In Vitro Probiotic Potential of Lactic Acid Bacteria Isolated from Aguamiel and Pulque and Antibacterial Activity Against Pathogens
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation of Lactic Acid Bacteria
2.2. Characterization of Isolates as Potential Probiotics
2.2.1. Resistance to Antibiotics of Isolated Strains
2.2.2. Survival Assessment of LAB Strains under Simulated Gastrointestinal Tract (GIT) Conditions
2.2.3. Antimicrobial Activity
2.3. Effect of LAB on the Growth of Helicobacter pylori Strains
2.4. Identification of Genus and Species using PCR
2.5. Statistical Analysis
3. Results
3.1. Isolation of LAB
3.2. Characterization of Isolates as Potential Probiotics
3.2.1. Resistance of LAB Isolated Strains to Antibiotics
3.2.2. Survival of LAB Strains under Simulated Human Gastrointestinal Tract (GIT) Conditions
3.2.3. Antimicrobial Activity
3.3. Effect of LAB on Helicobacter pylori Growth
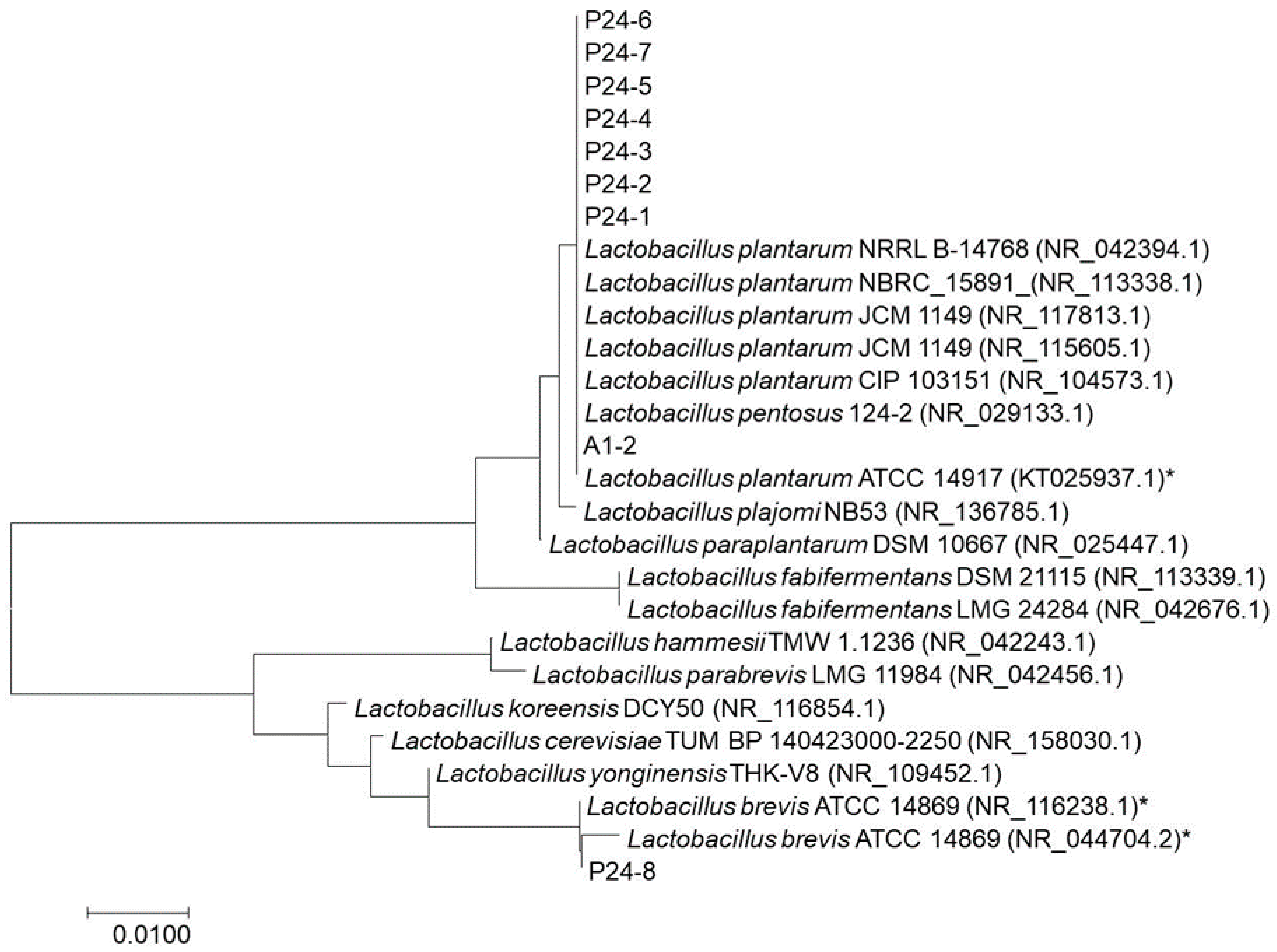
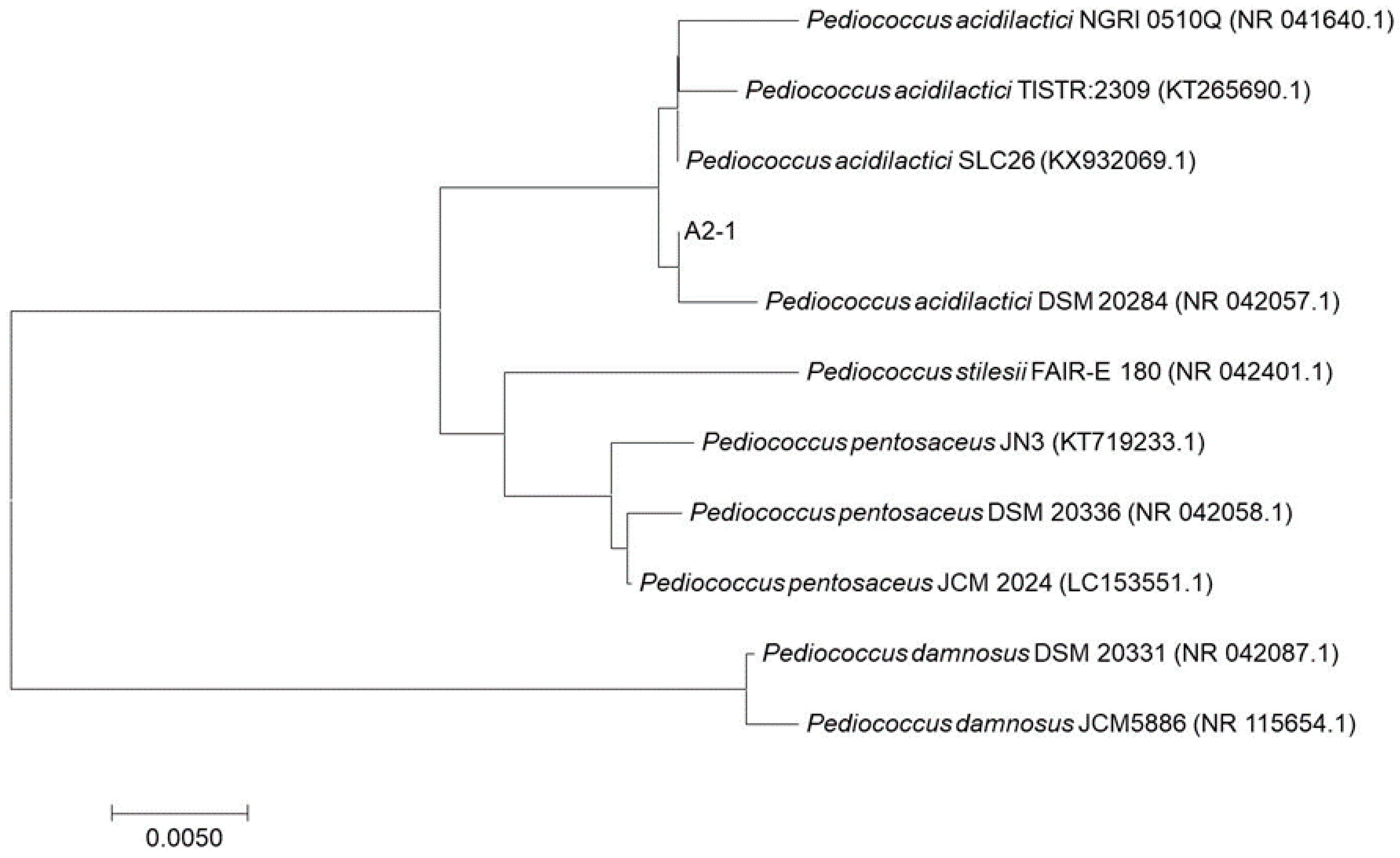
3.4. Genus and Species Identifications using PCR
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Escalante, A.; Giles-Gómez, M.; Hernández, G.; Córdova-Aguilar, M.S.; López-Munguía, A.; Gosset, G.; Bolívar, F. Analysis of bacterial community during the fermentation of pulque, a traditional Mexican alcoholic beverage, using a polyphasic approach. Int. J. Food Microbiol. 2008, 124, 126–134. [Google Scholar] [CrossRef]
- Valadez-Blanco, R.; Bravo-Villa, G.; Santos-Sánchez, N.F.; Velasco-Almendarez, S.I.; Montville, T.J. The artisanal production of pulque, a traditional beverage of the Mexican highlands. Probiotics Antimicro. Prot. 2012, 4, 140–144. [Google Scholar]
- Cruz-Guerrero, A.E.; Olvera, J.L.; García-Garibay, M.; Gómez-Ruiz, L. Inulinase-hyperproducing strains of Kluyveromyces sp. isolated from aguamiel (Agave sap) and pulque. World J. Microbiol. Biotechnol. 2006, 22, 115. [Google Scholar] [CrossRef]
- Escalante, A.; Flores, M.E.; Martínez, A.; López-Munguía, A.; Bolívar, F.; Gosset, G. Characterization of bacterial diversity in pulque, a traditional Mexican alcoholic fermented beverage, as determined by 16S rDNA analysis. FEMS Microbiol. Lett. 2004, 235, 273–279. [Google Scholar] [CrossRef]
- Angmo, K.; Kumari, A.; Bhalla, T.C. Probiotic characterization of lactic acid bacteria isolated from fermented foods and beverage of Ladakh. LWT-Food Sci. Technol. 2016, 66, 428–435. [Google Scholar] [CrossRef]
- Bengoa, A.A.; Llamas, M.G.; Iraporda, C.; Dueñas, M.T.; Abraham, A.G.; Garrote, G.L. Impact of growth temperature on exopolysaccharide production and probiotic properties of Lactobacillus paracasei strains isolated from kefir grains. Food Microbiol. 2018, 69, 212–218. [Google Scholar] [CrossRef]
- Guo, X.H.; Kim, J.M.; Nam, H.M.; Park, S.Y.; Kim, J.M. Screening lactic acid bacteria from swine origins for multistrain probiotics based on in vitro functional properties. Anaerobe 2010, 16, 321–326. [Google Scholar] [CrossRef]
- Bao, Y.; Zhang, Y.; Zhang, Y.; Liu, Y.; Wang, S.; Dong, X.; Wang, Y.; Zhang, H. Screening of potential probiotic properties of Lactobacillus fermentum isolated from traditional dairy products. Food Control 2010, 21, 695–701. [Google Scholar] [CrossRef]
- Lee, N.K.; Lee, E.K.; Paik, H.D. Potential probiotic properties of phytase-producing Lactobacillus salivarius FC113. Ann. Microbiol. 2013, 63, 555–560. [Google Scholar] [CrossRef]
- Pieniz, S.; Andreazza, R.; Anghinoni, T.; Camargo, F.; Brandelli, A. Probiotic potential, antimicrobial and antioxidant activities of Enterococcus durans strain LAB18s. Food Control 2014, 37, 251–256. [Google Scholar] [CrossRef]
- Cats, A.; Kuipers, E.J.; Bosschaert, M.A.R.; Pot, R.G.J.; Vandenbroucke-Grauls, C.M.J.E.; Kusters, J.G. Effect of frequent consumption of a Lactobacillus casei-containing milk drink in Helicobacter pylori-colonized subjects. Aliment. Pharm. Ther. 2003, 17, 429–435. [Google Scholar] [CrossRef]
- Chen, X.; Tian, F.; Liu, X.; Zhao, J.; Zhang, H.P.; Zhang, H.; Chen, W. In vitro screening of lactobacilli with antagonistic activity against Helicobacter pylori from traditionally fermented foods. J. Dairy Sci. 2010, 93, 5627–5634. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.Q.; Su, T.; Fan, J.G.; Lu, Y.X.; Zheng, P.; Li, X.H.; Guo, C.Y.; Xu, P.; Gong, Y.F.; Li, Z.S. Adjuvant probiotics improve the eradication effect of triple therapy for Helicobacter pylori infection. World J. Gastroenterol. 2012, 18, 6302. [Google Scholar] [CrossRef] [PubMed]
- Kao, C.Y.; Sheu, B.S.; Wu, J.J. Helicobacter pylori infection: An overview of bacterial virulence factors and pathogenesis. Biomedical J. 2016, 39, 14–23. [Google Scholar] [CrossRef] [PubMed]
- Emara, M.H.; Elhawari, S.A.; Yousef, S.; Radwan, M.I.; Abdel-Aziz, H.R. Emerging role of probiotics in the management of Helicobacter pylori infection: Histopathologic Perspectives. Helicobacter 2016, 21, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.W.; Shim, J.M.; Park, S.K.; Heo, H.J.; Kim, H.J.; Ham, K.S.; Kim, J.H. Isolation of lactic acid bacteria with probiotic potentials from kimchi, traditional Korean fermented vegetable. LWT-Food Sci. Technol. 2016, 71, 130–137. [Google Scholar] [CrossRef]
- Mojgani, N.; Hussaini, F.; Vaseji, N. Characterization of indigenous Lactobacillus strains for probiotic properties. Jundishapur J. Microb. 2015, 8, e17523. [Google Scholar] [CrossRef] [PubMed]
- D’Aimmo, M.R.; Modesto, M.; Biavati, B. Antibiotic resistance of lactic acid bacteria and Bifidobacterium spp. isolated from dairy and pharmaceutical products. Int. J. Food Microbiol. 2007, 115, 35–42. [Google Scholar] [CrossRef]
- Fraqueza, M.J. Antibiotic resistance of lactic acid bacteria isolated from dry-fermented sausages. Int. J. Food Microbiol. 2015, 212, 76–88. [Google Scholar] [CrossRef]
- Toomey, N.; Bolton, D.; Fanning, S. Characterisation and transferability of antibiotic resistance genes from lactic acid bacteria isolated from Irish pork and beef abattoirs. Res. Microbiol. 2010, 161, 127–135. [Google Scholar] [CrossRef]
- Zhao, T.; Doyle, M.P.; Zhao, P. Control of Listeria monocytogenes in a biofilm by competitive-exclusion microorganisms. Appl. Environ. Microb. 2004, 70, 3996–4003. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- Walsh, C. Antibiotics: Actions, Origins, Resistance; ASM Press: Washington, DC, USA, 2003; pp. 91–106. [Google Scholar]
- Amieva, M.; Peek, R.M., Jr. Pathobiology of Helicobacter pylori–induced gastric cancer. Gastroenterology 2016, 150, 64–78. [Google Scholar] [CrossRef] [PubMed]
- Shehata, M.G.; El Sohaimy, S.A.; El-Sahn, M.A.; Youssef, M.M. Screening of isolated potential probiotic lactic acid bacteria for cholesterol lowering property and bile salt hydrolase activity. Ann. Agric. Sci. 2016, 61, 65–75. [Google Scholar] [CrossRef] [Green Version]
- Blair, J.M.; Webber, M.A.; Baylay, A.J.; Ogbolu, D.O.; Piddock, L.J. Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 2015, 13, 42. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Oh, J.H.; Alexander, L.M.; Özçam, M.; Van Pijkeren, J.P. D-Ala-D-Ala ligase as a broad-host-range counterselection marker in vancomycin-resistant lactic acid bacteria. J. Bacteriol. 2018, 200, e00607-17. [Google Scholar] [CrossRef] [PubMed]
- Vinderola, C.G.; Reinheimer, J.A. Lactic acid starter and probiotic bacteria: A comparative “in vitro” study of probiotic characteristics and biological barrier resistance. Food Res. Int. 2003, 36, 895–904. [Google Scholar] [CrossRef]
- Margolles, A.; García, L.; Sánchez, B.; Gueimonde, M.; de los Reyes-Gavilán, C.G. Characterisation of a Bifidobacterium strain with acquired resistance to cholate-a preliminary study. Int. J. Food Microbiol. 2003, 82, 191–198. [Google Scholar] [CrossRef]
- Jacobsen, C.N.; Nielsen, V.R.; Hayford, A.E.; Møller, P.L.; Michaelsen, K.F.; Paerregaard, A.; Jakobsen, M. Screening of probiotic activities of forty-seven strains of Lactobacillus spp. by in vitro techniques and evaluation of the colonization ability of five selected strains in humans. Appl. Environ. Microb. 1999, 65, 4949–4956. [Google Scholar]
- Booth, I.R. Regulation of cytoplasmic pH in bacteria. Microbiol. Rev. 1985, 49, 359. [Google Scholar]
- Matsumoto, M.; Ohishi, H.; Benno, Y. H+-ATPase activity in Bifidobacterium with special reference to acid tolerance. Int. J. Food Microbiol. 2004, 93, 109–113. [Google Scholar] [CrossRef] [PubMed]
- Miwa, T.; Esaki, H.; Umemori, J.; Hino, T. Activity of H (+)-ATPase in ruminal bacteria with special reference to acid tolerance. Appl. Environ. Microb. 1997, 63, 2155–2158. [Google Scholar]
- Casado Muñoz, M.C.; Benomar, N.; Ennahar, S.; Horvatovich, P.; Lerma, L.L.; Knapp, C.W.; Galvez, A.; Abriouel, H. Comparative proteomic analysis of a potentially probiotic Lactobacillus pentosus MP-10 for the identification of key proteins involved in antibiotic resistance and biocide tolerance. Int. J. Food Microbiol. 2016, 222, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Pérez Montoro, B.; Benomar, N.; Gómez, N.C.; Ennahar, S.; Horvatovich, P.; Knapp, C.W.; Gálvez, A.; Abriouel, H. Proteomic analysis of Lactobacillus pentosus for the identification of potential markers involved in acid resistance and their influence on other probiotic features. Food Microbiol. 2018, 72, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.H.; Yu, B.; Jang, S.H.; Tsen, H.Y. Different probiotic properties for Lactobacillus fermentum strains isolated from swine and poultry. Anaerobe 2007, 13, 107–113. [Google Scholar] [CrossRef] [PubMed]
- Bengoa, A.A.; Zavala, L.; Carasi, P.; Trejo, S.A.; Bronsoms, S.; de los Ángeles Serradell, M.; Garrote, G.L.; Abraham, A.G. Simulated gastrointestinal conditions increase adhesion ability of Lactobacillus paracasei strains isolated from kefir to Caco-2 cells and mucin. Food Res. Int. 2017, 103, 462–467. [Google Scholar] [CrossRef] [PubMed]
- Makras, L.; Triantafyllou, V.; Fayol-Messaoudi, D.; Adriany, T.; Zoumpopoulou, G.; Tsakalidou, E.; Servin, A.; De Vuyst, L. Kinetic analysis of the antibacterial activity of probiotic lactobacilli towards Salmonella enterica serovar Typhimurium reveals a role for lactic acid and other inhibitory compounds. Res. Microbiol. 2006, 157, 241–247. [Google Scholar] [CrossRef] [PubMed]
- Hirshfield, I.N.; Terzulli, S.; O’Byrne, C. Weak organic acids: a panoply of effects on bacteria. Sci. Prog. 2003, 86, 245–269. [Google Scholar] [CrossRef]
- Zhang, Z.; Tao, X.; Shah, N.P.; Wei, H. Antagonistics against pathogenic Bacillus cereus in milk fermentation by Lactobacillus plantarum ZDY2013 and its anti-adhesion effect on Caco-2 cells against pathogens. J. Dairy Sci. 2016, 99, 2666–2674. [Google Scholar] [CrossRef]
- Biswas, K.; Upadhayay, S.; Rapsang, G.F.; Joshi, S.R. Antibacterial and Synergistic Activity Against b-Lactamase-Producing Nosocomial Bacteria by Bacteriocin of LAB Isolated From Lesser Known Traditionally Fermented Products of India. HAYATI J. Biosci. 2018, 24, 87–95. [Google Scholar] [CrossRef]
- Magnusson, J.; Ström, K.; Roos, S.; Sjögren, J.; Schnürer, J. Broad and complex antifungal activity among environmental isolates of lactic acid bacteria. FEMS Microbiol. Lett. 2003, 219, 129–135. [Google Scholar] [CrossRef] [Green Version]
- De Vrese, M.; Kristen, H.; Rautenberg, P.; Laue, C.; Schrezenmeir, J. Probiotic lactobacilli and bifidobacteria in a fermented milk product with added fruit preparation reduce antibiotic associated diarrhea and Helicobacter pylori activity. J. Dairy Res. 2011, 78, 396–403. [Google Scholar] [CrossRef] [PubMed]
- Westerik, N.; Reid, G.; Sybesma, W.; Kort, R. The probiotic Lactobacillus rhamnosus for alleviation of Helicobacter pylori-associated gastric pathology in East Africa. Front. Microbiol. 2018, 9, 1873. [Google Scholar] [CrossRef] [PubMed]
- Pan, M.; Wan, C.; Xie, Q.; Huang, R.; Tao, X.; Shah, N.P.; Wei, H. Changes in gastric microbiota induced by Helicobacter pylori infection and preventive effects of Lactobacillus plantarum ZDY 2013 against such infection. J. Dairy Sci. 2016, 99, 970–981. [Google Scholar] [CrossRef] [PubMed]
- Duary, R.K.; Bhausaheb, M.A.; Batish, V.K.; Grover, S. Anti-inflammatory and immunomodulatory efficacy of indigenous probiotic Lactobacillus plantarum Lp91 in colitis mouse model. Mol. Biol. Rep. 2012, 39, 4765–4775. [Google Scholar] [CrossRef] [PubMed]
| MIC (µg/mL) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Strain | Amk | Amx | Chl | Gnt | Lvf | Spc | Ttr | Vnc |
| P24-1 | ≤1.0 | ≤1.0 | ≤1.0 | ≤1.0 | ≤1.0 | ≤1.0 | ≤1.0 | ≤100 |
| P24-2 | ≤1.0 | ≤1.0 | ≤1.0 | ≤1.0 | ≤1.0 | ≤1.0 | ≤1.0 | ≤100 |
| P24-3 | ≤1.0 | ≤1.0 | ≤1.0 | ≤1.0 | ≤1.0 | ≤1.0 | ≤1.0 | ≤100 |
| P24-4 | ≤1.0 | ≤1.0 | ≤1.0 | ≤1.0 | ≤1.0 | ≤1.0 | ≤1.0 | ≤100 |
| P24-5 | ≤1.0 | ≤1.0 | ≤1.0 | ≤1.0 | ≤1.0 | ≤1.0 | ≤1.0 | ≤100 |
| P24-6 | ≤1.0 | ≤1.0 | ≤1.0 | ≤1.0 | ≤10 | ≤1.0 | ≤1.0 | ≤100 |
| P24-7 | ≤1.0 | ≤1.0 | ≤1.0 | ≤1.0 | ≤10 | ≤1.0 | ≤1.0 | ≤100 |
| P24-8 | ≤1.0 | ≤1.0 | ≤1.0 | ≤1.0 | ≤10 | ≤1.0 | ≤10 | ≤100 |
| A1-2 | ≤1.0 | ≤1.0 | ≤1.0 | ≤1.0 | ≤10 | ≤1.0 | ≤1.0 | ≤100 |
| A2-1 | ≤1.0 | ≤1.0 | ≤1.0 | ≤1.0 | ≤10 | ≤1.0 | ≤1.0 | ≤100 |
| L. casei | ≤1.0 | ≤1.0 | ≤1.0 | ≤1.0 | ≤10 | ≤10 | ≤1.0 | ≥250 |
| Strain | Resistance to Gastric Juice (Log CFU/mL) | Resistance to Bile Salts and Pancreatin (Log CFU/mL) | Antibacterial Activity (mm) | |||||
|---|---|---|---|---|---|---|---|---|
| Control * | pH 2 and Pepsin | Survival Rate (%) | Control * | Bile Salts and Pancreatin ** | Survival Rate (%) ** | E. coli | S. aureus | |
| P24-1 | 9.2 ± 0.06 | 5.9 ± 0.01 | 64.4 | 12.5 ± 0.06 | 8.4 ± 0.01 | 66.9 | 29.0 ± 2.1 ab | 26.0 ± 2.0 ab |
| P24-2 | 9.3 ± 0.02 | 5.9 ± 0.03 | 63.2 | 11.8 ± 0.02 | 7.5 ± 0.03 | 58.8 | 33.3 ± 3.5 a | 28.3 ± 2.5 a |
| P24-3 | 9.4 ± 0.10 | 7.6 ± 0.02 | 80.7 | 11.6 ± 0.10 | 7.1 ± 0.02 | 64.5 | 28.3 ± 2.8 ab | 24.3 ± 1.5 ab |
| P24-4 | 9.0± 0.03 | 5.5 ± 0.03 | 61.5 | 11.3 ± 0.03 | 6.2 ± 0.03 | 54.3 | 19.0 ± 6.8 c | 28.7 ± 1.5 a |
| P24-5 | 9.6 ± 0.05 | 5.9 ± 0.02 | 61.5 | 12.4 ± 0.05 | 10.2 ± 0.02 | 66.0 | - | - |
| P24-6 | 9.1 ± 0.03 | 7.0 ± 0.14 | 76.5 | 12.5 ± 0.03 | 10.1 ± 0.14 | 65.0 | - | - |
| P24-7 | 9.6 ± 0.02 | 9.1 ± 0.0 | 95.0 | 12.6 ± 0.02 | 9.6 ± 0.01 | 52.5 | - | - |
| P24-8 | 9.9 ± 0.04 | 9.5 ± 0.0 | 96.3 | 12.6 ± 0.04 | 11.2 ± 0.01 | 65.3 | - | - |
| A1-2 | 8.9 ± 0.02 | 7.5 ± 0.18 | 84.2 | 12.6 ± 0.02 | 7.6 ± 0.18 | 60.8 | 30.0 ± 5.4 ab | 22.3 ± 4.0 b |
| A2-1 | 8.8 ± 0.02 | 6.4 ± 0.02 | 73.3 | 12.7 ± 0.02 | 6.1 ± 0.02 | 55.7 | 24.0 ± 1.6 bc | 21.7 ± 3.2 b |
| L. casei Shirota | 8.6 ± 0.04 | 4.4 ± 0.09 | 85.0 | 12.3 ± 0.04 | 7.8 ± 0.09 | 62.5 | 22.3 ± 1.5 bc | - |
| Helicobacter pylori | |||||
|---|---|---|---|---|---|
| Strain | ATCC 43504 | ATCC 700392 | 1L | 1B | 2 |
| P24-1 | ± | - | - | - | - |
| P24-2 | ++ | - | - | - | - |
| P24-3 | ± | - | - | - | - |
| P24-4 | ± | - | - | - | - |
| P24-5 | ± | - | - | - | - |
| P24-6 | ++ | - | ++ | - | - |
| P24-7 | ± | - | - | - | - |
| P24-8 | ++ | - | - | - | - |
| A1-2 | ± | - | ± | - | - |
| A2-1 | ± | - | - | - | - |
| L. casei | ± | - | - | - | - |
| Strain | Species Identity | % Identity * | Accession Number |
|---|---|---|---|
| P24-1 | Lactobacillus plantarum ATCC 14917 | 100 | KT025937.1 |
| Lactobacillus plantarum JCM 1149 | 100 | NR_115605.1 | |
| P24-2 | Lactobacillus plantarum JCM 1149 | 99.9 | NR_117813.1 |
| Lactobacillus pentosus 124-2 | 99.9 | NR_029133.1 | |
| P24-3 | Lactobacillus plantarum JCM 1149 | 99.8 | NR_115605.1 |
| Lactobacillus plantarum NBRC 15891 | 99.8 | NR_113338.1 | |
| P24-4 | Lactobacillus plantarum JCM 1149 | 100 | NR_115605.1 |
| Lactobacillus plantarum NBRC 15891 | 99.8 | NR_113338.1 | |
| P24-5 | Lactobacillus plantarum ATCC 14917 | 100 | KT025937.1 |
| Lactobacillus plantarum JCM 1149 | 100 | NR_115605.1 | |
| P24-6 | Lactobacillus plantarum ATCC 14917 | 100 | KT025937.1 |
| Lactobacillus plantarum JCM 1149 | 100 | NR_115605.1 | |
| P24-7 | Lactobacillus plantarum CIP 103151 | 100 | NR_104573.1 |
| Lactobacillus plantarum JCM 1149 | 100 | NR_117813.1 | |
| P24-8 | Lactobacillus brevis ATCC 14869 | 100 | NR_116238.1 |
| Lactobacillus yonginensis THK-V8 | 97.8 | NR_109452.1 | |
| A1-2 | Lactobacillus plantarum CIP 103151 | 100 | NR_104573.1 |
| Lactobacillus plantarum JCM 1149 | 100 | NR_117813.1 | |
| A2-1 | Pediococcus acidilactici DSM 20284 | 99.6 | NR_042057.1 |
| Pediococcus acidilactici NGRI 0510Q | 98.3 | NR_041640.1 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cervantes-Elizarrarás, A.; Cruz-Cansino, N.d.S.; Ramírez-Moreno, E.; Vega-Sánchez, V.; Velázquez-Guadarrama, N.; Zafra-Rojas, Q.Y.; Piloni-Martini, J. In Vitro Probiotic Potential of Lactic Acid Bacteria Isolated from Aguamiel and Pulque and Antibacterial Activity Against Pathogens. Appl. Sci. 2019, 9, 601. https://doi.org/10.3390/app9030601
Cervantes-Elizarrarás A, Cruz-Cansino NdS, Ramírez-Moreno E, Vega-Sánchez V, Velázquez-Guadarrama N, Zafra-Rojas QY, Piloni-Martini J. In Vitro Probiotic Potential of Lactic Acid Bacteria Isolated from Aguamiel and Pulque and Antibacterial Activity Against Pathogens. Applied Sciences. 2019; 9(3):601. https://doi.org/10.3390/app9030601
Chicago/Turabian StyleCervantes-Elizarrarás, Alicia, Nelly del Socorro Cruz-Cansino, Esther Ramírez-Moreno, Vicente Vega-Sánchez, Norma Velázquez-Guadarrama, Quinatzin Yadira Zafra-Rojas, and Javier Piloni-Martini. 2019. "In Vitro Probiotic Potential of Lactic Acid Bacteria Isolated from Aguamiel and Pulque and Antibacterial Activity Against Pathogens" Applied Sciences 9, no. 3: 601. https://doi.org/10.3390/app9030601
APA StyleCervantes-Elizarrarás, A., Cruz-Cansino, N. d. S., Ramírez-Moreno, E., Vega-Sánchez, V., Velázquez-Guadarrama, N., Zafra-Rojas, Q. Y., & Piloni-Martini, J. (2019). In Vitro Probiotic Potential of Lactic Acid Bacteria Isolated from Aguamiel and Pulque and Antibacterial Activity Against Pathogens. Applied Sciences, 9(3), 601. https://doi.org/10.3390/app9030601






