Molecular Mechanism Research into the Replication Capability of Nanostructures Based on Rapid Heat Cycle Molding
Abstract
:1. Introduction
2. Establishment of Interface Model and Simulation Experiment
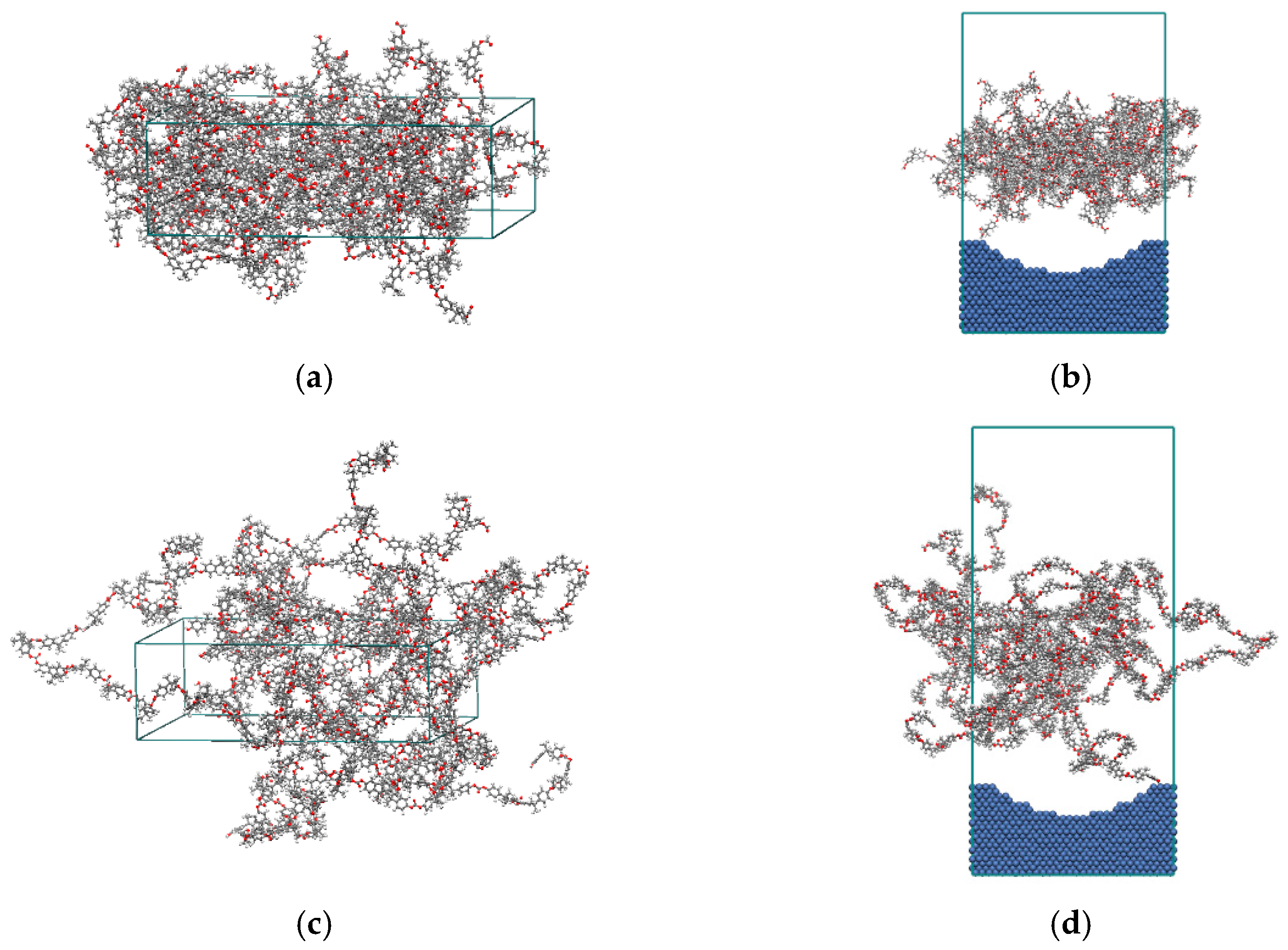
2.1. Establishment of Interface Model
2.2. Molecular Dynamics Simulation
3. Results and Discussion
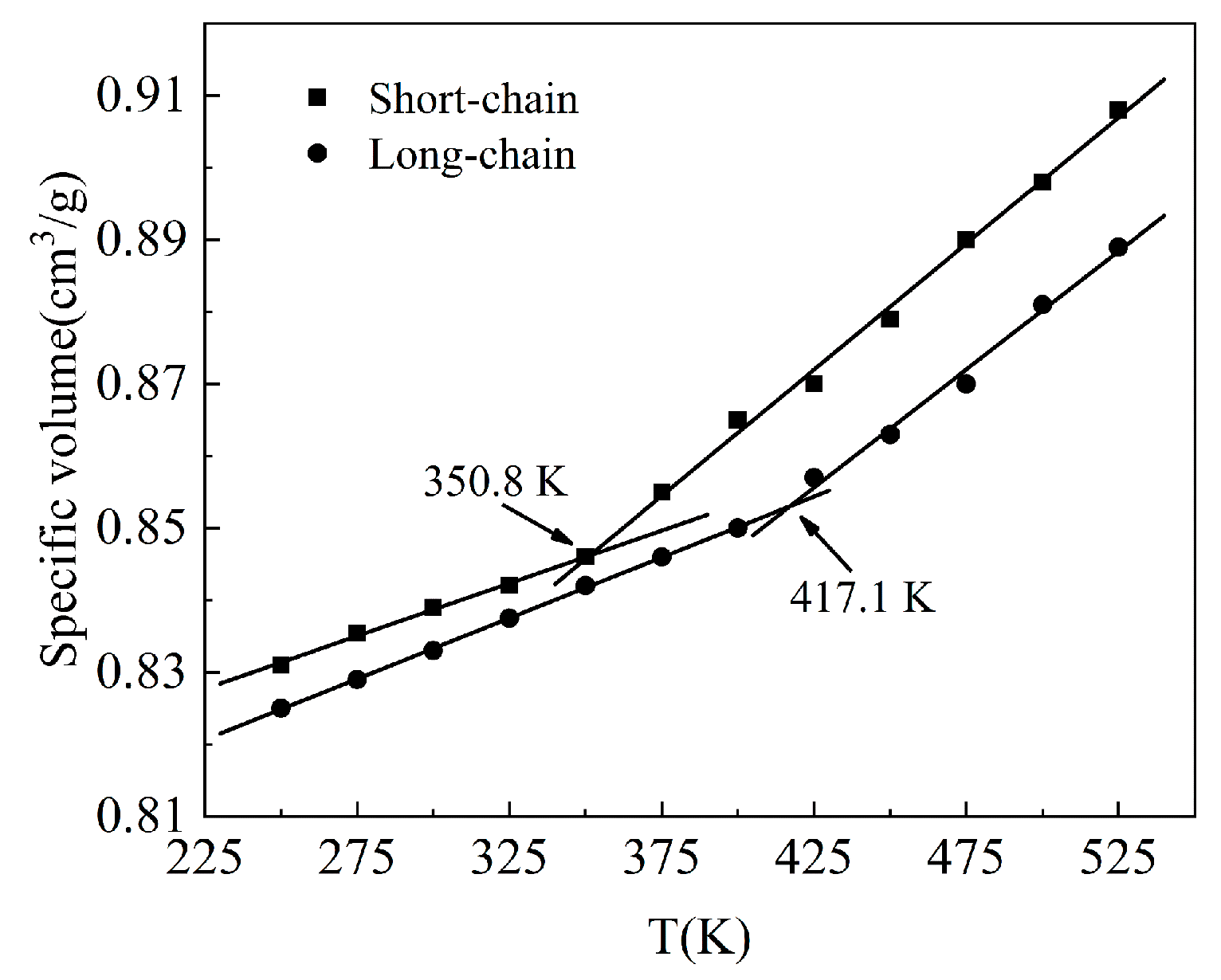
3.1. Determination of RHCM Mold Temperature
3.2. Concentration Profile
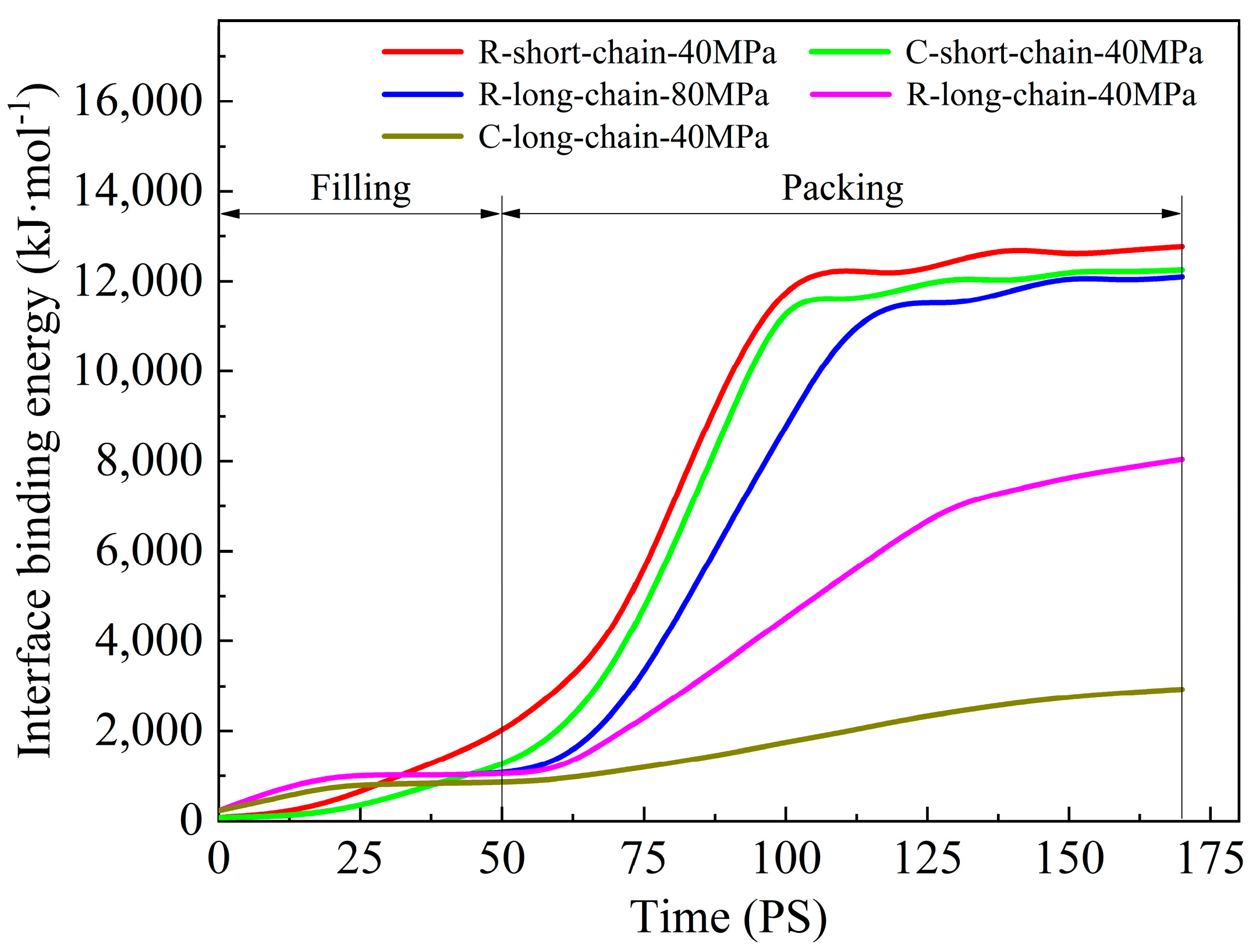
3.3. Interface Binding Energy
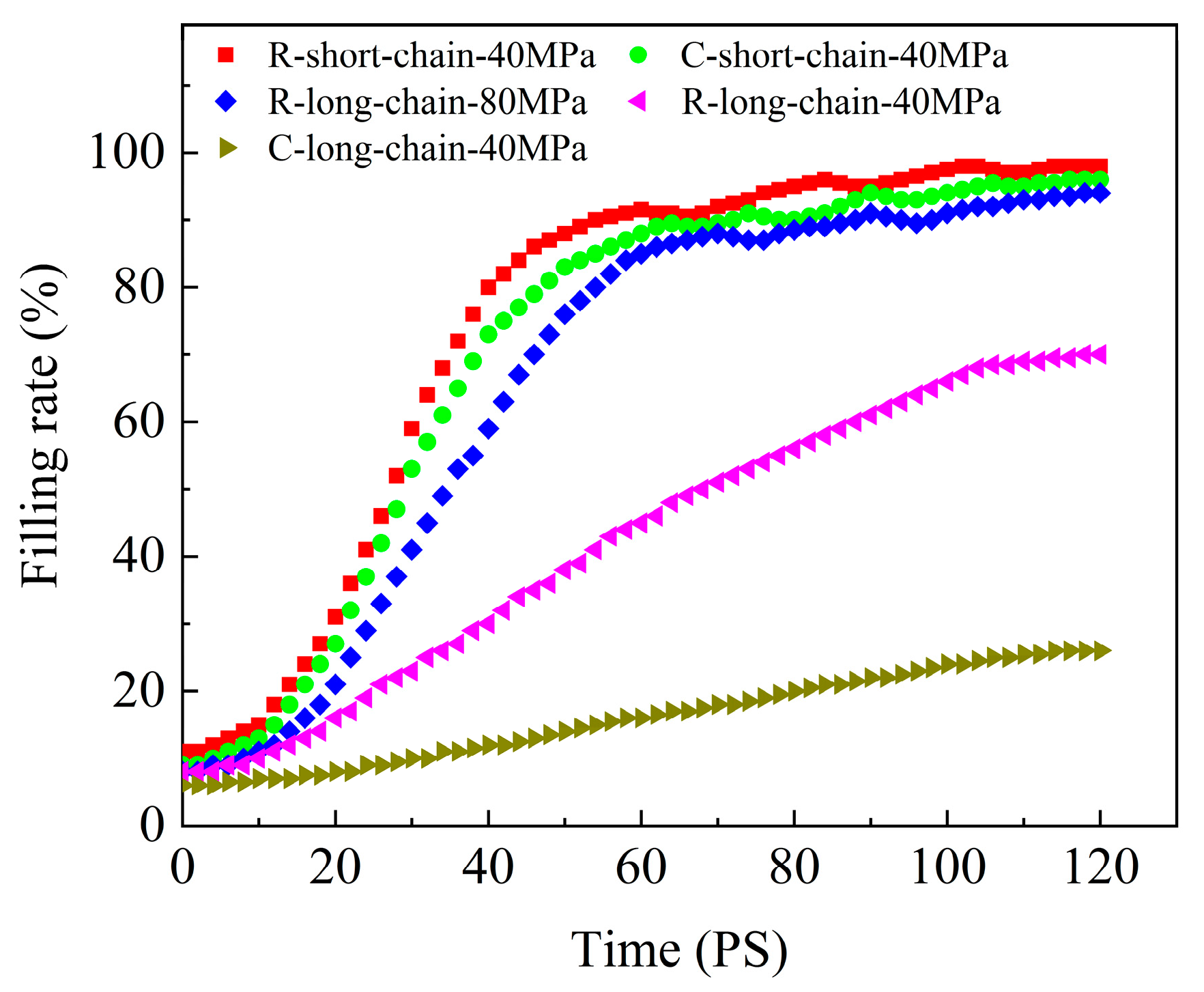
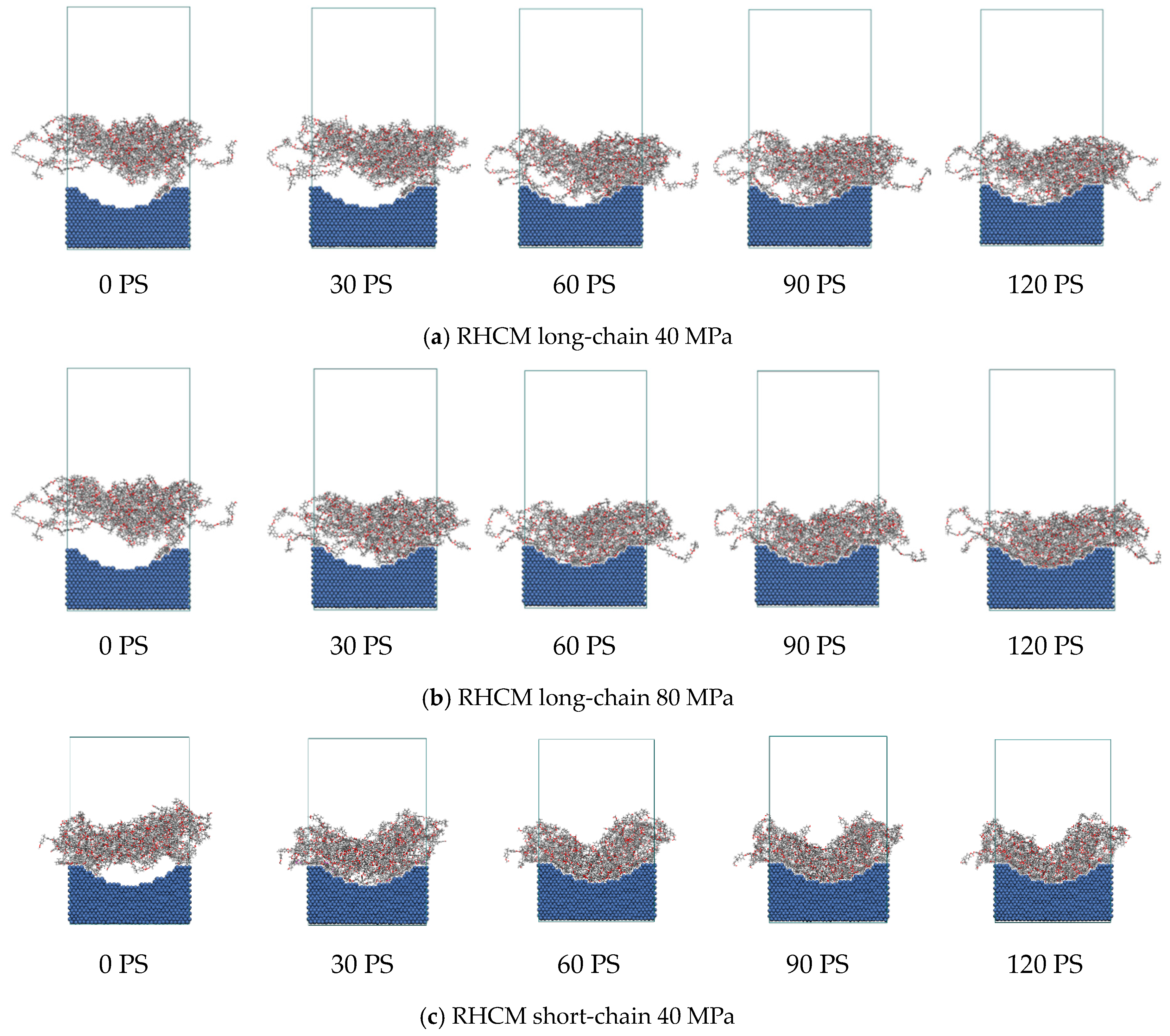
3.4. Replication Capability of Nanostructures
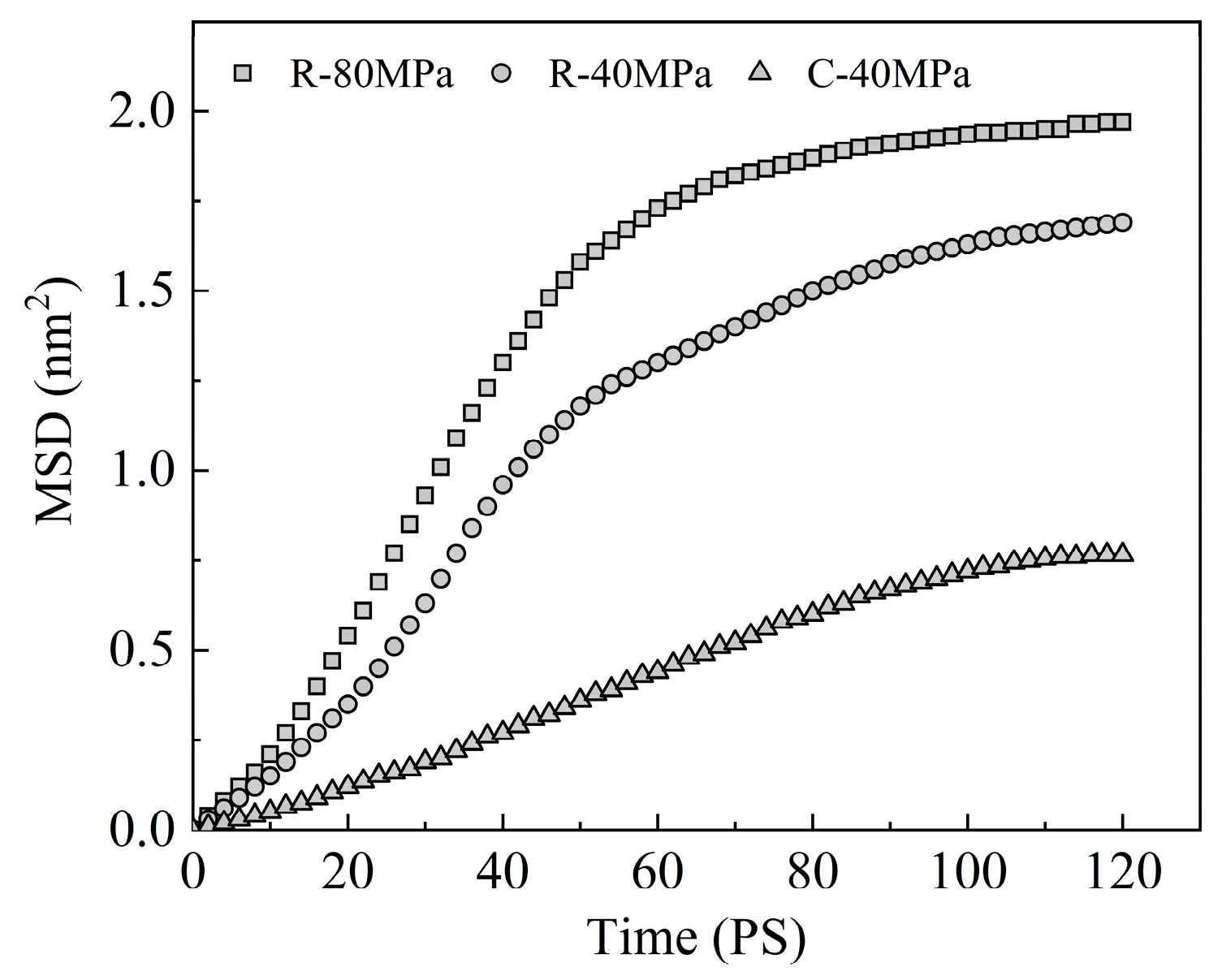
3.5. Molecular Chain Motion
4. Conclusions
- (1)
- The Tg of the amorphous PC long-chain simulation system is 417 K, which is basically consistent with the experimental result. Moreover, the Tg of the short-chain system is greatly reduced. Thus, the Tg results of the two simulation systems conform to Flory free volume theory, indicating that the simulation results are reliable.
- (2)
- In the filling stage, it is difficult for PC molecular chains to overcome the resistance, and only a few atoms fill a short distance into the nanostructures. The filling is mainly in the packing stage. Since the mold temperature is low in CIM, the long-chain PC system at the nanostructures cools rapidly and the viscosity increases. Thus, the molecular chain motion is blocked, the migration motion becomes weak, and it is difficult to cross the high potential energy region. The result is that the filling effect is poor, with low replication capability. However, for the short-chain system, the glass transition temperature and viscosity are lower, and it is easy to overcome resistance under a certain pressure, with a high filling rate and good replication capability.
- (3)
- The high temperature of the mold in RHCM can enable the polymer to maintain a high temperature for a long time, and the Brownian motion of the molecular chains is active. Under the packing pressure, molecular chains can overcome the restriction of the entanglement barrier and viscous flow activation energy, and realize the transition of position, with a good filling effect.
- (4)
- The evolution process of the polymer filling morphology can be regarded as the diffusion motion of the molecular chain along the direction of a tube composed of other chains around it, which is realized by the Brownian motion of chain segments under the packing pressure. Temperature and pressure play a key role in this process. In RHCM, increasing the pressure can promote the migration and diffusion of the molecular chains and enhance the filling degree of the polymer to the nanostructure. Consequently, the replication capability of nanostructure parts for mold cavities can be enhanced, and the molding precision of nanostructure parts is guaranteed.
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Jiang, B.; Zhou, M.; Weng, C.; Zhang, L.; Lv, H. Fabrication of nanopillar arrays by combining electroforming and injection molding. Int. J. Adv. Manuf. Technol. 2016, 86, 1319–1328. [Google Scholar] [CrossRef]
- Yang, C.; Su, L.; Huang, C.; Huang, H.-X.; Castro, J.M.; Yi, A.Y. Effect of packing pressure on refractive index variation in injection molding of precision plastic optical lens. Adv. Polym. Technol. 2011, 30, 51–61. [Google Scholar] [CrossRef]
- Wang, X.; Gu, J.; Shen, C.; Wang, X. Warpage optimization with dynamic injection molding technology and sequential optimization method. Int. J. Adv. Manuf. Technol. 2015, 78, 177–187. [Google Scholar] [CrossRef]
- Lin, H.Y.; Chang, C.H.; Young, W.B. Experimental and analytical study on filling of nano structures in micro injection molding. Int. Commun. Heat Mass Transf. 2010, 37, 1477–1486. [Google Scholar] [CrossRef]
- Rytka, C.; Kristiansen, P.M.; Neyer, A. Iso- and variothermal injection compression moulding of polymer micro- and nanostructures for optical and medical applications. J. Micromech. Microeng. 2015, 25, 065008. [Google Scholar] [CrossRef]
- Stormonth-Darling, J.M.; Saeed, A.; Reynolds, P.M.; Gadegaard, N. Injection Molding Micro- and Nanostructures in Thermoplastic Elastomers. Macromol. Mater. Eng. 2016, 301, 964–971. [Google Scholar] [CrossRef] [Green Version]
- Maghsoudi, K.; Jafari, R.; Momen, G.; Farzaneh, M. Micro-nanostructured polymer surfaces using injection molding: A review. Mater. Today Commun. 2017, 13, 126–143. [Google Scholar] [CrossRef]
- Madsen, J.S.; Jensen, S.A.; Nakotte, L.; Vogelsang, A.; Thamdrup, L.H.; Czolkos, I.; Johansson, A.; Garnaes, J.; Nielsen, T.; Nygard, J.; et al. Scatterometry for optimization of injection molded nanostructures at the fabrication line. Int. J. Adv. Manuf. Technol. 2018, 99, 2669–2676. [Google Scholar] [CrossRef]
- Zhou, M.; Xiong, X.; Jiang, B.; Weng, C. Fabrication of high aspect ratio nanopillars and micro/nano combined structures with hydrophobic surface characteristics by injection molding. Appl. Surf. Sci. 2018, 427, 854–860. [Google Scholar] [CrossRef]
- Macintyre, D.; Thoms, S. The fabrication of high resolution features by mould injection. Microelectron. Eng. 1998, 41–42, 211–214. [Google Scholar] [CrossRef]
- Pranov, H.; Rasmussen, H.K.; Larsen, N.B.; Gadegaard, N. On the injection molding of nanostructured polymer surfaces. Polym. Eng. Sci. 2006, 46, 160–171. [Google Scholar] [CrossRef]
- Wang, G.; Hui, Y.; Zhang, L.; Zhao, G. Research on temperature and pressure responses in the rapid mold heating and cooling method based on annular cooling channels and electric heating. Int. J. Heat Mass Transf. 2018, 116, 1192–1203. [Google Scholar] [CrossRef]
- Hattori, S.; Nagato, K.; Hamaguchi, T.; Nakao, M. Rapid injection molding of high-aspect-ratio nanostructures. Microelectron. Eng. 2010, 87, 1546–1549. [Google Scholar] [CrossRef]
- Lin, H.Y.; Chang, C.H.; Young, W.B. Experimental Study on the Filling of Nano Structures with Infrared Mold Surface Heating. Int. Polym. Proc. 2011, 26, 73–81. [Google Scholar] [CrossRef]
- Ling, C.; Liang, X.; Fan, F.; Yang, Z. Diffusion behavior of the model diesel components in different polymer membranes by molecular dynamic simulation. Chem. Eng. Sci. 2012, 84, 292–302. [Google Scholar] [CrossRef]
- Shi, M.; Zhang, Y.; Cheng, L.; Jiao, Z.; Yang, W.; Tan, J.; Ding, Y. Interfacial Diffusion and Bonding in Multilayer Polymer Films: A Molecular Dynamics Simulation. J. Phys. Chem. B 2016, 120, 10018–10029. [Google Scholar] [CrossRef]
- Yang, S.; Yu, S.; Cho, M. Influence of mold and substrate material combinations on nanoimprint lithography process: MD simulation approach. Appl. Surf. Sci. 2014, 301, 189–198. [Google Scholar] [CrossRef]
- Paul, W. Molecular dynamics simulations of the glass transition in polymer melts. Polymer 2004, 45, 3901–3905. [Google Scholar] [CrossRef]
- Zhou, M.; Jiang, B.; Weng, C. Molecular dynamics study on polymer filling into nano-cavity by injection molding. Comput. Mater. Sci. 2016, 120, 36–42. [Google Scholar] [CrossRef]
- Zhou, M.; Xiong, X.; Drummer, D.; Jiang, B. Interfacial interaction and joining property of direct injection-molded polymer-metal hybrid structures: A molecular dynamics simulation study. Appl. Surf. Sci. 2019, 478, 680–689. [Google Scholar] [CrossRef]
- Jiang, B.; Zhou, M.; Lu, L.; Weng, C. Molecular dynamic simulation of injection molded nanostructures. Polym. Mater. Sci. Eng. 2015, 31, 122–126. [Google Scholar]
- Sun, H. COMPASS: An ab Initio Force-Field Optimized for Condensed-Phase Applications Overview with Details on Alkane and Benzene Compounds. J. Phys. Chem. B 1998, 102, 7338–7364. [Google Scholar] [CrossRef]
- Fox, T.G.; Flory, P.J. Second-Order Transition Temperatures and Related Properties of Polystyrene. I. Influence of Molecular Weight. J. Appl. Phys. 1950, 21, 581–591. [Google Scholar] [CrossRef]
- Fox, T.G.; Flory, P.J. The glass temperature and related properties of polystyrene. Influence of molecular weight. J. Polym. Sci. 1954, 14, 315–319. [Google Scholar] [CrossRef] [Green Version]
- Rohling, J.H.; Medina, A.N.; Bento, A.C.; Pereira, J.R.D.; Rubira, A.F.; Baesso, M.L.; Miranda, L.C.M. Differential thermal lens temperature scanning approach to glass transition analysis in polymers: Application to polycarbonate. J. Phys. D Appl. Phys. 2001, 34, 407–412. [Google Scholar] [CrossRef]
- Sha, B.; Dimov, S.; Griffiths, C.; Packianather, M.S. Investigation of micro-injection moulding: Factors affecting the replication quality. J. Mater. Process. Technol. 2007, 183, 284–296. [Google Scholar] [CrossRef]
- Zhang, L.; Tian, S.; Peng, T. Molecular Simulations of Sputtering Preparation and Transformation of Surface Properties of Au/Cu Alloy Coatings Under Different Incident Energies. Metals 2019, 9, 259. [Google Scholar] [CrossRef]
- Pan, F.; Peng, F.; Jiang, Z. Diffusion behavior of benzene/cyclohexane molecules in poly (vinyl alcohol)-graphite hybrid membranes by molecular dynamics simulation. Chem. Eng. Sci. 2007, 62, 703–710. [Google Scholar] [CrossRef]
- Abiad, M.G.; Carvajal, M.T.; Campanella, O.H. A review on methods and theories to describe the glass transition phenomenon: Applications in food and pharmaceutical products. Food Eng. Rev. 2009, 1, 105–132. [Google Scholar] [CrossRef]
- Fang, X.; Vitrac, O. Predicting diffusion coefficients of chemicals in and through packaging materials. Crit. Rev. Food Sci. Nutr. 2015, 57, 275–312. [Google Scholar] [CrossRef]
- De Gennes, P.G. Kinetics of diffusion-controlled processes in dense polymer systems. II. Effects of entanglements. J. Chem. Phys. 1982, 76, 3322–3326. [Google Scholar] [CrossRef]
- De Gennes, P.G. Reptation of a Polymer Chain in the Presence of Fixed Obstacles. J. Chem. Phys. 1971, 55, 572–579. [Google Scholar] [CrossRef]
- Li, Y.; Abberton, B.C.; Kroeger, M.; Liu, W.K. Challenges in Multiscale Modeling of Polymer Dynamics. Polymers 2013, 5, 751–832. [Google Scholar] [CrossRef] [Green Version]
- Reis, R.A.; Silva, F.C.; Nobrega, R.; Oliveira, J.V.; Tavares, F.W. Molecular dynamics simulation data of self-diffusion coefficient for Lennard–Jones chain fluids. Fluid Phase Equilib. 2004, 221, 25–33. [Google Scholar] [CrossRef]
- Durand, M.; Meyer, H.; Benzerara, O.; Baschnagel, J.; Vitrac, O. Molecular dynamics simulations of the chain dynamics in monodisperse oligomer melts and of the oligomer tracer diffusion in an entangled polymer matrix. J. Chem. Phys. 2010, 132, 194902. [Google Scholar] [CrossRef]


| E | CIM | RHCM | ||||||
|---|---|---|---|---|---|---|---|---|
| PC/Ni | PC | Ni | Einter | PC/Ni | PC | Ni | Einter | |
| Etotal | −1,801,355 | 7602 | −1,808,100 | −857 | −1,775,840 | 19,366 | −1,794,160 | −1046 |
| Enonbond | −1,820,032 | −11,075 | −1,808,100 | −857 | −1,804,925 | −9719 | −1,794,160 | −1046 |
| EvdW | −1,803,136 | 5821 | −1,808,100 | −857 | −1,788,213 | 6993 | −1,794,160 | −1046 |
| Eelectrostatic | −16,897 | −16,897 | 0 | 0 | −16,712 | −16,712 | 0 | 0 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, M.; Xin, Y. Molecular Mechanism Research into the Replication Capability of Nanostructures Based on Rapid Heat Cycle Molding. Appl. Sci. 2019, 9, 1683. https://doi.org/10.3390/app9081683
Zhang M, Xin Y. Molecular Mechanism Research into the Replication Capability of Nanostructures Based on Rapid Heat Cycle Molding. Applied Sciences. 2019; 9(8):1683. https://doi.org/10.3390/app9081683
Chicago/Turabian StyleZhang, Meili, and Yong Xin. 2019. "Molecular Mechanism Research into the Replication Capability of Nanostructures Based on Rapid Heat Cycle Molding" Applied Sciences 9, no. 8: 1683. https://doi.org/10.3390/app9081683




