The Role of the Human Brain Neuron–Glia–Synapse Composition in Forming Resting-State Functional Connectivity Networks
Abstract
:1. Introduction
2. Materials and Methods
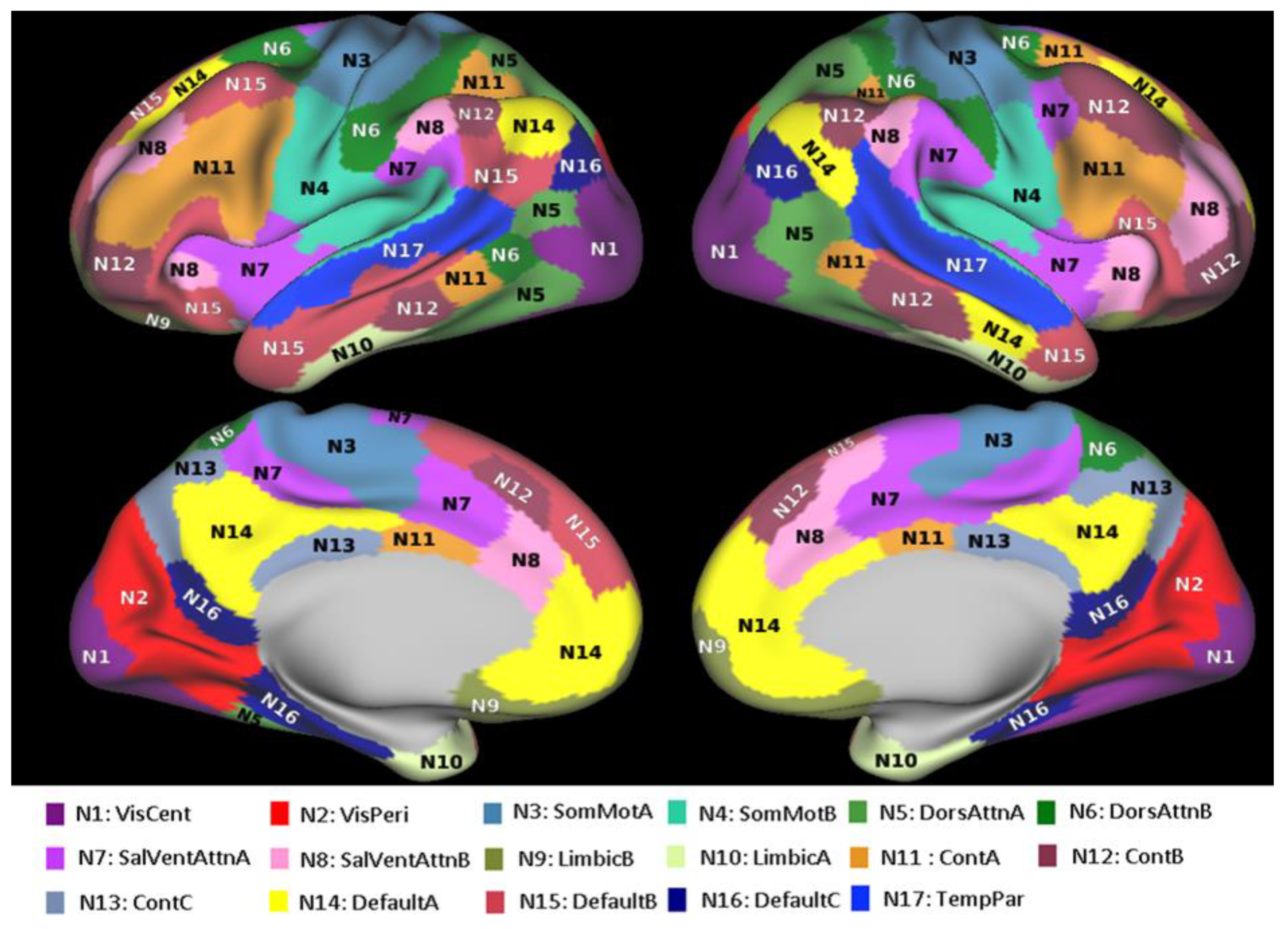
2.1. Resting-State Functional Connectivity Data
2.2. Quantitative Gradient-Recalled Echo (qGRE) MRI Data Analysis
2.3. Structural Connectivity Analysis Based on qGRE-Defined Brain Cellular Structure
2.4. Analysis Based on T1w/T2w Approach
3. Results
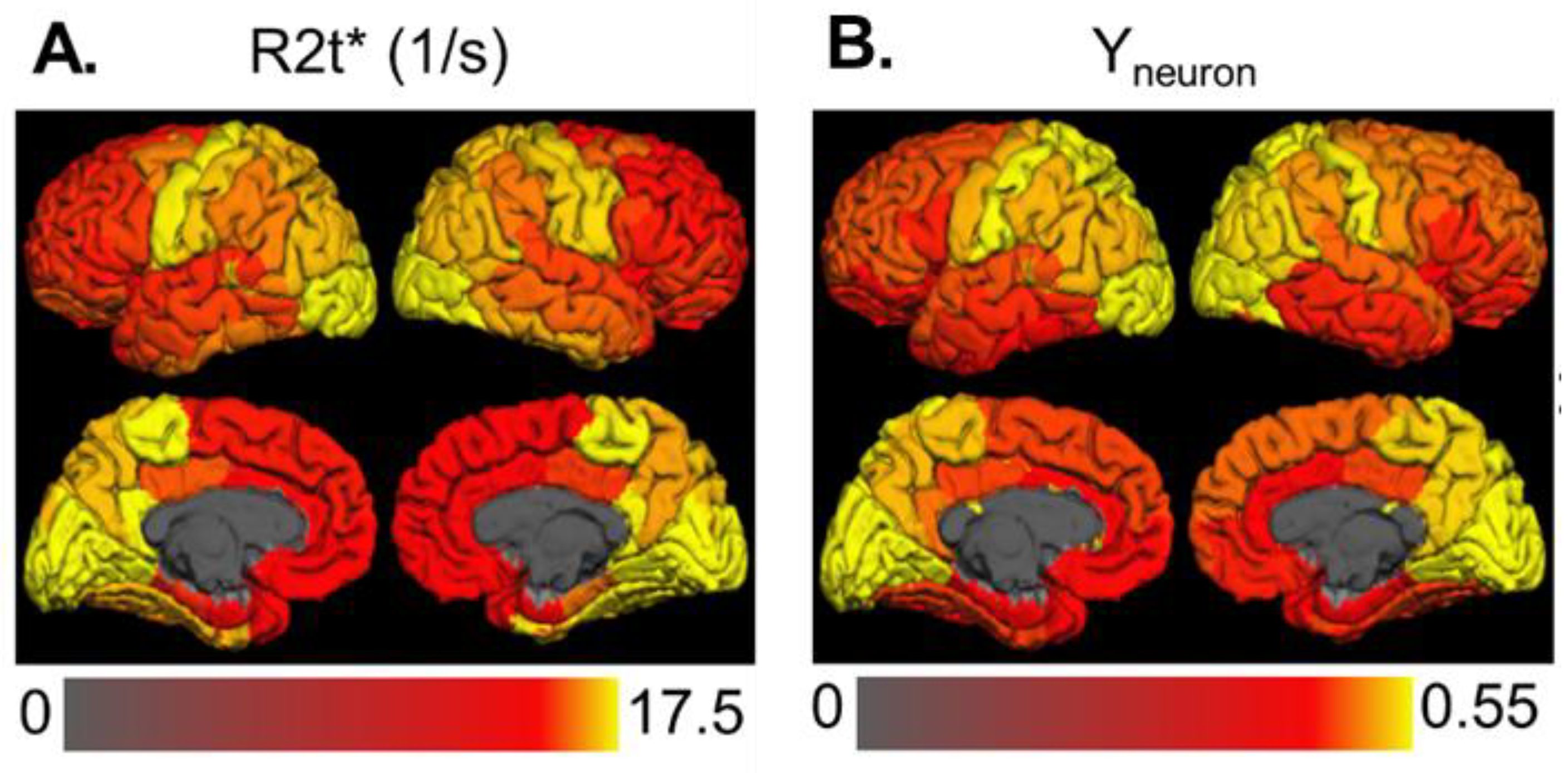
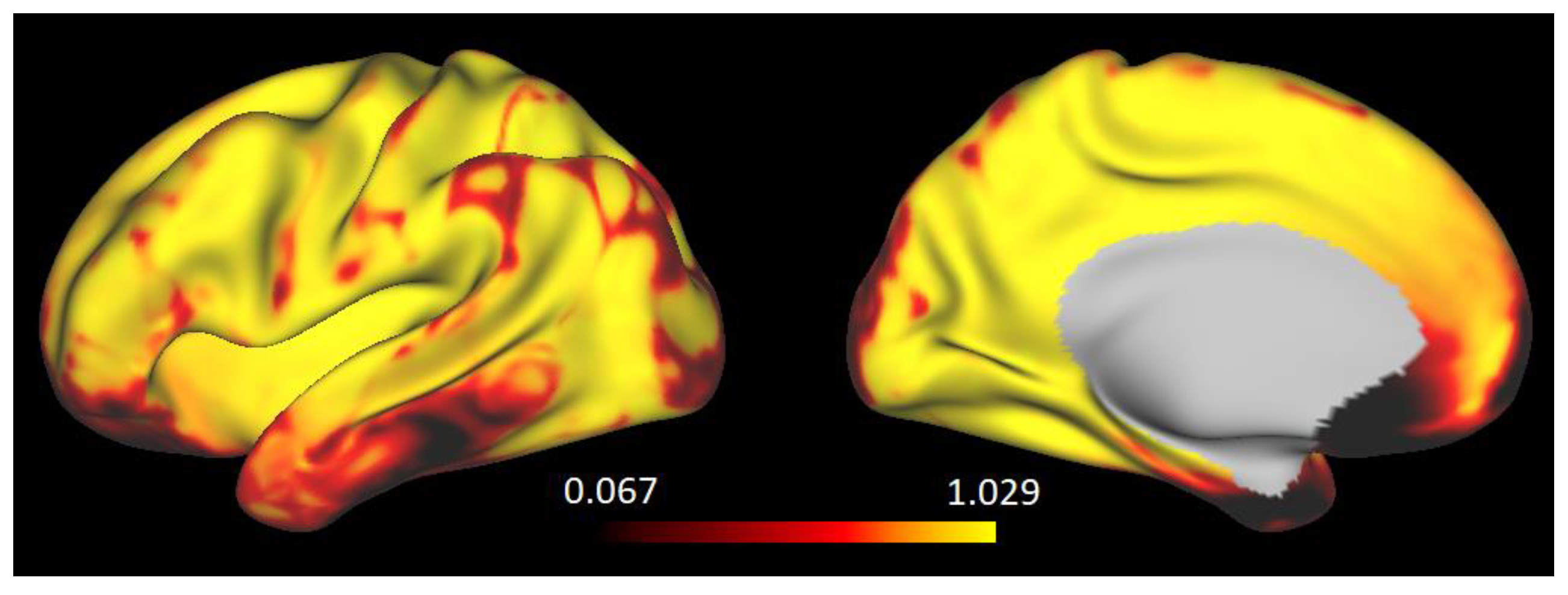
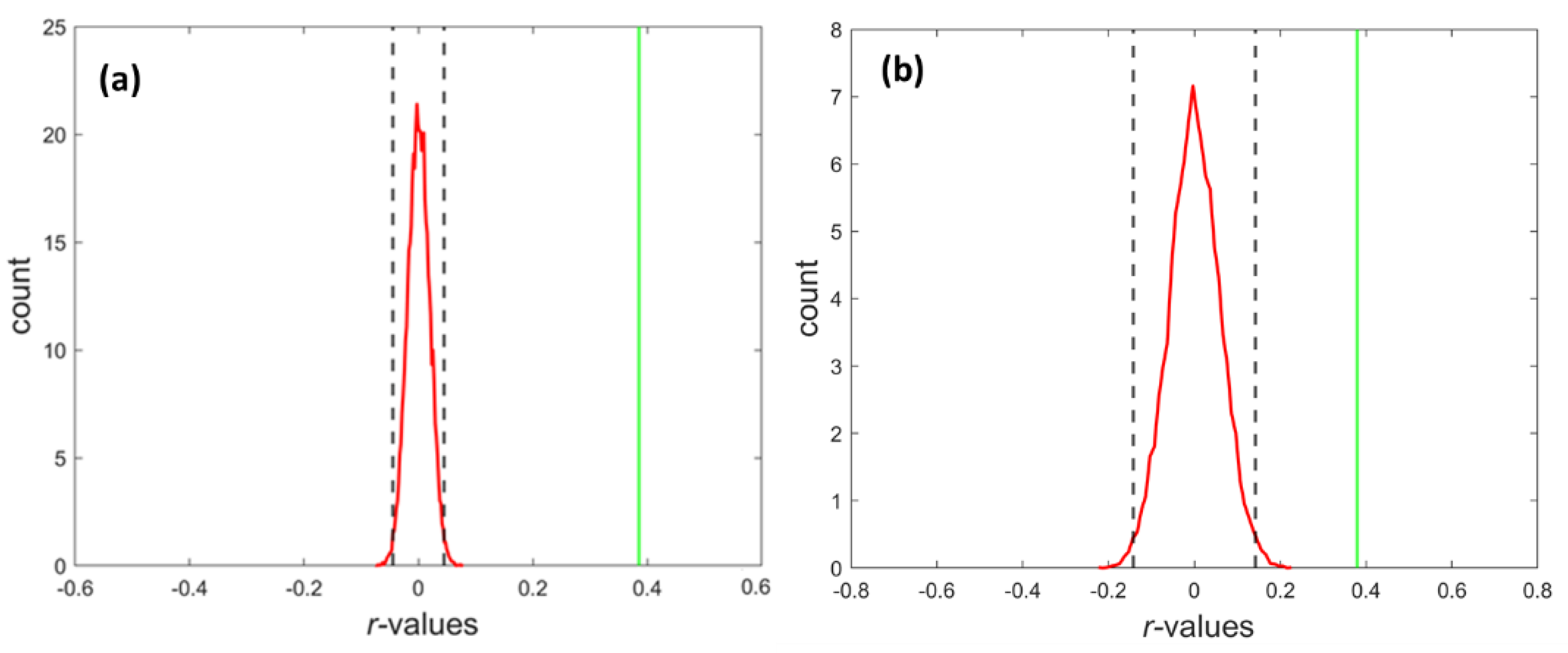
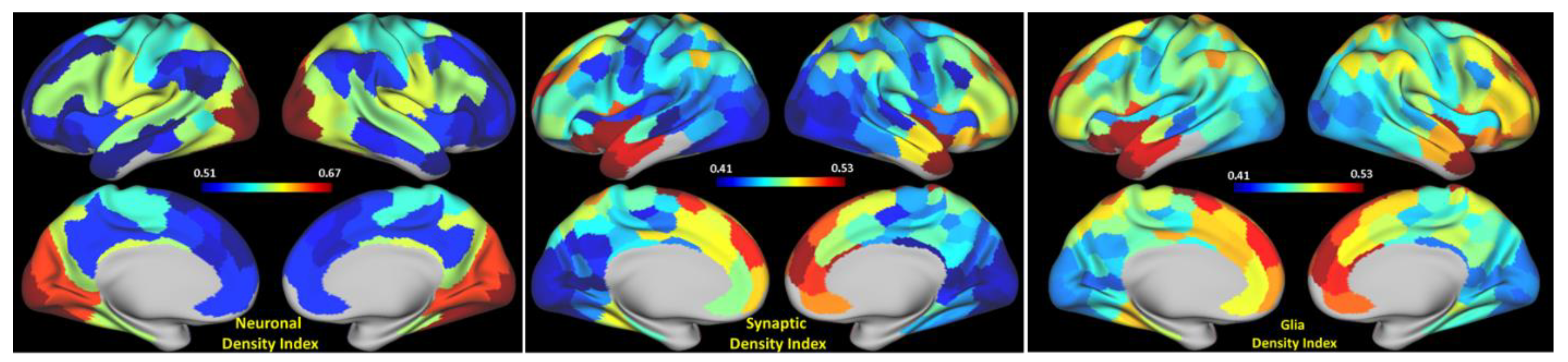
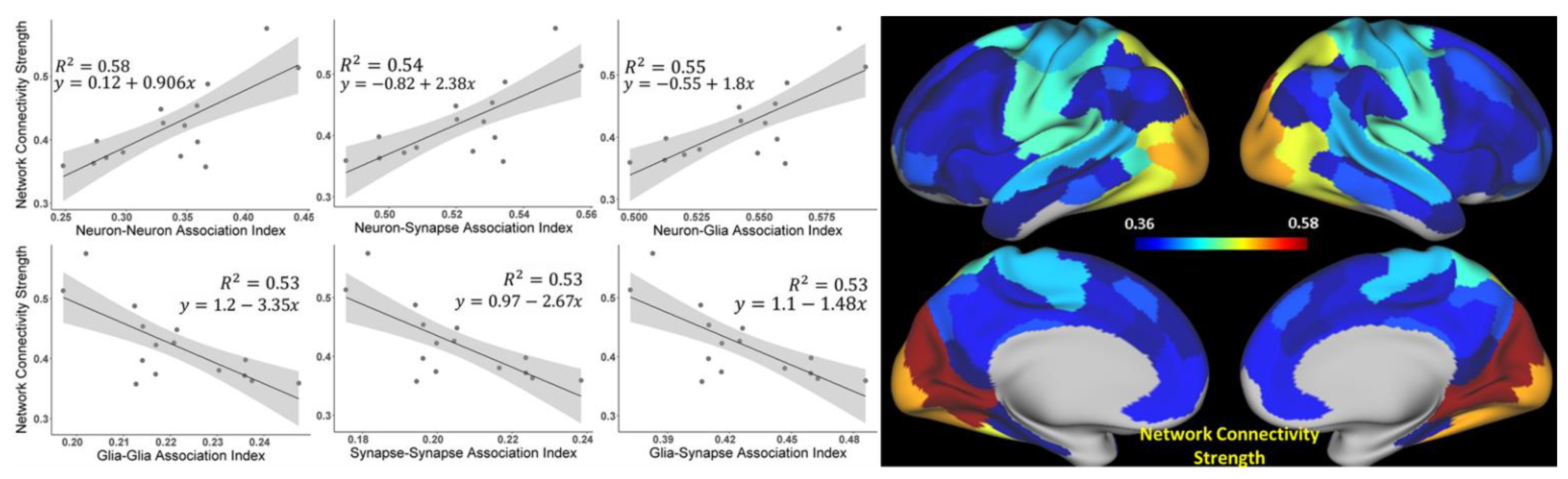
3.1. The Strength of Resting-State Functional Networks Is Significantly Associated with the Neuron–Neuron, Neuron–Glia, and Neuron–Synaptic Structural Circuits in the Human Brain Cortex
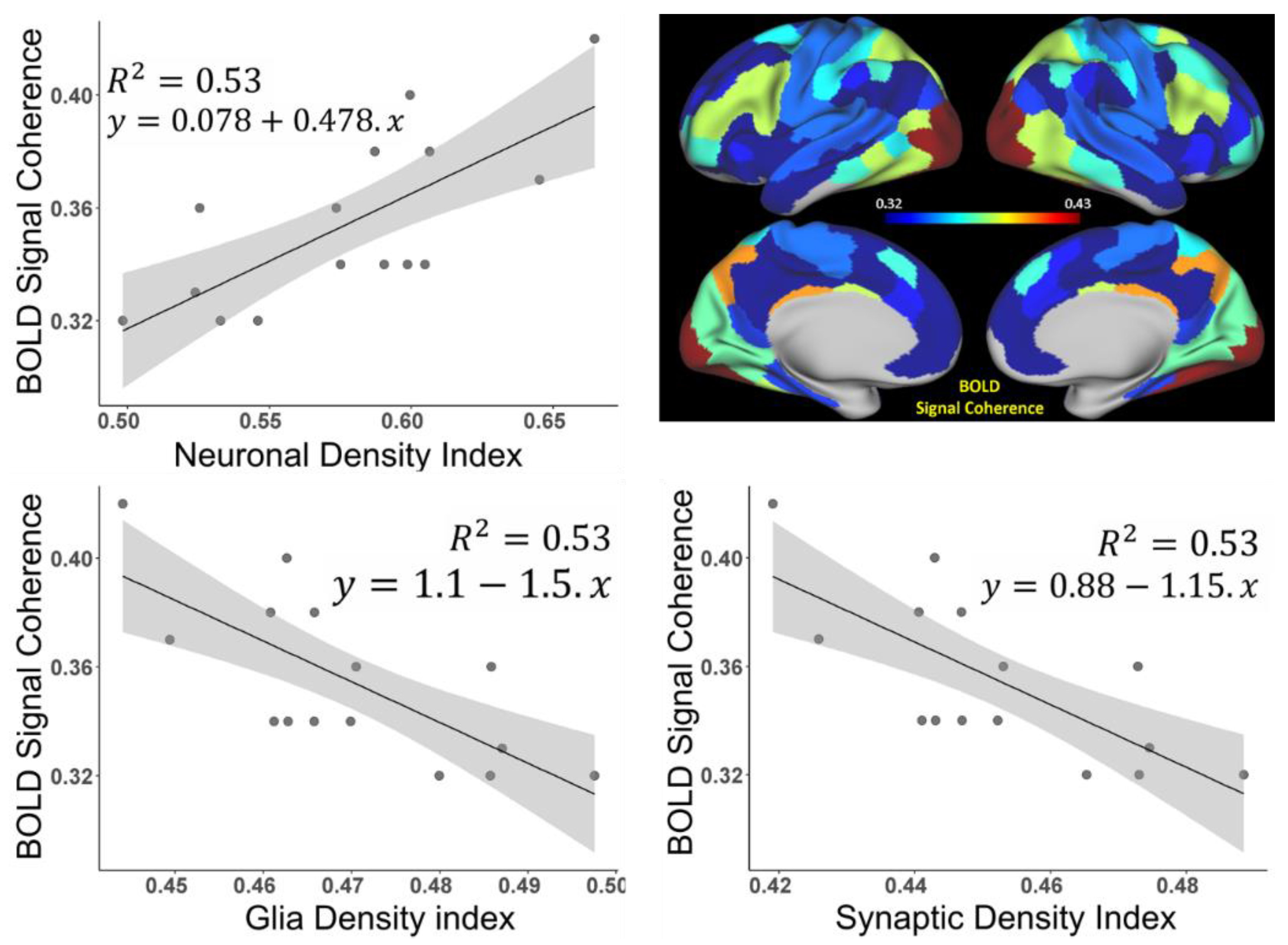
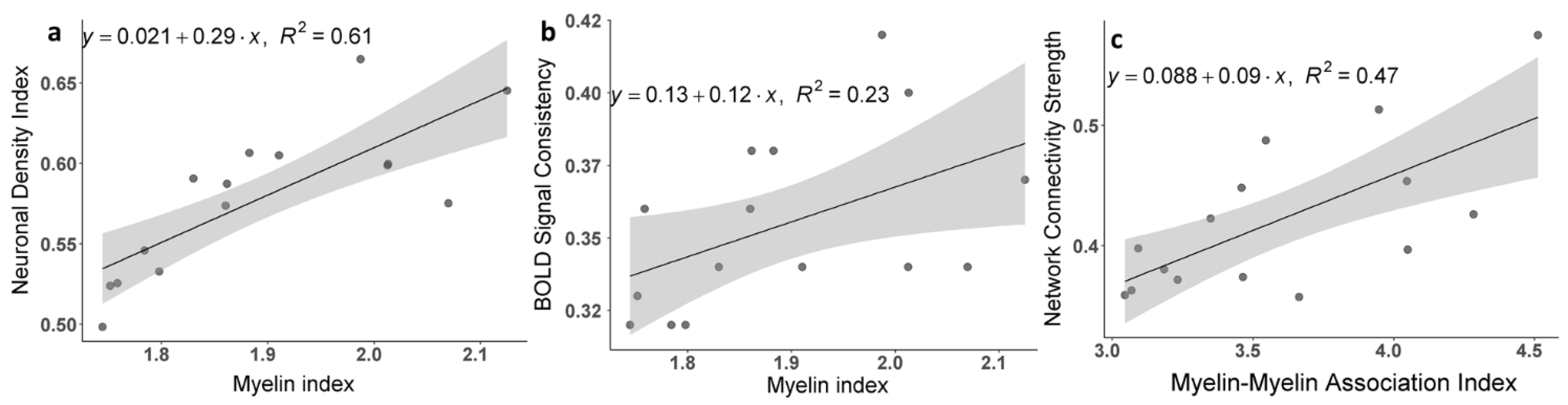
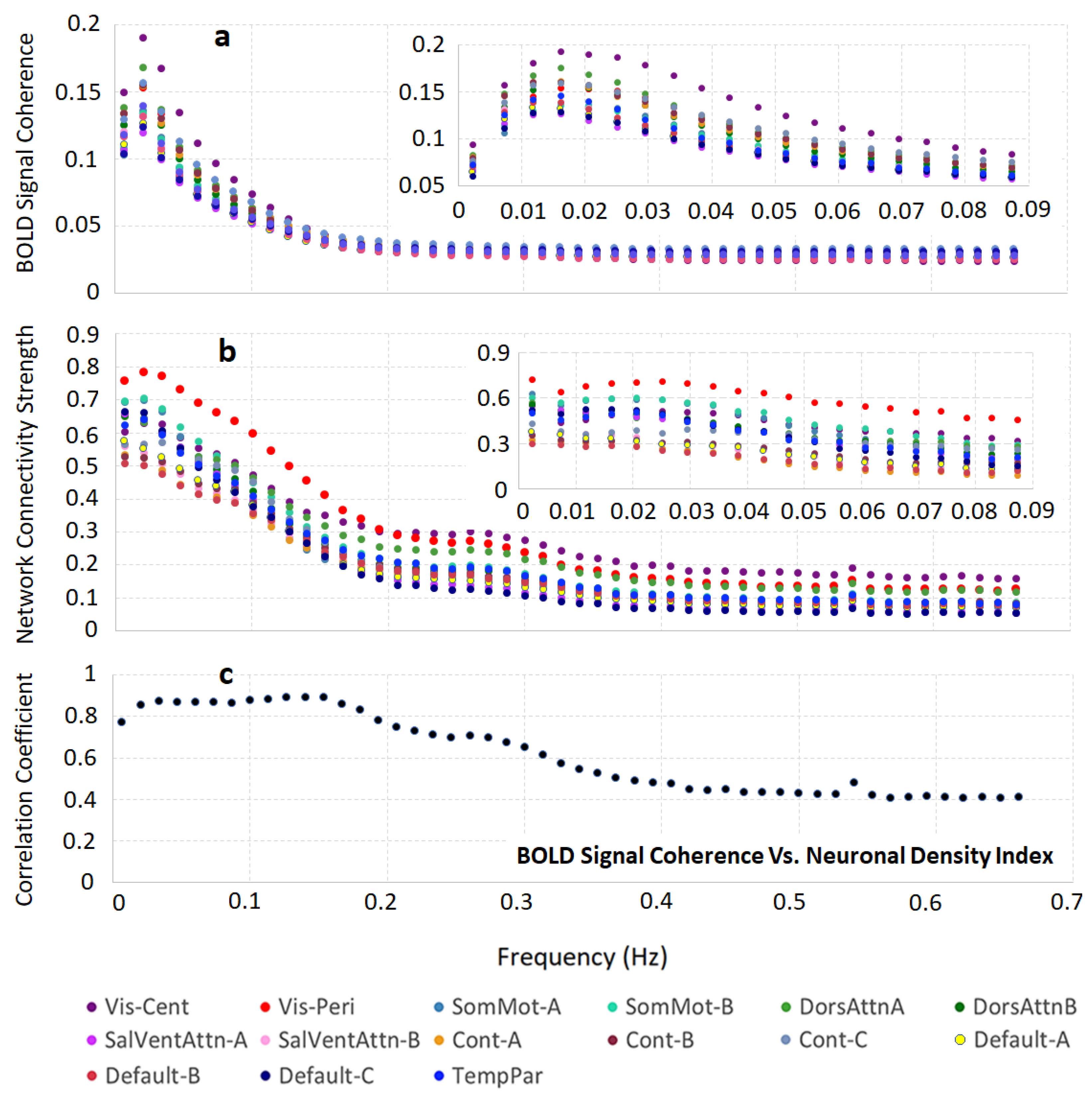
3.2. Brain Cortical Cellular Composition Shows the Strongest Association with the Resting-State BOLD Signal Coherence and Network Connectivity in the Infra-Slow Frequency Range of Neuronal Activity
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A


| NETWORKS | R2t*Mean | R2t*STD | NDI | GDI | SDI | BOLD Coherence | FCS |
|---|---|---|---|---|---|---|---|
| Vis-Cent | 19.67 | 0.8 | 0.66 | 0.44 | 0.42 | 0.51 | 0.42 |
| Vis-Peri | 18.86 | 0.48 | 0.65 | 0.45 | 0.43 | 0.57 | 0.37 |
| SomMot-A | 16.95 | 0.82 | 0.58 | 0.47 | 0.45 | 0.43 | 0.34 |
| SomMot-B | 18.02 | 0.78 | 0.6 | 0.46 | 0.44 | 0.45 | 0.34 |
| DorsAttnA | 17.88 | 0.67 | 0.61 | 0.46 | 0.44 | 0.49 | 0.38 |
| DorsAttnB | 17.13 | 0.79 | 0.57 | 0.47 | 0.45 | 0.45 | 0.36 |
| SalVentAttn-A | 16.61 | 0.68 | 0.53 | 0.49 | 0.47 | 0.37 | 0.32 |
| SalVentAttn-B | 16.34 | 0.71 | 0.52 | 0.49 | 0.47 | 0.36 | 0.33 |
| Cont-A | 17.69 | 0.73 | 0.59 | 0.47 | 0.45 | 0.37 | 0.38 |
| Cont-B | 16.55 | 0.74 | 0.53 | 0.49 | 0.47 | 0.40 | 0.36 |
| Cont-C | 17.87 | 0.96 | 0.6 | 0.46 | 0.44 | 0.40 | 0.40 |
| Default-A | 16.6 | 0.71 | 0.55 | 0.48 | 0.47 | 0.38 | 0.32 |
| Default-B | 16.29 | 0.83 | 0.5 | 0.5 | 0.49 | 0.36 | 0.32 |
| Default-C | 18.06 | 0.71 | 0.6 | 0.49 | 0.44 | 0.36 | 0.34 |
| TempPar | 17.81 | 0.68 | 0.59 | 0.47 | 0.45 | 0.42 | 0.34 |
References
- Biswal, B.; Yetkin, F.Z.; Haughton, V.M.; Hyde, J.S. Functional connectivity in the motor cortex of resting human brain using echo-planar mri. Magn. Reson. Med. 1995, 34, 537–541. [Google Scholar] [CrossRef]
- Buckner, R.L.; Krienen, F.M.; Yeo, B.T. Opportunities and limitations of intrinsic functional connectivity MRI. Nat. Neurosci. 2013, 16, 832–837. [Google Scholar] [CrossRef]
- Fox, M.D.; Raichle, M.E. Spontaneous fluctuations in brain activity observed with functional magnetic resonance imaging. Nat. Rev. Neurosci. 2007, 8, 700–711. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, S.; Lee, T.M.; Kay, A.R.; Tank, D.W. Brain magnetic resonance imaging with contrast dependent on blood oxygenation. Proc. Natl. Acad. Sci. USA 1990, 87, 9868–9872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Essen, D.C.; Smith, S.M.; Barch, D.M.; Behrens, T.E.J.; Yacoub, E.; Ugurbil, K.; Consortium, W.-M.H. The WU-Minn Human Con-nectome Project: An overview. Neuroimage 2013, 80, 62–79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Damoiseaux, J.S.; Rombouts, S.A.R.B.; Barkhof, F.; Scheltens, P.; Stam, C.J.; Smith, S.M.; Beckmann, C.F. Consistent resting-state networks across healthy subjects. Proc. Natl. Acad. Sci. USA 2006, 103, 13848–13853. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomas Yeo, B.T.; Krienen, F.M.; Sepulcre, J.; Sabuncu, M.R.; Lashkari, D.; Hollinshead, M.; Roffman, J.L.; Smoller, J.W.; Zöllei, L.; Polimeni, J.R.; et al. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J. Neurophysiol. 2011, 106, 1125–1165. [Google Scholar] [CrossRef]
- Andrews-Hanna, J.R.; Snyder, A.Z.; Vincent, J.L.; Lustig, C.; Head, D.; Raichle, M.E.; Buckner, R.L. Disruption of large-scale brain systems in advanced aging. Neuron 2007, 56, 924–935. [Google Scholar] [CrossRef] [Green Version]
- Buckner, R.L.; Snyder, A.Z.; Shannon, B.J.; LaRossa, G.; Sachs, R.; Fotenos, A.F.; Sheline, Y.I.; Klunk, W.E.; Mathis, C.A.; Morris, J.C.; et al. Molecular, structural, and functional characterization of Alzheimer’s disease: Evidence for a relationship between default activity, amyloid, and memory. J. Neurosci. 2005, 25, 7709–7717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Luca, M.; Beckmann, C.F.; De Stefano, N.; Matthews, P.M.; Smith, S.M. fMRI resting state networks define distinct modes of long-distance interactions in the human brain. Neuroimage 2006, 29, 1359–1367. [Google Scholar] [CrossRef]
- Greicius, M.D.; Supekar, K.; Menon, V.; Dougherty, R.F. Resting-State Functional Connectivity Reflects Structural Connectivity in the Default Mode Network. Cereb. Cortex 2008, 19, 72–78. [Google Scholar] [CrossRef]
- Toosy, A.T.; Ciccarelli, O.; Parker, G.J.; Wheeler-Kingshott, C.A.; Miller, D.H.; Thompson, A.J. Characterizing function–structure relationships in the human visual system with functional MRI and diffusion tensor imaging. NeuroImage 2004, 21, 1452–1463. [Google Scholar] [CrossRef] [PubMed]
- Uddin, L.Q. Complex relationships between structural and functional brain connectivity. Trends Cogn. Sci. 2013, 17, 600–602. [Google Scholar] [CrossRef] [Green Version]
- Honey, C.; Sporns, O.; Cammoun, L.; Gigandet, X.; Thiran, J.-P.; Meuli, R.; Hagmann, P. Predicting human resting-state functional connectivity from structural connectivity. Proc. Natl. Acad. Sci. USA 2009, 106, 2035–2040. [Google Scholar] [CrossRef] [Green Version]
- Ding, Z.; Huang, Y.; Bailey, S.K.; Gao, Y.; Cutting, L.E.; Rogers, B.P.; Newton, A.T.; Gore, J.C. Detection of synchronous brain activity in white matter tracts at rest and under functional loading. Proc. Natl. Acad. Sci. USA 2017, 115, 595–600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fields, R.D.; Woo, D.H.; Basser, P.J. Glial Regulation of the Neuronal Connectome through Local and Long-Distant Communication. Neuron 2015, 86, 374–386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pannasch, U.; Vargová, L.; Reingruber, J.; Ezan, P.; Holcman, D.; Giaume, C.; Syková, E.; Rouach, N. Astroglial networks scale synaptic activity and plasticity. Proc. Natl. Acad. Sci. USA 2011, 108, 8467–8472. [Google Scholar] [CrossRef] [Green Version]
- Ullian, E.M.; Sapperstein, S.K.; Christopherson, K.S.; Barres, B.A. Control of Synapse Number by Glia. Science 2001, 291, 657–661. [Google Scholar] [CrossRef]
- Pellerin, L.; Magistretti, P.J. Sweet sixteen for ANLS. J. Cereb. Blood Flow Metab. 2012, 32, 1152–1166. [Google Scholar] [CrossRef] [PubMed]
- Stogsdill, J.; Eroglu, C. The interplay between neurons and glia in synapse development and plasticity. Curr. Opin. Neurobiol. 2016, 42, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Araque, A.; Navarrete, M. Glial cells in neuronal network function. Philos. Trans. R Soc. Lond. B Biol. Sci. 2010, 365, 2375–2381. [Google Scholar] [CrossRef]
- Ulrich, X.; Yablonskiy, D.A. Separation of cellular and BOLD contributions to T2* signal relaxation. Magn. Reson. Med. 2016, 75, 606–615. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yablonskiy, D.A.; Haacke, E.M. Theory of NMR signal behavior in magnetically inhomogeneous tissues: The static dephasing regime. Magn. Reson. Med. 1994, 32, 749–763. [Google Scholar] [CrossRef]
- Yablonskiy, D. Quantitation of intrinsic magnetic susceptibility-related effects in a tissue matrix. Phantom study. Magn. Reson. Med. 1998, 39, 417–428. [Google Scholar] [CrossRef]
- Yablonskiy, D.A.; Sukstanskii, A.L.; Luo, J.; Wang, X. Voxel spread function method for correction of magnetic field inhomogeneity effects in quantitative gradient-echo-based MRI. Magn. Reson. Med. 2012, 70, 1283–1292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wen, J.; Goyal, M.S.; Astafiev, S.; Raichle, M.E.; Yablonskiy, D.A. Genetically defined cellular correlates of the baseline brain MRI signal. Proc. Natl. Acad. Sci. USA 2018, 115, E9727–E9736. [Google Scholar] [CrossRef] [Green Version]
- Herculano-Houzel, S. The glia/neuron ratio: How it varies uniformly across brain structures and species and what that means for brain physiology and evolution. Glia 2014, 62, 1377–1391. [Google Scholar] [CrossRef] [PubMed]
- Cullen, D.K.; Gilroy, M.E.; Irons, H.R.; LaPlaca, M.C. Synapse-to-neuron ratio is inversely related to neuronal density in mature neuronal cultures. Brain Res. 2010, 1359, 44–55. [Google Scholar] [CrossRef] [Green Version]
- Glasser, M.F.; Van Essen, D.C. Mapping Human Cortical Areas In Vivo Based on Myelin Content as Revealed by T1- and T2-Weighted MRI. J. Neurosci. 2011, 31, 11597–11616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schaefer, A.; Kong, R.; Gordon, E.M.; Laumann, T.O.; Zuo, X.-N.; Holmes, A.J.; Eickhoff, S.B.; Yeo, B.T.T. Local-Global Parcellation of the Human Cerebral Cortex from Intrinsic Functional Connectivity MRI. Cereb. Cortex 2018, 28, 3095–3114. [Google Scholar] [CrossRef]
- Van Essen, D.C.; Glasser, M.F.; Dierker, D.L.; Harwell, J.; Coalson, T. Parcellations and hemispheric asymmetries of human cerebral cortex analyzed on surface-based atlases. Cereb. Cortex 2012, 22, 2241–2262. [Google Scholar] [CrossRef]
- Han, Y.; Wang, J.; Zhao, Z.; Min, B.; Lu, J.; Li, K.; He, Y.; Jia, J. Frequency-dependent changes in the amplitude of low-frequency fluctuations in amnestic mild cognitive impairment: A resting-state fMRI study. NeuroImage 2011, 55, 287–295. [Google Scholar] [CrossRef]
- Mitra, A.; Kraft, A.; Wright, P.; Acland, B.; Snyder, A.Z.; Rosenthal, Z.; Czerniewski, L.; Bauer, A.; Snyder, L.; Culver, J.; et al. Spontaneous Infra-slow Brain Activity Has Unique Spatiotemporal Dynamics and Laminar Structure. Neuron 2018, 98, 297–305.e6. [Google Scholar] [CrossRef] [Green Version]
- Palva, J.M.; Palva, S. Infra-slow fluctuations in electrophysiological recordings, blood-oxygenation-level-dependent signals, and psychophysical time series. NeuroImage 2012, 62, 2201–2211. [Google Scholar] [CrossRef] [PubMed]
- Raut, R.V.; Snyder, A.Z.; Raichle, M.E. Hierarchical dynamics as a macroscopic organizing principle of the human brain. Proc. Natl. Acad. Sci. USA 2020, 117, 20890–20897. [Google Scholar] [CrossRef]
- Zhang, D.Y.; Raichle, M.E. Disease and the brain’s dark energy. Nat. Rev. Neurol. 2010, 6, 15–28. [Google Scholar] [CrossRef] [PubMed]
- Glasser, M.F.; Sotiropoulos, S.N.; Wilson, J.A.; Coalson, T.S.; Fischl, B.; Andersson, J.L.; Xu, J.; Jbabdi, S.; Webster, M.; Polimeni, J.R.; et al. The minimal preprocessing pipelines for the Human Connectome Project. NeuroImage 2013, 80, 105–124. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Wen, J.; Cross, A.; Yablonskiy, D.A. On the relationship between cellular and hemodynamic properties of the human brain cortex throughout adult lifespan. NeuroImage 2016, 133, 417–429. [Google Scholar] [CrossRef] [Green Version]
- Wen, J.; Cross, A.; Yablonskiy, D.A. On the role of physiological fluctuations in quantitative gradient echo MRI: Implications for GEPCI, QSM, and SWI. Magn. Reson. Med. 2014, 73, 195–203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yablonskiy, D.A.; Sukstanskii, A.L.; He, X. Blood oxygenation level-dependent (BOLD)-based techniques for the quantification of brain hemodynamic and metabolic properties—Theoretical models and experimental approaches. NMR Biomed. 2012, 26, 963–986. [Google Scholar] [CrossRef] [Green Version]
- Lazari, A.; Lipp, I. Can MRI measure myelin? Systematic review, qualitative assessment, and meta-analysis of studies validating microstructural imaging with myelin histology. NeuroImage 2021, 230, 117744. [Google Scholar] [CrossRef] [PubMed]
- Glasser, M.F.; Coalson, T.S.; Robinson, E.C.; Hacker, C.D.; Harwell, J.; Yacoub, E.; Ugurbil, K.; Andersson, J.; Beckmann, C.F.; Jenkinson, M.; et al. A multi-modal parcellation of human cerebral cortex. Nature 2016, 536, 171–178. [Google Scholar] [CrossRef] [Green Version]
- Craddock, R.C.; James, G.A.; Iii, P.E.H.; Hu, X.P.; Mayberg, H.S. A whole brain fMRI atlas generated via spatially constrained spectral clustering. Hum. Brain Mapp. 2011, 33, 1914–1928. [Google Scholar] [CrossRef] [Green Version]
- Gordon, E.M.; Laumann, T.O.; Adeyemo, B.; Huckins, J.; Kelley, W.M.; Petersen, S.E. Generation and Evaluation of a Cortical Area Parcellation from Resting-State Correlations. Cereb. Cortex 2014, 26, 288–303. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Tokoglu, F.; Papademetris, X.; Constable, R. Groupwise whole-brain parcellation from resting-state fMRI data for network node identification. NeuroImage 2013, 82, 403–415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raichle, M.E. The restless brain: How intrinsic activity organizes brain function. Philos. Trans. R. Soc. B Biol. Sci. 2015, 370, 20140172. [Google Scholar] [CrossRef]
- Richiardi, J.; Altmann, A.; Milazzo, A.-C.; Chang, C.; Chakravarty, M.M.; Banaschewski, T.; Barker, G.J.; Bokde, A.L.; Bromberg, U.; Büchel, C.; et al. Correlated gene expression supports synchronous activity in brain networks. Science 2015, 348, 1241–1244. [Google Scholar] [CrossRef] [Green Version]
- Hawrylycz, M.; Miller, J.A.; Menon, V.; Feng, D.; A Dolbeare, T.; Guillozet-Bongaarts, A.L.; Jegga, A.G.; Aronow, B.J.; Lee, C.-K.; Bernard, A.; et al. Canonical genetic signatures of the adult human brain. Nat. Neurosci. 2015, 18, 1832–1844. [Google Scholar] [CrossRef] [Green Version]
- Goyal, M.S.; Hawrylycz, M.; Miller, J.A.; Snyder, A.Z.; Raichle, M.E. Aerobic Glycolysis in the Human Brain Is Associated with Development and Neotenous Gene Expression. Cell Metab. 2014, 19, 49–57. [Google Scholar] [CrossRef] [Green Version]
- Collins, C.E.; Airey, D.C.; Young, N.A.; Leitch, D.B.; Kaas, J.H. Neuron densities vary across and within cortical areas in primates. Proc. Natl. Acad. Sci. USA 2010, 107, 15927–15932. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elston, G.N.; Piccione, R.B.; Defelipe, J. The Pyramidal Cell in Cognition: A Comparative Study in Human and Monkey. J. Neurosci. 2001, 21, RC163. [Google Scholar] [CrossRef] [PubMed]
- Magistretti, P.J.; Allaman, I. A Cellular Perspective on Brain Energy Metabolism and Functional Imaging. Neuron 2015, 86, 883–901. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzuki, A.; Stern, S.A.; Bozdagi, O.; Huntley, G.W.; Walker, R.H.; Magistretti, P.J.; Alberini, C.M. Astrocyte-Neuron Lactate Transport Is Required for Long-Term Memory Formation. Cell 2011, 144, 810–823. [Google Scholar] [CrossRef] [Green Version]
- Poskanzer, K.E.; Yuste, R. Astrocytic regulation of cortical UP states. Proc. Natl. Acad. Sci. USA 2011, 108, 18453–18458. [Google Scholar] [CrossRef] [Green Version]
- Burt, J.B.; Demirtas, M.; Eckner, W.J.; Navejar, N.M.; Ji, J.L.; Martin, W.J.; Bernacchia, A.; Anticevic, A.; Murray, J.D. Hierarchy of tran-scriptomic specialization across human cortex captured by structural neuroimaging topography. Nat. Neurosci. 2018, 21, 1251–1259. [Google Scholar] [CrossRef]
- Margulies, D.S.; Ghosh, S.S.; Goulas, A.; Falkiewicz, M.; Huntenburg, J.M.; Langs, G.; Bezgin, G.; Eickhoff, S.B.; Castellanos, F.; Petrides, M.; et al. Situating the default-mode network along a principal gradient of macroscale cortical organization. Proc. Natl. Acad. Sci. USA 2016, 113, 12574–12579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palmqvist, S.; Schöll, M.; Strandberg, O.; Mattsson-Carlgren, N.; Stomrud, E.; Zetterberg, H.; Blennow, K.; Landau, S.; Jagust, W.; Hansson, O. Earliest accumulation of β-amyloid occurs within the default-mode network and concurrently affects brain connectivity. Nat. Commun. 2017, 8, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Huntenburg, J.M.; Bazin, P.-L.; Margulies, D.S. Large-Scale Gradients in Human Cortical Organization. Trends Cogn. Sci. 2018, 22, 21–31. [Google Scholar] [CrossRef] [Green Version]
- Glasser, M.F.; Goyal, M.S.; Preuss, T.M.; Raichle, M.E.; Van Essen, D.C. Trends and properties of human cerebral cortex: Correlations with cortical myelin content. NeuroImage 2013, 93, 165–175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vaishnavi, S.N.; Vlassenko, A.G.; Rundle, M.M.; Snyder, A.Z.; Mintun, M.A.; Raichle, M.E. Regional aerobic glycolysis in the human brain. Proc. Natl. Acad. Sci. USA 2010, 107, 17757–17762. [Google Scholar] [CrossRef] [Green Version]
- Chaudhuri, R.; Knoblauch, K.; Gariel, M.-A.; Kennedy, H.; Wang, X.-J. A Large-Scale Circuit Mechanism for Hierarchical Dynamical Processing in the Primate Cortex. Neuron 2015, 88, 419–431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kahali, S.; Raichle, M.E.; Yablonskiy, D.A. The Role of the Human Brain Neuron–Glia–Synapse Composition in Forming Resting-State Functional Connectivity Networks. Brain Sci. 2021, 11, 1565. https://doi.org/10.3390/brainsci11121565
Kahali S, Raichle ME, Yablonskiy DA. The Role of the Human Brain Neuron–Glia–Synapse Composition in Forming Resting-State Functional Connectivity Networks. Brain Sciences. 2021; 11(12):1565. https://doi.org/10.3390/brainsci11121565
Chicago/Turabian StyleKahali, Sayan, Marcus E. Raichle, and Dmitriy A. Yablonskiy. 2021. "The Role of the Human Brain Neuron–Glia–Synapse Composition in Forming Resting-State Functional Connectivity Networks" Brain Sciences 11, no. 12: 1565. https://doi.org/10.3390/brainsci11121565






