
- KISTI site:
- ○
- DTNs: DTN2, DTN3, and DTN4.
- ▪
- Data transfer nodes equipped with: a 10GE NIC and an Intel NVMe storage drive.
- ○
- Switches: one SDN enabled switch (Z91000 HP running PicaOS).
- ○
- ONOS SDN controller along with AmoebaNet solution.
- Task 1: We setup 1 Gpbs dedicated bandwidth for QoS-guaranteed end-to-end path and transferred a 372.5 GB data set from DTN2 at KISTI to BDE1 at the first logical site of Fermilab (i.e., FNAL).
- Task 2: We configured 1 Gbps dedicated bandwidth for QoS-guaranteed end-to-end path and transferred a 20 GB data set from DTN2 at KISTI to BDE3 at the first logical site of Fermilab (i.e., FNAL).
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Allcock, W.; Bresnahan, J.; Kettimuthu, R.; Link, M.; Dumitrescu, C.; Raicu, I.; Foster, I. The Globus striped GridFTP framework and server. In Proceedings of the ACM/IEEE Conference on Supercomputing; IEEE Computer Society: Washington, DC, USA, 2005; p. 54. [Google Scholar]
- BBCP. Available online: http://www.slac.stanford.edu/~abh/bbcp/ (accessed on 15 July 2018).
- Globus Online. Available online: https://www.globus.org/ (accessed on 15 July 2018).
- Dart, E.; Rotman, L.; Tierney, B.; Hester, M.; Zurawski, J. The science dmz: A network design pattern for data-intensive science. Sci. Program. 2014, 22, 173–185. [Google Scholar] [CrossRef]
- Energy Sciences Network (ESNet). Science DMZ. Available online: https://fasterdata.es.net/science-dmz/ (accessed on 5 September 2018).
- Energy Sciences Network (ESNet). Data Transfer Nodes. Available online: https://fasterdata.es.net/science-dmz/DTN/ (accessed on 5 September 2018).
- Internet2. Advanced Layer 2 Service. Available online: https://www.internet2.edu/products-services/advanced-networking/layer-2-services/ (accessed on 10 September 2018).
- Guok, C. ESNet Network Engineer, and David Robertson. In ESNet on-Demand Secure Circuits and Advance Reservation System (OSCARS); Internet2 Joint Techs Workshop: Salt Lake City, UT, USA, 2005. [Google Scholar]
- Shenker, S.; Casado, M.; Koponen, T.; McKeown, N. The future of networking, and the past of protocols. Open Netw. Summit 2011, 20, 1–30. [Google Scholar]
- McKeown, N. Software-defined networking. INFOCOM Keynote Talk 2009, 17, 30–32. [Google Scholar]
- Scott-Hayward, S.; O’Callaghan, G.; Sezer, S. SDN security: A survey. In IEEE SDN For Future Networks and Services (SDN4FNS); IEEE: Piscataway, NJ, USA, 2013; pp. 1–7. [Google Scholar]
- Shah, S.A.R.; Wu, W.; Lu, Q.; Zhang, L.; Sasidharan, S.; DeMar, P.; Noh, S.Y. AmoebaNet: An SDN-enabled network service for big data science. J. Netw. Comput. Appl. 2018, 119, 70–82. [Google Scholar] [CrossRef]
- BigData Express (BDE). Available online: http://bigdataexpress.fnal.gov/ (accessed on 5 April 2019).
- Vinoski, S. Advanced message queuing protocol. IEEE Internet Comput. 2006, 6, 87–89. [Google Scholar] [CrossRef]
- Guok, C.P.; Robertson, D.W.; Chaniotakis, E.; Thompson, M.R.; Johnston, W.; Tierney, B. A user driven dynamic circuit network implementation. In Proceedings of the 2008 IEEE Globecom Workshops, New Orleans, LA, USA, 30 November–4 December 2008; pp. 1–5. [Google Scholar]
- Roberts, G.; Kudoh, T.; Monga, I.; Sobieski, J.; MacAuley, J.; Guok, C. Nsi connection service v2.0. GFD 2014, 212, 1–119. [Google Scholar]
- Berde, P.; Gerola, M.; Hart, J.; Higuchi, Y.; Kobayashi, M.; Koide, T.; Parulkar, G. ONOS: Towards an open, distributed SDN OS. In Proceedings of the 3rd Workshop on Hot Topics in Software Defined Networking, Chicago, IL, USA, 22 August 2014; ACM: New York, NY, USA, 2014; pp. 1–6. [Google Scholar]
- Open Networking Foundation. ONOS Project. Available online: http://onosproject.org/ (accessed on 5 August 2018).
- Kreutz, D.; Ramos, F.; Verissimo, P.; Rothenberg, C.E.; Azodolmolky, S.; Uhlig, S. Software-defined networking: A comprehensive survey. arXiv, 2014; arXiv:1406.0440. [Google Scholar] [CrossRef]
- Nunes, B.A.A.; Mendonca, M.; Nguyen, X.N.; Obraczka, K.; Turletti, T. A survey of software-defined networking: Past, present, and future of programmable networks. IEEE Commun. Surv. Tutor. 2014, 16, 1617–1634. [Google Scholar] [CrossRef]
- Al-Fares, M.; Radhakrishnan, S.; Raghavan, B.; Huang, N.; Vahdat, A. Hedera: Dynamic flow scheduling for data center networks. In Nsdi; USENIX: Berkley, CA, USA, 2010; Volume 10. [Google Scholar]
- Niranjan Mysore, R.; Pamboris, A.; Farrington, N.; Huang, N.; Miri, P.; Radhakrishnan, S.; Vahdat, A. Portland: A scalable fault-tolerant layer 2 data center network fabric. In ACM SIGCOMM Computer Communication Review; No. 4; ACM: New York, NY, USA, 2009; Volume 39, pp. 39–50. [Google Scholar]
- Wang, R.; Butnariu, D.; Rexford, J. OpenFlow-Based Server Load Balancing Gone Wild. Hot-ICE 2011, 11, 12. [Google Scholar]
- Curtis, A.R.; Mogul, J.C.; Tourrilhes, J.; Yalagandula, P.; Sharma, P.; Banerjee, S. DevoFlow: Scaling flow management for high-performance networks. In ACM SIGCOMM Computer Communication Review; No. 4; ACM: New York, NY, USA, 2011; Volume 41, pp. 254–265. [Google Scholar]
- Yeganeh, S.H.; Tootoonchian, A.; Ganjali, Y. On scalability of software-defined networking. IEEE Commun. Mag. 2013, 51, 136–141. [Google Scholar] [CrossRef]
- Hong, C.Y.; Kandula, S.; Mahajan, R.; Zhang, M.; Gill, V.; Nanduri, M.; Wattenhofer, R. Achieving high utilization with software-driven WAN. In ACM SIGCOMM Computer Communication Review; No. 4; ACM: New York, NY, USA, 2013; Volume 43, pp. 15–26. [Google Scholar]
- Jain, S.; Kumar, A.; Mandal, S.; Ong, J.; Poutievski, L.; Singh, A.; Zolla, J. B4: Experience with a globally-deployed software defined WAN. In ACM SIGCOMM Computer Communication Review; No. 4; ACM: New York, NY, USA, 2013; Volume 43, pp. 3–14. [Google Scholar]
- Yan, Z.; Zhang, P.; Vasilakos, A.V. A security and trust framework for virtualized networks and software-defined networking. Secur. Commun. Netw. 2016, 9, 3059–3069. [Google Scholar] [CrossRef]
- Shu, Z.; Wan, J.; Li, D.; Lin, J.; Vasilakos, A.V.; Imran, M. Security in software-defined networking: Threats and countermeasures. Mob. Netw. Appl. 2016, 21, 764–776. [Google Scholar] [CrossRef]
- Shin, S.W.; Porras, P.; Yegneswara, V.; Fong, M.; Gu, G.; Tyson, M. Fresco: Modular composable security services for software-defined networks. In Proceedings of the 20th Annual Network & Distributed System Security Symposium, San Diego, CA, USA, 25–27 February 2013. [Google Scholar]
- Kreutz, D.; Ramos, F.; Verissimo, P. Towards secure and dependable software-defined networks. In Proceedings of the 2nd ACM SIGCOMM Workshop on Hot Topics in Software Defined Networking, Hong Kong, China, 16 August 2013; ACM: New York, NY, USA, 2013; pp. 55–60. [Google Scholar]
- Bosshart, P.; Daly, D.; Gibb, G.; Izzard, M.; McKeown, N.; Rexford, J.; Walker, D. P4: Programming protocol-independent packet processors. ACM SIGCOMM Comput. Commun. Rev. 2014, 44, 87–95. [Google Scholar] [CrossRef]
- Kim, C.; Sivaraman, A.; Katta, N.; Bas, A.; Dixit, A.; Wobker, L.J. In-band network telemetry via programmable dataplanes. In ACM SIGCOMM; ACM: New York, NY, USA, 2015. [Google Scholar]
- Popescu, D.A.; Antichi, G.; Moore, A.W. Enabling fast hierarchical heavy hitter detection using programmable data planes. SOSR 2017, 191–192. [Google Scholar] [CrossRef]
- Sivaraman, A.; Kim, C.; Krishnamoorthy, R.; Dixit, A.; Budiu, M. Dc. p4: Programming the forwarding plane of a data-center switch. In Proceedings of the 1st ACM SIGCOMM Symposium on Software Defined Networking Research, Santa Clara, CA, USA, 17–18 June 2015; ACM: New York, NY, USA, 2015; p. 2. [Google Scholar]
- Laki, S.; Horpácsi, D.; Vörös, P.; Kitlei, R.; Leskó, D.; Tejfel, M. High speed packet forwarding compiled from protocol independent data plane specifications. In Proceedings of the ACM SIGCOMM Conference, Florianopolis, Brazil, 22–26 August 2016; ACM: New York, NY, USA, 2016; pp. 629–630. [Google Scholar]
- Pacific Research Platform (PRP). Available online: http://prp.ucsd.edu/ (accessed on 15 September 2018).
- Geant AutoBAHN. Available online: https://forge.geant.net/forge/display/autobahn/Home (accessed on 15 October 2018).
- Zurawski, J.; Boyd, E.; Lehman, T.; McKee, S.; Mughal, A.; Newman, H.; Yang, X. Scientific data movement enabled by the DYNES instrument. In Proceedings of the First International Workshop on Network-Aware Data Management, Seattle, DC, USA, 14 November 2011; ACM: New York, NY, USA, 2011; pp. 41–48. [Google Scholar]
- Hazlewood, V.; Benninger, K.; Peterson, G.; Charcalla, J.; Sparks, B.; Hanley, J.; Lappa, J. Developing applications with networking capabilities via end-to-end SDN (DANCES). In Proceedings of the XSEDE16 Conference on Diversity, Big Data, and Science at Scale, Miami, FL, USA, 17–21 July 2016; ACM: New York, NY, USA, 2016; p. 29. [Google Scholar]
- Liu, Y.; Liu, Z.; Kettimuthu, R.; Rao, N.S.; Chen, Z.; Foster, I. Data transfer between scientific facilities–bottleneck analysis, insights, and optimizations. In Proceedings of the 19th IEEE/ACM International Symposium on Cluster, Cloud and Grid Computing (CCGRID), Larnaca, Cyprus, 14–17 May 2019; pp. 122–131. [Google Scholar]
- Liu, Z.; Kettimuthu, R.; Foster, I.; Rao, N.S. Cross-geography scientific data transferring trends and behavior. In Proceedings of the 27th International Symposium on High-Performance Parallel and Distributed Computing, Tempe, Arizona, 11–15 June 2018; ACM: New York, NY, USA, 2018; pp. 267–278. [Google Scholar]



© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shah, S.A.R.; Noh, S.-Y. A Dynamic Programmable Network for Large-Scale Scientific Data Transfer Using AmoebaNet. Appl. Sci. 2019, 9, 4541. https://doi.org/10.3390/app9214541
Shah SAR, Noh S-Y. A Dynamic Programmable Network for Large-Scale Scientific Data Transfer Using AmoebaNet. Applied Sciences. 2019; 9(21):4541. https://doi.org/10.3390/app9214541
Chicago/Turabian StyleShah, Syed Asif Raza, and Seo-Young Noh. 2019. "A Dynamic Programmable Network for Large-Scale Scientific Data Transfer Using AmoebaNet" Applied Sciences 9, no. 21: 4541. https://doi.org/10.3390/app9214541
APA StyleShah, S. A. R., & Noh, S.-Y. (2019). A Dynamic Programmable Network for Large-Scale Scientific Data Transfer Using AmoebaNet. Applied Sciences, 9(21), 4541. https://doi.org/10.3390/app9214541