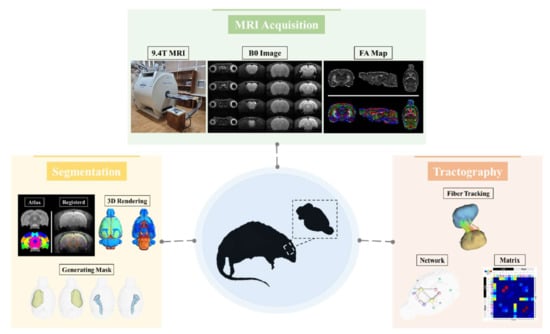
Deterministic Tractography Analysis of Rat Brain Using SIGMA Atlas in 9.4T MRI
Abstract
1. Introduction
2. Materials and Methods
2.1. Preparation of Animals
2.2. Animal Models
2.3. MRI Acquisition
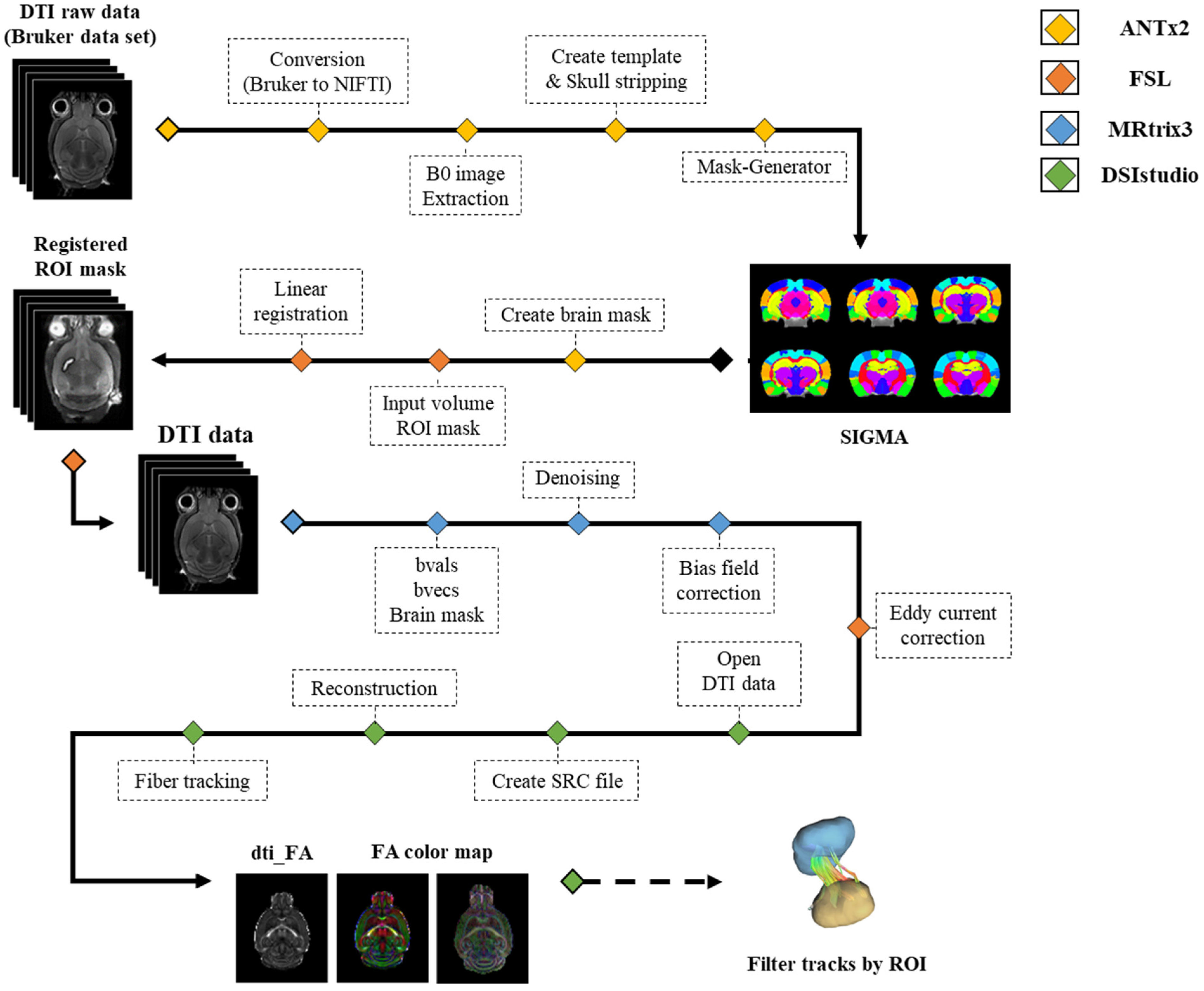
2.4. Image Data Processing
2.5. Deterministic Tractography
3. Results
3.1. SIGMA Atlas-Based Whole Brain Segmentation and Registration
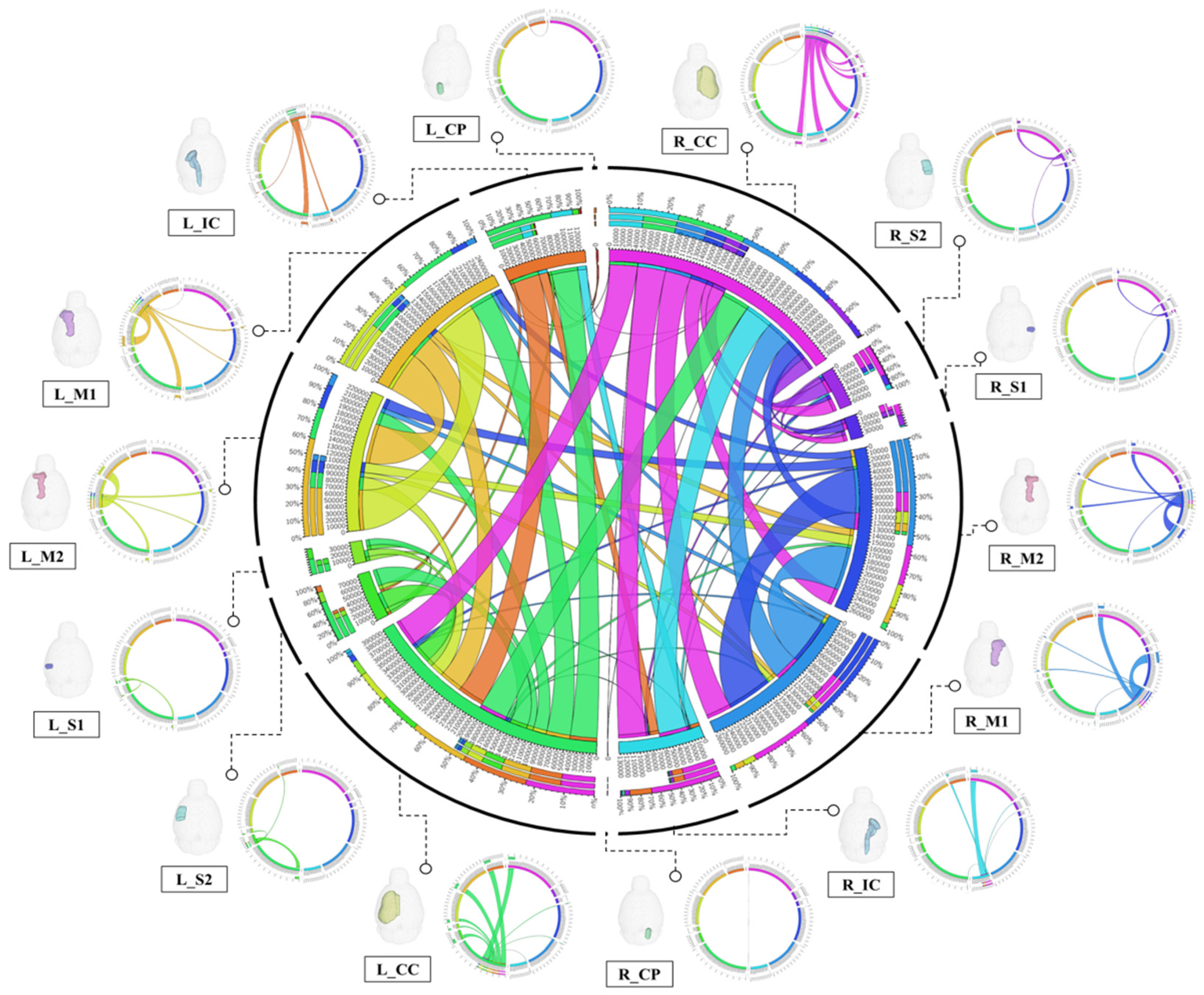
3.2. Deterministic Tractographic Analysis
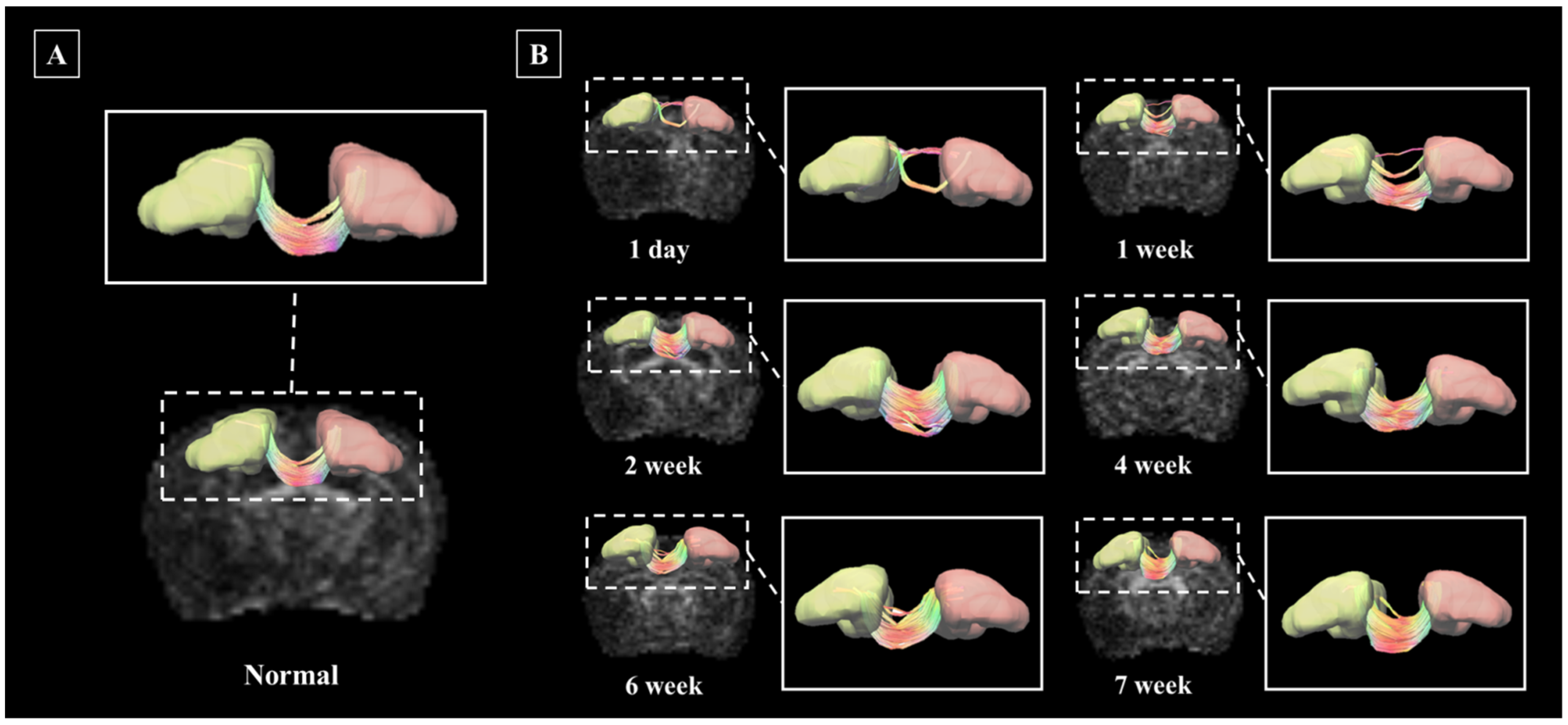
3.3. Application of Deterministic Tractographic Analysis of Stroke Model
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cozzi, J.; Fraichard, A.; Thiam, K. Use of genetically modified rat models for translational medicine. Drug Discov. Today 2008, 13, 488–494. [Google Scholar] [CrossRef] [PubMed]
- Veraart, J.; Leergaard, T.B.; Antonsen, B.T.; Van Hecke, W.; Blockx, I.; Jeurissen, B.; Jiang, Y.; Van der Linden, A.; Jonhnson, G.A.; Verhoye, M.; et al. Population-averaged diffusion tensor imaging atlas of the Sprague Dawley rat brain. Neuroimage 2011, 58, 975–983. [Google Scholar] [CrossRef] [PubMed]
- Pallast, N.; Diedenhofen, M.; Blaschke, S.; Wieters, F.; Wiedermann, D.; Hoehn, M.; Fink, G.R.; Aswendt, M. Processing pipeline for atlas-based imaging data analysis of structural and functional mouse brain MRI (AIDAmri). Front. Neuroinform. 2019, 13, 42. [Google Scholar] [CrossRef] [PubMed]
- Badhwar, A.; Lerch, J.P.; Hamel, E.; Sled, J.G. Impaired structural correlates of memory in Alzheimer’s disease mice. NeuroImage Clin. 2013, 3, 290–300. [Google Scholar] [CrossRef]
- Hébert, F.; Grand’Maison, M.; Ho, M.K.; Lerch, J.P.; Hamel, E.; Bedell, B.J. Cortical atrophy and hypoperfusion in a transgenic mouse model of Alzheimer’s disease. Neurobiol. Aging 2013, 34, 1644–1652. [Google Scholar] [CrossRef] [PubMed]
- Pearn, M.L.; Niesman, I.R.; Egawa, J.; Sawada, A.; Almenar-Queralt, A.; Shah, S.B.; Duckworth, J.L.; Head, B.P. Pathophysiology associated with traumatic brain injury: Current treatments and potential novel therapeutics. Cell. Mol. Neurobiol. 2017, 37, 571–585. [Google Scholar] [CrossRef]
- Xue, K.; Luo, C.; Zhang, D.; Yang, T.; Li, J.; Gong, D.; Chen, L.; Medina, Y.I.; Gotman, J.; Zhou, D.; et al. Diffusion tensor tractography reveals disrupted structural connectivity in childhood absence epilepsy. Epilepsy Res. 2014, 108, 125–138. [Google Scholar] [CrossRef]
- Ciccarelli, O.; Toosy, A.T.; Parker, G.J.; Wheeler-Kingshott, C.A.M.; Barker, G.J.; Miller, D.H.; Thompson, A.J. Diffusion tractography based group mapping of major white-matter pathways in the human brain. Neuroimage 2003, 19, 1545–1555. [Google Scholar] [CrossRef]
- Dauguet, J.; Peled, S.; Berezovskii, V.; Delzescaux, T.; Warfield, S.K.; Born, R.; Westin, C.F. Comparison of fiber tracts derived from in-vivo DTI tractography with 3D histological neural tract tracer reconstruction on a macaque brain. Neuroimage 2007, 37, 530–538. [Google Scholar] [CrossRef] [PubMed]
- Tournier, J.D.; Calamante, F.; Connelly, A. MRtrix: Diffusion tractography in crossing fiber regions. Int. J. Imaging Syst. Technol. 2012, 22, 53–66. [Google Scholar] [CrossRef]
- Yeh, F.C.; Verstynen, T.D.; Wang, Y.; Fernández-Miranda, J.C.; Tseng, W.Y.I. Deterministic diffusion fiber tracking improved by quantitative anisotropy. PLoS ONE 2013, 8, e80713. [Google Scholar]
- Jones, D.K. Challenges and limitations of quantifying brain connectivity in vivo with diffusion MRI. Imaging Med. 2010, 2, 341. [Google Scholar] [CrossRef]
- Roberts, J.A.; Perry, A.; Roberts, G.; Mitchell, P.B.; Breakspear, M. Consistency-based thresholding of the human connectome. NeuroImage 2017, 145, 118–129. [Google Scholar] [CrossRef]
- Sotiropoulos, S.N.; Zalesky, A. Building connectomes using diffusion MRI: Why, how and but. NMR Biomed. 2019, 32, e3752. [Google Scholar] [CrossRef] [PubMed]
- Behrens, T.E.; Berg, H.J.; Jbabdi, S.; Rushworth, M.F.; Woolrich, M.W. Probabilistic diffusion tractography with multiple fibre orientations: What can we gain? Neuroimage 2007, 34, 144–155. [Google Scholar] [CrossRef]
- Sherbondy, A.J.; Dougherty, R.F.; Napel, S.; Wandell, B.A. Identifying the human optic radiation using diffusion imaging and fiber tractography. J. Vis. 2008, 8, 12. [Google Scholar] [CrossRef]
- Chen, J.L.; Kumar, S.; Williamson, V.J.; Scholz, J.; Griffiths, T.D.; Stewart, L. Detection of the arcuate fasciculus in congenital amusia depends on the tractography algorithm. Front. Psychol. 2015, 6, 9. [Google Scholar] [CrossRef]
- Hu, B.; Ye, B.; Yang, Y.; Zhu, K.; Kang, Z.; Kuang, S.; Kuang, S.; Luo, L.; Shan, H. Quantitative diffusion tensor deterministic and probabilistic fiber tractography in relapsing–remitting multiple sclerosis. Eur. J. Radiol. 2011, 79, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Bucci, M.; Mandelli, M.L.; Berman, J.I.; Amirbekian, B.; Nguyen, C.; Berger, M.S.; Henry, R.G. Quantifying diffusion MRI tractography of the corticospinal tract in brain tumors with deterministic and probabilistic methods. NeuroImage Clin. 2013, 3, 361–368. [Google Scholar] [CrossRef]
- Petersen, M.V.; Lund, T.E.; Sunde, N.; Frandsen, J.; Rosendal, F.; Juul, N.; Østergaard, K. Probabilistic versus deterministic tractography for delineation of the cortico-subthalamic hyperdirect pathway in patients with Parkinson disease selected for deep brain stimulation. J. Neurosurg. 2017, 126, 1657–1668. [Google Scholar] [CrossRef]
- Sarwar, T.; Ramamohanarao, K.; Zalesky, A. Mapping connectomes with diffusion MRI: Deterministic or probabilistic tractography? Magn. Reson. Med. 2019, 81, 1368–1384. [Google Scholar] [CrossRef]
- Papp, E.A.; Leergaard, T.B.; Calabrese, E.; Johnson, G.A.; Bjaalie, J.G. Waxholm Space atlas of the Sprague Dawley rat brain. Neuroimage 2014, 97, 374–386. [Google Scholar] [CrossRef]
- Wijetunge, C.D.; Saeed, I.; Boughton, B.A.; Spraggins, J.M.; Caprioli, R.M.; Bacic, A.; Roessner, U.; Halgamuge, S.K. EXIMS: An improved data analysis pipeline based on a new peak picking method for EXploring Imaging Mass Spectrometry data. Bioinformatics 2015, 31, 3198–3206. [Google Scholar] [CrossRef][Green Version]
- Immonen, R.; Smith, G.; Brady, R.D.; Wright, D.; Johnston, L.; Harris, N.G.; Manninen, E.; Salo, R.; Branch, C.; Duncan, D.; et al. Harmonization of pipeline for preclinical multicenter MRI biomarker discovery in a rat model of post-traumatic epileptogenesis. Epilepsy Res. 2019, 150, 46–57. [Google Scholar] [CrossRef]
- Goerzen, D.; Fowler, C.; Devenyi, G.A.; Germann, J.; Madularu, D.; Chakravarty, M.M.; Near, J. An MRI-derived neuroanatomical atlas of the Fischer 344 rat brain. Sci. Rep. 2020, 10, 1–13. [Google Scholar] [CrossRef]
- Diao, Y.; Yin, T.; Gruetter, R.; Jelescu, I.O. PIRACY: An optimized pipeline for functional connectivity analysis in the rat brain. Front. Neurosci. 2021, 15, 285. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.J.; Kovacevic, N.; Lobaugh, N.J.; Sled, J.G.; Henkelman, R.M.; Henderson, J.T. Neuroanatomical differences between mouse strains as shown by high-resolution 3D MRI. Neuroimage 2006, 29, 99–105. [Google Scholar] [CrossRef] [PubMed]
- Lancelot, S.; Roche, R.; Slimen, A.; Bouillot, C.; Levigoureux, E.; Langlois, J.B.; Zimmer, L.; Costes, N. A multi-atlas based method for automated anatomical rat brain MRI segmentation and extraction of PET activity. PLoS ONE 2014, 9, e109113. [Google Scholar] [CrossRef]
- Barrière, D.A.; Magalhães, R.; Novais, A.; Marques, P.; Selingue, E.; Geffroy, F.; Marques, F.; Cerqueira, J.; Sousa, J.C.; Boumezbeur, F.; et al. The SIGMA rat brain templates and atlases for multimodal MRI data analysis and visualization. Nat. Commun. 2019, 10, 5699. [Google Scholar] [CrossRef]
- Shin, J.A.; Park, E.M.; Choi, J.S.; Seo, S.M.; Kang, J.L.; Lee, K.E.; Cho, S. Ischemic preconditioning-induced neuroprotection is associated with differential expression of IL-1β and IL-1 receptor antagonist in the ischemic cortex. J. Neuroimmunol. 2009, 217, 14–19. [Google Scholar] [CrossRef] [PubMed]
- Lein, E.S.; Hawrylycz, M.J.; Ao, N.; Ayres, M.; Bensinger, A.; Bernard, A.; Boe, A.F.; Boguski, M.S.; Brockway, K.S.; Byrnes, E.J.; et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature 2007, 445, 168–176. [Google Scholar] [CrossRef]
- Huebner, N.S.; Mechling, A.E.; Lee, H.L.; Reisert, M.; Bienert, T.; Hennig, J.; von Elverfeldt, D.; Harsan, L.A. The connectomics of brain demyelination: Functional and structural patterns in the cuprizone mouse model. Neuroimage 2017, 146, 1–18. [Google Scholar] [CrossRef]
- Koch, S.; Mueller, S.; Foddis, M.; Bienert, T.; von Elverfeldt, D.; Knab, F.; Farr, T.D.; Bernard, R.; Dopatka, M.; Rex, A.; et al. Atlas registration for edema-corrected MRI lesion volume in mouse stroke models. J. Cereb. Blood Flow Metab. 2019, 39, 313–323. [Google Scholar] [CrossRef]
- Jenkinson, M.; Bannister, P.; Brady, M.; Smith, S. Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage 2002, 17, 825–841. [Google Scholar] [CrossRef] [PubMed]
- Jenkinson, M.; Pechaud, M.; Smith, S. BET2: MR-based estimation of brain, skull and scalp surfaces. In Proceedings of the Eleventh Annual Meeting of the Organization for Human Brain Mapping, Toronto, ON, Canada, 12–16 June 2005; Volume 17, p. 167. [Google Scholar]
- Cordero-Grande, L.; Christiaens, D.; Hutter, J.; Price, A.N.; Hajnal, J.V. Complex diffusion-weighted image estimation via matrix recovery under general noise models. Neuroimage 2019, 200, 391–404. [Google Scholar] [CrossRef] [PubMed]
- Graham, M.S.; Drobnjak, I.; Zhang, H. Realistic simulation of artefacts in diffusion MRI for validating post-processing correction techniques. NeuroImage 2016, 125, 1079–1094. [Google Scholar] [CrossRef]
- Huber, M.; Trattnig, S.; Lintner, F. Anatomy, biochemistry, and physiology of articular cartilage. Investig. Radiol. 2000, 35, 573–580. [Google Scholar] [CrossRef] [PubMed]
- Straube, A. Basic description of computed tomography and magnetic resonance imaging of the brain. Sven. Veterinärtidning 2019, 71, 28–31. [Google Scholar]
- Guida, F.; De Gregorio, D.; Palazzo, E.; Ricciardi, F.; Boccella, S.; Belardo, C.; Iannotta, M.; Infantino, R.; Formato, F.; Marabese, I.; et al. Behavioral, biochemical and electrophysiological changes in spared nerve injury model of neuropathic pain. Int. J. Mol. Sci. 2020, 21, 3396. [Google Scholar] [CrossRef] [PubMed]
- Steward, C.A.; Marsden, C.A.; Prior, M.J.W.; Morris, P.G.; Shah, Y. Methodological considerations in rat brain BOLD contrast pharmacological MRI. Psychopharmacology 2005, 180, 687–704. [Google Scholar] [CrossRef]
- Crawley, J.N. Behavioral phenotyping strategies for mutant mice. Neuron 2008, 57, 809–818. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Rane, P.; Huang, W.; Liang, Z.; Kennedy, D.; Frazier, J.A.; King, J. Mapping resting-state brain networks in conscious animals. J. Neurosci. Methods 2010, 189, 186–196. [Google Scholar] [CrossRef] [PubMed]
- Evans, P.G.; Sokolska, M.; Alves, A.; Harrison, I.F.; Ohene, Y.; Nahavandi, P.; Ismail, O.; Miranda, E.; Lythgoe, M.F.; Thomas, D.L.; et al. Non-Invasive MRI of Blood–Cerebrospinal Fluid Barrier Function. Nat. Commun. 2020, 11, 2081. [Google Scholar] [CrossRef]
- Rubinov, M.; Sporns, O. Complex network measures of brain connectivity: Uses and interpretations. Neuroimage 2010, 52, 1059–1069. [Google Scholar] [CrossRef] [PubMed]
- Cui, Z.; Zhong, S.; Xu, P.; Gong, G.; He, Y. PANDA: A pipeline toolbox for analyzing brain diffusion images. Front. Hum. Neurosci. 2013, 7, 42. [Google Scholar] [CrossRef]
- De Guzman, A.E.; Wong, M.D.; Gleave, J.A.; Nieman, B.J. Variations in post-perfusion immersion fixation and storage alter MRI measurements of mouse brain morphometry. Neuroimage 2016, 142, 687–695. [Google Scholar] [CrossRef] [PubMed]
- Calabrese, E.; Badea, A.; Cofer, G.; Qi, Y.; Johnson, G.A. A diffusion MRI tractography connectome of the mouse brain and comparison with neuronal tracer data. Cereb. Cortex 2015, 25, 4628–4637. [Google Scholar] [CrossRef]
- Cacciola, A.; Calamuneri, A.; Milardi, D.; Mormina, E.; Chillemi, G.; Marino, S.; Naro, A.; Rizzo, G.; Anastasi, G.; Quartarone, A. A connectomic analysis of the human basal ganglia network. Front. Neuroanat. 2017, 11, 85. [Google Scholar] [CrossRef] [PubMed]
- Po, C.; Kalthoff, D.; Kim, Y.B.; Nelles, M.; Hoehn, M. White matter reorganization and functional response after focal cerebral ischemia in the rat. PLoS ONE 2012, 7, e45629. [Google Scholar] [CrossRef]
- Liu, Z.; Xin, H.; Chopp, M. Axonal remodeling of the corticospinal tract during neurological recovery after stroke. Neural Regen. Res. 2021, 16, 939. [Google Scholar] [CrossRef]
- Andersson, G.; Orädd, G.; Sultan, F.; Novikov, L.N. In vivo diffusion tensor imaging, diffusion kurtosis imaging, and tractography of a sciatic nerve injury model in rat at 9.4 T. Sci. Rep. 2018, 8, 12911. [Google Scholar] [CrossRef]
- Pallast, N.; Wieters, F.; Nill, M.; Fink, G.R.; Aswendt, M. Graph theoretical quantification of white matter reorganization after cortical stroke in mice. NeuroImage 2020, 217, 116873. [Google Scholar] [CrossRef]
- Veksler, R.; Vazana, U.; Serlin, Y.; Prager, O.; Ofer, J.; Shemen, N.; Fisher, A.M.; Minaeva, O.; Hua, N.; Saar-Ashkenazy, R.; et al. Slow blood-to-brain transport underlies enduring barrier dysfunction in American football players. Brain 2020, 143, 1826–1842. [Google Scholar] [CrossRef]
- Gao, Y.; Choe, A.S.; Stepniewska, I.; Li, X.; Avison, M.J.; Anderson, A.W. Validation of DTI tractography-based measures of primary motor area connectivity in the squirrel monkey brain. PLoS ONE 2013, 8, e75065. [Google Scholar] [CrossRef]
- Sinke, M.R.; Otte, W.M.; van Meer, M.P.; van der Toorn, A.; Dijkhuizen, R.M. Modified structural network backbone in the contralesional hemisphere chronically after stroke in rat brain. J. Cereb. Blood Flow Metab. 2018, 38, 1642–1653. [Google Scholar] [CrossRef] [PubMed]
- Jeurissen, B.; Descoteaux, M.; Mori, S.; Leemans, A. Diffusion MRI fiber tractography of the brain. NMR Biomed. 2019, 32, e3785. [Google Scholar] [CrossRef]
- Trinkle, S.; Foxley, S.; Wildenberg, G.; Kasthuri, N.; La Rivière, P. The role of spatial embedding in mouse brain networks constructed from diffusion tractography and tracer injections. NeuroImage 2021, 244, 118576. [Google Scholar] [CrossRef]
- Schilling, K.G.; Nath, V.; Hansen, C.; Parvathaneni, P.; Blaber, J.; Gao, Y.; Neher, P.; Aydogan, D.B.; Shi, Y.; Ocampo-Pineda, M.; et al. Limits to anatomical accuracy of diffusion tractography using modern approaches. NeuroImage 2019, 185, 1–11. [Google Scholar] [CrossRef]
- Poulin, P.; Jörgens, D.; Jodoin, P.M.; Descoteaux, M. Tractography and machine learning: Current state and open challenges. Magn. Reson. Imaging 2019, 64, 37–48. [Google Scholar] [CrossRef]
- Sarwar, T.; Ramamohanarao, K.; Zalesky, A. A critical review of connectome validation studies. NMR Biomed. 2021, 34, e4605. [Google Scholar] [CrossRef]
- Rheault, F.; Poulin, P.; Caron, A.V.; St-Onge, E.; Descoteaux, M. Common misconceptions, hidden biases and modern challenges of dMRI tractography. J. Neural Eng. 2020, 17, 011001. [Google Scholar] [CrossRef]
- Schilling, K.G.; Petit, L.; Rheault, F.; Remedios, S.; Pierpaoli, C.; Anderson, A.W.; Landman, B.A.; Descoteaux, M. Brain connections derived from diffusion MRI tractography can be highly anatomically accurate—if we know where white matter pathways start, where they end, and where they do not go. Brain Struct. Funct. 2020, 225, 2387–2402. [Google Scholar] [CrossRef]
- Morales, H. Current and Future Challenges of Functional MRI and Diffusion Tractography in the Surgical Setting: From Eloquent Brain Mapping to Neural Plasticity. In Seminars in Ultrasound, CT and MRI; WB Saunders: Philadelphia, PA, USA, 2021; Volume 42, pp. 474–489. [Google Scholar]
- Huang, H.; Zhang, J.; Van Zijl, P.C.; Mori, S. Analysis of noise effects on DTI-based tractography using the brute-force and multi-ROI approach. Magn. Reson. Med. Off. J. Int. Soc. Magn. Reson. Med. 2004, 52, 559–565. [Google Scholar] [CrossRef] [PubMed]
- Chou, M.C.; Lin, Y.R.; Huang, T.Y.; Wang, C.Y.; Chung, H.W.; Juan, C.J.; Chen, C.Y. FLAIR diffusion-tensor MR tractography: Comparison of fiber tracking with conventional imaging. Am. J. Neuroradiol. 2005, 26, 591–597. [Google Scholar] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Im, S.-J.; Suh, J.-Y.; Shim, J.-H.; Baek, H.-M. Deterministic Tractography Analysis of Rat Brain Using SIGMA Atlas in 9.4T MRI. Brain Sci. 2021, 11, 1656. https://doi.org/10.3390/brainsci11121656
Im S-J, Suh J-Y, Shim J-H, Baek H-M. Deterministic Tractography Analysis of Rat Brain Using SIGMA Atlas in 9.4T MRI. Brain Sciences. 2021; 11(12):1656. https://doi.org/10.3390/brainsci11121656
Chicago/Turabian StyleIm, Sang-Jin, Ji-Yeon Suh, Jae-Hyuk Shim, and Hyeon-Man Baek. 2021. "Deterministic Tractography Analysis of Rat Brain Using SIGMA Atlas in 9.4T MRI" Brain Sciences 11, no. 12: 1656. https://doi.org/10.3390/brainsci11121656
APA StyleIm, S.-J., Suh, J.-Y., Shim, J.-H., & Baek, H.-M. (2021). Deterministic Tractography Analysis of Rat Brain Using SIGMA Atlas in 9.4T MRI. Brain Sciences, 11(12), 1656. https://doi.org/10.3390/brainsci11121656