Deep Learning-Based Studies on Pediatric Brain Tumors Imaging: Narrative Review of Techniques and Challenges
Abstract
:1. Introduction
2. Related Works
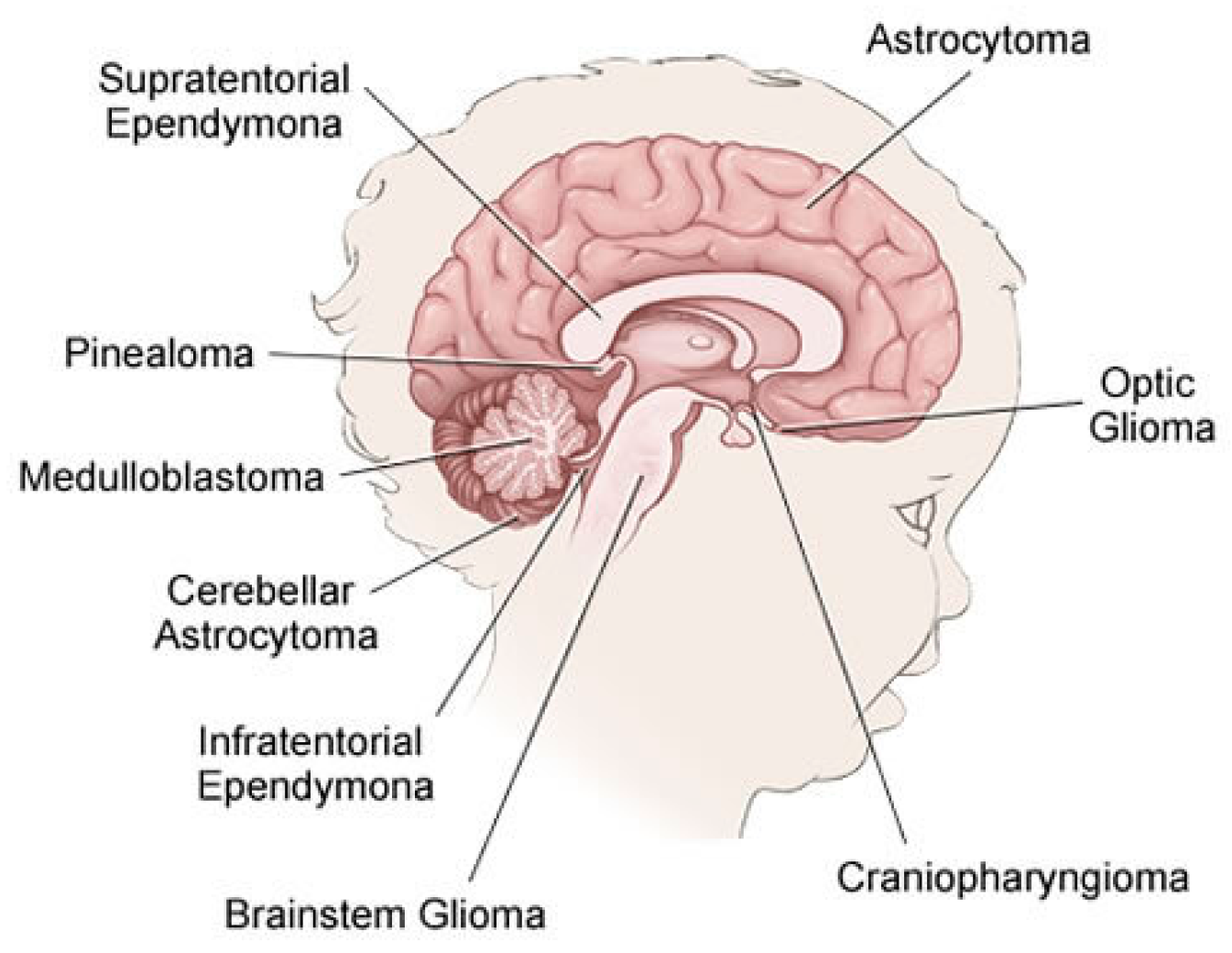
2.1. Brain Tumor in Childhood
- Gliomas: Is a generic name for a number of cancers, including:
- Astrocytomas (which include glioblastomas): From a specific type of glial cells called astrocytes, these types of tumors are usually started. They are often grouped by grade. Low-grade astrocytomas include pilocytic astrocytomas, subependymal giant cell astrocytomas (SEGAs), diffuse astrocytomas, pleomorphic xanthoastrocytomas (PXAs) and optic gliomas. High-grade astrocytomas include glioblastomas and anaplastic astrocytomas.
- Oligodendrogliomas: From a specific type of cerebral cells called oligodendrocytes, these types of tumor are usually started. Oligodendrogliomas have been categorized as Grade II tumors that account for over 1% of children’s brain tumors.
- Ependymomas: From the ependymal cells which line the spinal cord, begins this type of tumors which responsible for around 5% of brain tumors in children. They can vary from Grade I tumors to Grade III tumors (anaplastic ependymomas).
- Brainstem gliomas: This tumor is a glioma that develops in brain stem and is responsible for around 10% to 20% of brain tumors in children. They are common in two types: focal brain stem gliomas or diffuse midline gliomas.
- Embryonal tumors: These tumors begin in the central nervous system, in early forms of nerve cells. In children, they are common among younger children rather than older children. Embryonic tumors account for around 10–20% of brain tumors, including the most frequent type: medulloblastomas, and less common types such as medulloepithelioma, and atypical teratoid (ATRT).
- Pineal tumors: Are there any types of tumors that could be found in the pineal gland? The most popular, fastest growing and difficult to treat type of these forms is called pineoblastomas.
- Craniopharyngiomas: Craniopharyngiomas account for approximately 4% of children’s brain tumors. They occur over the pituitary gland, but it is under the brain itself that these slow-growing tumors begin.
- Mixed neuronal and glial tumors: This type of tumor combined between neuronal and glial tumors. They include dysembryoplastic neuroepithelial tumors (DNETs) and gangliogliomais.
- Choroid plexus tumors: They are a rare tumor, many of which are benign and some are malignant.
- Schwannomas: They begin in cells that surround and separate the cranial nerves and other nerves. These rare tumors are usually benign.
- In or near brain tumors: These include chordomas, tumors of germ cells, neuroblastomas, pituitary tumors, meningiomas (Grade I to Grade III) and lymphomas.
- Metastatic or secondary brain tumors: The tumors that begin in other organs and then spread to the brain are metastatic or secondary brain tumors. They are often less frequent than primary brain tumors and often treated differently.
2.2. Pediatric Brain Imaging Technique
2.3. Reading MRI Sequences
2.4. Available Pediatric Brain Datasets
- dHCP: The Developing Human Connectome Project (dHCP) [28] is an ERC-funded collaboration between King’s College London, Imperial College London and the University of Oxford. dBCP has two data releases as to date. The first open access data release consists of images of 40 representative term neonatal subjects. The imaging data includes structural imaging, structural connectivity data (diffusion MRI) and functional connectivity data (resting-state fMRI). The second open access data release consists of images of 558 neonatal subjects. The released dataset includes T1w and T2w structural data supplied as initial image data and after pipeline preprocessing. The images included in this release were obtained from infants born and imaged between 24–45 weeks of age. Using a dedicated neonatal imaging device which included a neonatal 32 channel phased array head coil, imaging was carried out on 3T Philips Achieva.
- PBTA: Pediatric Brain Tumor Atlas (PBTA) [29] is a collaborative effort, which is led by the Children’s Brain Tumor Tissue Consortium (CBTTC), to accelerate discoveries for therapeutic intervention for children brain tumors diagnosed. The first release of the Pediatric Brain Tumor Atlas (PBTA) dataset, which comprises over 30 different types of pediatric brain tumors covering over 1000 subjects, occurred on September, 2018. Data types include match tumor/normal, whole genome data (WGS), RNAseq, proteomics, longitudinal clinical data, imaging data (including MRIs and radiology reports), histology slide images and pathology reports.
- HCP: The Lifespan Human Connectome Project Development [30] lunch Lifespan HCP Release 1.0 in May 2019 for HCP-Development and HCP-Aging. All HCP-development (ages 5–21) data is shared in the NIMH Data Archive, NDA Collection. Lifespan HCP Release 1.0 data includes unprocessed data of all modalities (structural MRI, resting state fMRI, task fMRI, and diffusion MRI) for 655 HCP-D subjects, minimally preprocessed structural MRI data (only) for 84 subjects, and basic demographic data (age, sex, race/ethnicity, and handedness) for all released HCP-D subjects.
- PING: Pediatric Imaging, Neurocognition, and Genetics [31] data of 1400 children aged between 3 and 20 years are included in this genetics data resource. PING data access is thoroughly handled by the NIMH Data Repository.
- iSeg-2017 and iSeg-2019: Challenge data six-month infant brain MRI segmentation (iSeg-2017) [32]. Comparing (semi-)automatic algorithms for the segmentation of 6-month infant brain tissues and the calculation of corresponding structures was its goal of the iSeg-2017 competition. On a Siemens head-only 3 T scanner with a circular polarized head coil, all scans for the 10 infant subjects were obtained. The six-month infant brain MRI segmentation (iSeg-2017) [33] aims to facilitate automated six-month infant brain MRI segmentation algorithms from multiple sites. They offered iSeg-2017 data for training datasets. For the validation dataset, 13 T1 and T2 subject MR images are given. T1- and T2-weighted MR images from three different sites are used in the test dataset.
- IBSR: Internet Brain Segmentation Repository [34]. Along with magnetic resonance brain image data, IBSR provides manually-guided expert segmentation results. Its aim is to promote the assessment and development of methods of segmentation. This dataset contains eighteen currently available subjects aged 7–71 years.
- ABIDE I and ABIDE II: The first ABIDE [35] project launched in August 2012 reflects Autism Brain Imaging Data Sharing (ABIDE I). Seventeen foreign sites were interested in ABIDE I, exchanging previously acquired resting state functional magnetic resonance imaging (R-fMRI) data. ABIDE I is comprised of 1112 datasets, including 539 from ASD individuals and 573 from typical controls. ABIDE II [36]. In order to further encourage research on the brain connectome in ASD, ABIDE II was released in 2016. There are 19 sites in ABIDEII, donating a total of 1114 datasets from 521 ASD individuals and 593 typical controls.
- CoRR: The Consortium for Reliability and Reproducibility [37]. The goal was to create an open science database for the imaging community to facilitate the assessment of the reliability and reproducibility of functional and structural connectomics studies. CoRR contains 33 datasets, 32 of which are available for download at present. Four of these datasets contains pediatric brain MIR images. IPCAS 2 includes 35 typically developing children. Each participant underwent two scanning sessions one month apart. Three modalities (T1/EPI (echo planar imaging)/DTI (diffusion tensor imaging)) of brain images were acquired for all subjects. IPCAS 7 includes 74 typically developing children. Each participant was scanned twice within a session. Three modalities (T1/T2/EPI) of brain images were acquired for all subjects.
2.5. Data Acquisition and Analysis Methods for Human Brain Activity
3. Pediatric Brain Tumor Deep Learning-Based Studies
3.1. Pediatric Brain Tumor Detection and Classification
3.2. Pediatric Brain Tumor Segmentation
3.3. Related Pediatric Brain Tumor Studies
| Authors | Tumor Subject | Methodology | Modality | Dataset | Results |
|---|---|---|---|---|---|
| Ladefoged, Claes Nøhr, et al. (2018) [68] | Air, soft tissue and bone tissue | DeepUTE | PET/MRI (vendor-provided UTE images) | 79 children (aged between 2–14 years) | Jaccard index 0.74/0.79 in soft tissue, 0.53/0.70 in bone tissue, 0.57/0.62 in air |
| Wang, Geliang, et al. (2020) [44] | Brain region volume Small-world properties Properties of brain structural network | BET, iBEAT and iBEAT with manual correction | 3D T1WI | 22 neonates (13 boys and 9 girls) | Brain regions analysis: significant differences in 50 brain region with iBEAT with manual correction showed the more accurate brain segmentation |
| Chang, Alex, et al. (2020) [69] | Whole body | DCGAN, StyleGAN, PGStyleGAN, StyleGAN2 + FID/DFD VAE for evaluation | 360 wbMRI slices | 90 healthy patients (ages 4 to 18) | FID, DFD, false positive rate: (457.30, 23.72, 0%) for DCGAN, (481.3, 19.378, 0%) for StyleGAN, (442.61, 18.56, 20%) for PGStyleGAN, (497.09, 17.234, 30%) for StyleGAN2 |
4. Medical and Technical Challenges
5. Conclusions and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kleihues, P.; Burger, P.C.; Scheithauer, B.W. The New WHO Classification of Brain Tumours. Brain Pathol. 1993, 3, 255–268. [Google Scholar] [CrossRef]
- Schroeder, A.; Heller, D.A.; Winslow, M.M.; Dahlman, J.E.; Pratt, G.W.; Langer, R.; Jacks, T.; Anderson, D.G. Treating metastatic cancer with nanotechnology. Nat. Rev. Cancer 2011, 12, 39–50. [Google Scholar] [CrossRef]
- Rehman, A.; Saba, T. An intelligent model for visual scene analysis and compression. Int. Arab. J. Inf. Technol. 2013, 10, 126–136. [Google Scholar]
- Dang, M.; Phillips, P.C. Pediatric Brain Tumors. Contin. Lifelong Learn. Neurol. 2017, 23, 1727–1757. [Google Scholar] [CrossRef]
- Montemurro, N. Glioblastoma Multiforme and Genetic Mutations: The Issue Is Not Over Yet. An Overview of the Current Literature. J. Neurol. Surg. Part A Central Eur. Neurosurg. 2020, 81, 064–070. [Google Scholar] [CrossRef] [Green Version]
- DeAngelis, L.M. Brain Tumors. N. Engl. J. Med. 2001, 344, 114–123. [Google Scholar] [CrossRef] [Green Version]
- Wen, P.Y.; Macdonald, D.R.; Reardon, D.A.; Cloughesy, T.F.; Sorensen, A.G.; Galanis, E.; DeGroot, J.; Wick, W.; Gilbert, M.R.; Lassman, A.B.; et al. Updated Response Assessment Criteria for High-Grade Gliomas: Response Assessment in Neuro-Oncology Working Group. J. Clin. Oncol. 2010, 28, 1963–1972. [Google Scholar] [CrossRef]
- Liang, Z.; Lauterbur, P. Principles of Magnetic Resonance Imaging: A Signal Processing Perspective; IEEE Press: New York, NY, USA, 2002; Volume 19, pp. 86–87. [Google Scholar]
- Neurosurgery, B.T.C.J.H.M. Types of Brain and Spinal Cord Tumors in Children. Available online: https://www.hopkinsmedicine.org/neurology_neurosurgery/centers_clinics/brain_tumor/specialty-centers/pediatric/tumors/ (accessed on 17 April 2021).
- Pruitt, D.W.; Bolikal, P.D.; Bolger, A.K. Rehabilitation Considerations in Pediatric Brain Tumors. Current Physical Medicine and Rehabilitation Reports 2019, 7, 81–88. [Google Scholar] [CrossRef]
- About Brain and Spinal Cord Tumors in Children. Available online: https://www.cancer.org/cancer/brain-spinal-cord-tumors-children/about/types-of-brain-and-spinal-tumors.html (accessed on 28 May 2021).
- Chen, W. Clinical Applications of PET in Brain Tumors. J. Nucl. Med. 2007, 48, 1468–1481. [Google Scholar] [CrossRef] [Green Version]
- Prasad, P.V. Magnetic Resonance Imaging: Methods and Biologic Applications; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2006; Volume 124. [Google Scholar]
- Saad, N.M.; Abu Bakar, S.A.R.S.; Muda, A.S.; Mokji, M.M. Review of Brain Lesion Detection and Classification using Neuroimaging Analysis Techniques. J. Teknol. 2015, 74. [Google Scholar] [CrossRef] [Green Version]
- Ortiz, A.; Górriz, J.M.; Ramírez, J.; Salas-Gonzalez, D. Improving MRI segmentation with probabilistic GHSOM and multiobjective optimization. Neurocomputing 2013, 114, 118–131. [Google Scholar] [CrossRef]
- Preston, D.C.; Shapiro, B.E. Neuroimaging in Neurology: An Interactive Approach; Elsevier Science Health Science Division: Amsterdam, The Netherlands, 2007. [Google Scholar]
- Luts, J.; Heerschap, A.; Suykens, J.A.; Van Huffel, S. A combined MRI and MRSI based multiclass system for brain tumour recognition using LS-SVMs with class probabilities and feature selection. Artif. Intell. Med. 2007, 40, 87–102. [Google Scholar] [CrossRef]
- Guzmán-De-Villoria, J.A.; Mateos-Pérez, J.M.; Fernández-García, P.; Castro, E.; Desco, M. Added value of advanced over conventional magnetic resonance imaging in grading gliomas and other primary brain tumors. Cancer Imaging 2014, 14, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Drevelegas, A. Imaging of Brain Tumors with Histological Correlations; Springer: Berlin/Heidelberg, Germany, 2011. [Google Scholar]
- Koley, S.; Sadhu, A.K.; Mitra, P.; Chakraborty, B.; Chakraborty, C. Delineation and diagnosis of brain tumors from post contrast T1-weighted MR images using rough granular computing and random forest. Appl. Soft Comput. 2016, 41, 453–465. [Google Scholar] [CrossRef]
- Zhang, N.; Ruan, S.; Lebonvallet, S.; Liao, Q.; Zhu, Y. Kernel feature selection to fuse multi-spectral MRI images for brain tumor segmentation. Comput. Vis. Image Underst. 2011, 115, 256–269. [Google Scholar] [CrossRef]
- Havaei, M.; Davy, A.; Warde-Farley, D.; Biard, A.; Courville, A.; Bengio, Y.; Pal, C.; Jodoin, P.-M.; Larochelle, H. Brain tumor segmentation with Deep Neural Networks. Med. Image Anal. 2017, 35, 18–31. [Google Scholar] [CrossRef] [Green Version]
- Sachdeva, J.; Kumar, V.; Gupta, I.; Khandelwal, N.; Ahuja, C.K. A package-SFERCB-“Segmentation, feature extraction, reduction and classification analysis by both SVM and ANN for brain tumors. ” Appl. Soft Comput. 2016, 47, 151–167. [Google Scholar] [CrossRef]
- Dunkl, V.; Cleff, C.; Stoffels, G.; Judov, N.; Sarikaya-Seiwert, S.; Law, I.; Bøgeskov, L.; Nysom, K.; Andersen, S.B.; Steiger, H.-J.; et al. The Usefulness of Dynamic O-(2-18F-Fluoroethyl)-L-Tyrosine PET in the Clinical Evaluation of Brain Tumors in Children and Adolescents. J. Nucl. Med. 2014, 56, 88–92. [Google Scholar] [CrossRef] [Green Version]
- Misch, M.; Guggemos, A.; Driever, P.H.; Koch, A.; Grosse, F.; Steffen, I.G.; Plotkin, M.; Thomale, U.-W. 18F-FET-PET guided surgical biopsy and resection in children and adolescence with brain tumors. Child’s Nerv. Syst. 2014, 31, 261–267. [Google Scholar] [CrossRef]
- Chukwueke, U.N.; Wen, P.Y. Use of the Response Assessment in Neuro-Oncology (RANO) criteria in clinical trials and clinical practice. CNS Oncol. 2019, 8, CNS28. [Google Scholar] [CrossRef] [Green Version]
- Sun, C.; Shrivastava, A.; Singh, S.; Gupta, A. Revisiting unreasonable effectiveness of data in deep learning era. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 843–852. [Google Scholar]
- The Developing Human Connectome Project (dHCP). Available online: http://www.developingconnectome.org/project/ (accessed on 15 April 2021).
- Pediatric Brain Tumor Atlas (PBTA). Available online: https://cbttc.org/pediatric-brain-tumor-atlas/ (accessed on 15 April 2021).
- The Lifespan Human Connectome Project Development (HCP). Available online: Https://www.humanconnectome.org/article/data-release-10-available-hcp-lifespan-aging-and-development (accessed on 15 April 2021).
- NITRC. Pediatric Imaging, Neurocognition, and Genetics (PING). Available online: Https://www.nitrc.org/projects/ping/ (accessed on 15 April 2021).
- iSeg. Challenge Data 6-month Infant Brain MRI Segmentation (iSeg-2017). Available online: Http://iseg2017.web.unc.edu/ (accessed on 15 April 2021).
- iSeg. 6-month Infant Brain MRI Segmentation (iSeg-2019). Available online: Https://iseg2019.web.unc.edu/ (accessed on 15 April 2021).
- NITRC. Internet Brain Segmentation Repository (IBSR). Available online: Https://www.nitrc.org/projects/ibsr/ (accessed on 15 April 2021).
- Autism Brain Imaging Data Exchange I (ABIDE I). Available online: http://fcon_1000.projects.nitrc.org/indi/abide/abide_I.html (accessed on 15 April 2021).
- Autism Brain Imaging Data Exchange II (ABIDE II). Available online: http://fcon_1000.projects.nitrc.org/indi/abide/abide_II.html (accessed on 15 April 2021).
- Consortium for Reliability and Reproducibility (CoRR). Available online: http://fcon_1000.projects.nitrc.org/indi/CoRR/html/ (accessed on 15 April 2021).
- Rüegg, J.; Gries, C.; Bond-Lamberty, B.; Bowen, G.J.; Felzer, B.S.; McIntyre, N.; A Soranno, P.; Vanderbilt, K.L.; Weathers, K.C.; Bond-Lamberty, B. Completing the data life cycle: Using information management in macrosystems ecology research. Front. Ecol. Environ. 2014, 12, 24–30. [Google Scholar] [CrossRef] [Green Version]
- Nielsen, H.J.; Hjørland, B. Curating research data: The potential roles of libraries and information professionals. J. Doc. 2014, 70, 221–240. [Google Scholar] [CrossRef]
- Berger, H. Über das Elektrenkephalogramm des Menschen. Eur. Arch. Psychiatry Clin. Neurosci. 1929, 87, 527–570. [Google Scholar] [CrossRef]
- Bear, M.; Connors, B.; Paradiso, M. Neuroscience: Exploring the Brain, 3rd ed.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2006; Volume 928. [Google Scholar]
- Paszkiel, S. Data Acquisition Methods for Human Brain Activity. In Analysis and Classification of EEG Signals for Brain-Computer Interfaces. Studies in Computational Intelligence; Springer: Berlin/Heidelberg, Germany, 2020; Volume 852, pp. 3–9. [Google Scholar]
- Paszkiel, S.; Szpulak, P. Methods of acquisition, archiving and biomedical data analysis of brain functioning. In Proceedings of the International Scientific Conference BCI 2018, Opole, Poland, 13–14 March 2018; pp. 158–171. [Google Scholar]
- Wang, G.; Hu, Y.; Li, X.; Wang, M.; Liu, C.; Yang, J.; Jin, C. Impacts of skull stripping on construction of three-dimensional T1-weighted imaging-based brain structural network in full-term neonates. Biomed. Eng. Online 2020, 19, 41. [Google Scholar] [CrossRef] [PubMed]
- Valente, G.; Kaas, A.L.; Formisano, E.; Goebel, R. Optimizing fMRI experimental design for MVPA-based BCI control: Combining the strengths of block and event-related designs. NeuroImage 2019, 186, 369–381. [Google Scholar] [CrossRef] [PubMed]
- Ovaysikia, S.; Tahir, K.A.; Chan, J.L.; DeSouza, J.F.X. Word Wins Over Face: Emotional Stroop Effect Activates the Frontal Cortical Network. Front. Hum. Neurosci. 2011, 4, 234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paszkiel, S. The population modeling of neuronal cell fractions for the use of controlling a mobile robot. Pomiary Autom. Robot. 2013, 17, 254–259. [Google Scholar]
- Paszkiel, S. Characteristics of question of blind source separation using Moore-Penrose pseudoinversion for reconstruction of EEG signal. In Proceedings of the International Conference Automation, Warsaw, Poland, 15–17 March 2017; pp. 393–400. [Google Scholar]
- Arle, J.E.; Morriss, C.; Wang, Z.J.; Zimmerman, R.A.; Phillips, P.G.; Sutton, L.N. Prediction of posterior fossa tumor type in children by means of magnetic resonance image properties, spectroscopy, and neural networks. J. Neurosurg. 1997, 86, 755–761. [Google Scholar] [CrossRef] [Green Version]
- Bidiwala, S.; Pittman, T. Neural Network Classification of Pediatric Posterior Fossa Tumors Using Clinical and Imaging Data. Pediatr. Neurosurg. 2004, 40, 8–15. [Google Scholar] [CrossRef]
- Quon, J.; Bala, W.; Chen, L.; Wright, J.; Kim, L.; Han, M.; Shpanskaya, K.; Lee, E.; Tong, E.; Iv, M.; et al. Deep Learning for Pediatric Posterior Fossa Tumor Detection and Classification: A Multi-Institutional Study. Am. J. Neuroradiol. 2020, 41, 1718–1725. [Google Scholar] [CrossRef]
- Ye, Z.; Srinivasa, K.; Lin, J.; Viox, J.D.; Song, C.; Wu, A.T.; Sun, P.; Song, S.-K.; Dahiya, S.; Rubin, J.B. Diffusion Basis Spectrum Imaging with Deep Neural Network Differentiates Distinct Histology in Pediatric Brain Tumors. bioRxiv 2020. [Google Scholar] [CrossRef]
- Prince, E.W.; Whelan, R.; Mirsky, D.M.; Stence, N.; Staulcup, S.; Klimo, P.; Anderson, R.C.E.; Niazi, T.N.; Grant, G.; Souweidane, M.; et al. Robust deep learning classification of adamantinomatous craniopharyngioma from limited preoperative radiographic images. Sci. Rep. 2020, 10, 1–13. [Google Scholar] [CrossRef]
- Zhang, W.; Li, R.; Deng, H.; Wang, L.; Lin, W.; Ji, S.; Shen, D. Deep convolutional neural networks for multi-modality isointense infant brain image segmentation. NeuroImage 2015, 108, 214–224. [Google Scholar] [CrossRef] [Green Version]
- Cui, Z.; Yang, J.; Qiao, Y. Brain MRI segmentation with patch-based CNN approach. In Proceedings of the 2016 35th Chinese Control Conference (CCC), Chengdu, China, 27–29 July 2016; pp. 7026–7031. [Google Scholar]
- Moeskops, P.; Viergever, M.A.; Mendrik, A.M.; De Vries, L.S.; Benders, M.J.N.L.; Isgum, I. Automatic Segmentation of MR Brain Images With a Convolutional Neural Network. IEEE Trans. Med. Imaging 2016, 35, 1252–1261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nie, D.; Wang, L.; Gao, Y.; Shen, D. Fully convolutional networks for multi-modality isointense infant brain image segmentation. In Proceedings of the 2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI), Prague, Czech Republic, 13–16 April 2016; 2016; Volume 2016, pp. 1342–1345. [Google Scholar]
- Rajchl, M.; Lee, M.C.H.; Oktay, O.; Kamnitsas, K.; Passerat-Palmbach, J.; Bai, W.; Damodaram, M.; Rutherford, M.A.; Hajnal, J.V.; Kainz, B.; et al. DeepCut: Object Segmentation From Bounding Box Annotations Using Convolutional Neural Networks. IEEE Trans. Med. Imaging 2017, 36, 674–683. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Géraud, T.; Bloch, I. From neonatal to adult brain MR image segmentation in a few seconds using 3D-like fully convolutional network and transfer learning. In Proceedings of the 2017 IEEE International Conference on Image Processing (ICIP), Beijing, China, 17–20 September 2017; pp. 4417–4421. [Google Scholar]
- Zeng, G.; Zheng, G. Multi-stream 3D FCN with multi-scale deep supervision for multi-modality isointense infant brain MR image segmentation. In Proceedings of the 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), Washington, DC, USA, 4–7 April 2018; pp. 136–140. [Google Scholar] [CrossRef] [Green Version]
- Nie, D.; Wang, L.; Adeli, E.; Lao, C.; Lin, W.; Shen, D. 3-D Fully Convolutional Networks for Multimodal Isointense Infant Brain Image Segmentation. IEEE Trans. Cybern. 2019, 49, 1123–1136. [Google Scholar] [CrossRef] [PubMed]
- Khalili, N.; Lessmann, N.; Turk, E.; Claessens, N.; de Heus, R.; Kolk, T.; Viergever, M.; Benders, M.; Išgum, I. Automatic brain tissue segmentation in fetal MRI using convolutional neural networks. Magn. Reson. Imaging 2019, 64, 77–89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dolz, J.; Gopinath, K.; Yuan, J.; Lombaert, H.; Desrosiers, C.; Ben Ayed, I. HyperDense-Net: A Hyper-Densely Connected CNN for Multi-Modal Image Segmentation. IEEE Trans. Med. Imaging 2019, 38, 1116–1126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dolz, J.; Desrosiers, C.; Wang, L.; Yuan, J.; Shen, D.; Ben Ayed, I. Deep CNN ensembles and suggestive annotations for infant brain MRI segmentation. Comput. Med. Imaging Graph. 2020, 79, 101660. [Google Scholar] [CrossRef]
- Bermudez, C.; Blaber, J.; Remedios, S.W.; Reynolds, J.E.; Lebel, C.; McHugo, M.; Heckers, S.; Huo, Y.; Landman, B.A. Generalizing deep whole brain segmentation for pediatric and post-contrast MRI with augmented transfer learning. In Proceedings of the Medical Imaging: Image Processing, Houston, TX, USA, 15–20 February 2020. [Google Scholar]
- Ding, Y.; Acosta, R.; Enguix, V.; Suffren, S.; Ortmann, J.; Luck, D.; Dolz, J.; Lodygensky, G.A. Using Deep Convolutional Neural Networks for Neonatal Brain Image Segmentation. Front. Neurosci. 2020, 14, 207. [Google Scholar] [CrossRef] [PubMed]
- Ladefoged, C.N.; Benoit, D.; Law, I.; Holm, S.; Kjær, A.; Højgaard, L.; E Hansen, A.; Andersen, F.L. Region specific optimization of continuous linear attenuation coefficients based on UTE (RESOLUTE): Application to PET/MR brain imaging. Phys. Med. Biol. 2015, 60, 8047–8065. [Google Scholar] [CrossRef]
- Ladefoged, C.N.; Marner, L.; Hindsholm, A.; Law, I.; Højgaard, L.; Andersen, F.L. Deep Learning Based Attenuation Correction of PET/MRI in Pediatric Brain Tumor Patients: Evaluation in a Clinical Setting. Front. Neurosci. 2019, 12, 1005. [Google Scholar] [CrossRef] [PubMed]
- Chang, A.; Suriyakumar, V.; Moturu, A.; Tewattanarat, N.; Doria, A.; Goldenberg, A. Using Generative Models for Pediatric wbMRI. arXiv 2020, arXiv:2006.00727. [Google Scholar]
- Knab, B.; Connell, P.P. Radiotherapy for pediatric brain tumors: When and how. Expert Rev. Anticancer Ther. 2007, 7, S69–S77. [Google Scholar] [CrossRef] [PubMed]
- Silva, A.H.D.; Aquilina, K. Surgical approaches in pediatric neuro-oncology. Cancer Metastasis Rev. 2019, 38, 723–747. [Google Scholar] [CrossRef]
- Ashraf, O.; Patel, N.V.; Hanft, S.; Danish, S.F. Laser-Induced Thermal Therapy in Neuro-Oncology: A Review. World Neurosurg. 2018, 112, 166–177. [Google Scholar] [CrossRef]
- Montemurro, N.; Anania, Y.; Cagnazzo, F.; Perrini, P. Survival outcomes in patients with recurrent glioblastoma treated with Laser Interstitial Thermal Therapy (LITT): A systematic review. Clin. Neurol. Neurosurg. 2020, 195, 105942. [Google Scholar] [CrossRef]
- Suh, J.H.; Barnett, G.H. Stereotactic radiosurgery for brain tumors in pediatric patients. Technol. Cancer Res. Treat. 2003, 2, 141–146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mostapha, M.; Styner, M. Role of deep learning in infant brain MRI analysis. Magn. Reson. Imaging 2019, 64, 171–189. [Google Scholar] [CrossRef]
- Younus, Z.S.; Mohamad, D.; Saba, T.; Alkawaz, M.H.; Rehman, A.; Al-Rodhaan, M.; Al-Dhelaan, A. Content-based image retrieval using PSO and k-means clustering algorithm. Arab. J. Geosci. 2015, 8, 6211–6224. [Google Scholar] [CrossRef]
- Al-Ameen, Z.; Sulong, G.; Rehman, A.; Al-Dhelaan, A.; Saba, T.; Al-Rodhaan, M. An innovative technique for contrast enhancement of computed tomography images using normalized gamma-corrected contrast-limited adaptive histogram equalization. EURASIP J. Adv. Signal Process. 2015, 2015, 32. [Google Scholar] [CrossRef]
- Afshar, P.; Plataniotis, K.N.; Mohammadi, A. Capsule Networks for Brain Tumor Classification Based on MRI Images and Coarse Tumor Boundaries. In Proceedings of the ICASSP 2019—2019 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Brighton, UK, 12–17 May 2019; pp. 1368–1372. [Google Scholar]
- Byrne, D.M. Recommendations for Cross-Sectional Imaging in Cancer Management; The Royal College of Radiologists: London, UK, 2014. [Google Scholar]
- Akshulakov, S.K.; Kerimbayev, T.T.; Biryuchkov, M.Y.; Urunbayev, Y.A.; Farhadi, D.S.; Byvaltsev, V.A. Current Trends for Improving Safety of Stereotactic Brain Biopsies: Advanced Optical Methods for Vessel Avoidance and Tumor Detection. Front. Oncol. 2019, 9, 947. [Google Scholar] [CrossRef] [PubMed]
- Lorenzen, A.; Groeschel, S.; Ernemann, U.; Wilke, M.; Schuhmann, M.U. Role of presurgical functional MRI and diffusion MR tractography in pediatric low-grade brain tumor surgery: A single-center study. Child’s Nerv. Syst. 2018, 34, 2241–2248. [Google Scholar] [CrossRef] [PubMed]




| Authors | Tumor Location/Type | Methodology | Modality | Dataset | Results |
|---|---|---|---|---|---|
| Arle, Jeffrey E., et al. (1997) [49] | Posterior fossa (astrocytomas, PNETs, and ependymoma) | Four back-propagation neural networks | MRS + MR + Metadata | Self-acquired dataset (33 children 6 months–14 years) | Classification accuracy rate 58–95% |
| Bidiwala, S. and Pittman (2004) [50] | Posterior fossa (astrocytom, ependymom, and medulloblastoma) | Neural networks | CT + MRI (T1WI, T2WI) + Metadata | Self-acquired dataset (33 Children) | Classification accuracy rate 72.7–85.7% |
| Quon, J.L., et al. (2020) [51] | Posterior fossa (diffuse midline glioma, medulloblastoma, pilocytic astrocytoma, and ependymoma) | Modified 2D ResNeXt-50-32x4d deep learning architecture | T2-weighted MRIs | Multi-institutional study (617 children) | Detection accuracy was AUROC of 0.99 Classification accuracy was 92% |
| Ye, Zezhong, et al. (2020) [52] | Several histologic elements of tumors of pediatric high-grade brain tumors | DHI model (DBSI + DNN) | Diffusion basis spectrum imaging (DBSI) | 9 pediatric brain tumor post-mortem specimens | Overall classification accuracy rate—83.3% |
| Prince, Eric W., et al. (2020) [53] | Adamantinomatous craniopharyngioma | CNN + genetic algorithm as a meta-heuristic optimizer | CT + MRI + combined CT and MRI | Multi-institutional study (39 children) | Classification accuracies 85.3%, 83.3%, and 87.8%, in respect to modality. |
| Authors | Segmented Subject | Methodology | Modality | Dataset | Results |
|---|---|---|---|---|---|
| Zhang, Wenlu, et al. (2015) [54] | Segmenting all three types of brain tissues (CSF, GM, WM) | Four 2D CNN | T1, T2, fractional anisotropy (FA) MRI | Self-acquired (10 infants, 6–8 months of age) | Overall dice ratios CFS 83.55% GM 85.18% WM 86.37% |
| Cui, Zhipeng, et al. (2016) [55] | Patch-based CNN segmentation of brain structure | Three different CN Ns | Manually segmented MRIs | Public dataset (CANDI neuroimaging access point 103 MRIs) small sets 4–5 MRI from each subject (6 to 17 year old age group) | Accuracy rate of 90% |
| Moeskops, Pim, et al. (2016) [56] | 8 subjects: CB, mWM, BGT, vCSF, uWM, BS, cGM, and eCSF. | CNNs | T1-weighted and T2-weighted MRI | Self-acquired (10 images at 30 weeks, 12 images at 40 weeks, 15 images at 23 years, 20 images at 70 years) | Average dice ratios 0.87 (coronal T2w 30 weeks), 0.82 (coronal T2w 40 weeks), 0.84 (axial T2w 40 weeks), 0.86 (axial T1w 70 years) and 0.91 (sagittal T1w 23 years). |
| Nie, Dong, et al. (2016) [57] | Segmenting all three types of brain tissues (CSF, GM, WM) | FCNs + multi-FCNs (mFCNs) | T1, T2, fractional anisotropy (FA) MRI | Self-acquired 10 healthy infants (6–8 months of age) | Average dice ratios FCNs (0.838 for CSF 0.861 for GM 0.885 for WM) mFCNs (0.855 for CSF 0.873 for GM 0.887 for WM) |
| Rajchl, Martin, et al. (2016) [58] | Whole brain pixel-wise segmentation | CNNs + fully connected conditional random field (CRF) | T2-weighted ssFSE sequence | Public dataset (55 fetal MRI subject) | DSC (%) CNNnaïve (74.0), DCBB (86.6), DCPS (90.3), CNNFS (94.1) |
| Xu, Yongchao, et al. (2017) [59] | Neonatal (CoGM, BGT, UWM, BS, CB, Vent, CSF) adults (CSF, WM, GM) | FCN + TL (VGG 16 network) | T1, T1-IR, FLAIR MRI | NeoBrainS12 + MRBrainS13 | Dice coefficient Neonatal: CoGM (0.79–0.87), BGT (0.89–0.93), UWM (0.91–0.95), BS (0.76–0.86), CB (0.91–0.94), Vent (0.85–0.88), CSF (0.82–0.89) Adults GM (85.40), WM (88.98), CSF (84.13) |
| Zeng, Guodong, and Guoyan Zheng (2018) [60] | Segment isointense infant brain MRI (CSF, GM, WM) | 3D FCNNs | T1 and T2 weighted MRI | Public dataset (MICCAI iSEG-2017) | Dice overlap coefficient CSF (0.954), GM (0.916), WM (0.896) |
| Nie, Dong, et al. (2019) [61] | Segment isointense infant brain MRI (CSF, GM, WM) | 3D FCNNs | T1, T2, fractional anisotropy (FA) MRI | Self-acquired (11 healthy infants MRIs) | Dice ratios 0.9190 for WM, 0.9401 for GM, 0.9610 for CSF |
| Khalili, Nadieh, et al. (2019) [62] | Segment of seven brain tissue classes: cerebellum, basal ganglia and thalami, ventricular cerebrospinal fluid, white matter, brain stem, cortical gray matter and extracerebral cerebrospinal fluid. | 2D FCN with identical U-net architecture | T2-weighted MRI | Self-acquired 12 fetuses (22.9–34.6 weeks post menstrual age) + neonatal MRI (40 weeks of post menstrual age) From the NeoBrainS12 dataset | DC over all tissue classes increases to 0.88 and MSD decrease to 0.37 mm |
| Dolz, Jose, et al. (2019) [63] | Segmenting all three types of brain tissues (CSF, GM, WM) | 3D FCNNs | Integrated T1 and T2 MRI | iSEG-2017 + MRBrainS-2013 | Baselines results with DSC (CSF 0.9580, WM 0.9183 and GM 0.9035) |
| Dolz, Jose, et al. (2020) [64] | Segment isointense infant brain MRI (CSF, GM, WM) | 3D FCNNs | T1-weighted and T2-weighted MRI | Public dataset (MICCAI iSEG-2017) | Accuracy rate 92–96% Ranked as first or second in most metrics in the MICCAI iSEG-2017 challenge |
| Bermudez, Camilo, et al. (2020) [65] | Whole brain segmentation | SLANT + TL | T1-weighted brain MRI with and without intravenous contrast | Public dataset—Open Access Series on Imaging Studies (OASIS) 45 subjects aged 18–96 years old, 30 pediatric subjects (aged 2.34–4.31 years old) 36 subjects paired | DSC Pediatric: 0.89 Contrast: 0.80. |
| Ding, Yang, et al. (2020) [66] | Three types of brain tissues (CSF, GM, WM) | LiviaNET and HyperDense-Net CNNs architectures | T1-weighted and T2-weighted MRI | Publicly dataset DHCP (Developing Human Connectome Project), 40 healthy neonates born | Dual-modality HyperDense-Net accuracy rate: 92–95% Single-modality LiviaNET accuracy rate: 88–90% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shaari, H.; Kevrić, J.; Jukić, S.; Bešić, L.; Jokić, D.; Ahmed, N.; Rajs, V. Deep Learning-Based Studies on Pediatric Brain Tumors Imaging: Narrative Review of Techniques and Challenges. Brain Sci. 2021, 11, 716. https://doi.org/10.3390/brainsci11060716
Shaari H, Kevrić J, Jukić S, Bešić L, Jokić D, Ahmed N, Rajs V. Deep Learning-Based Studies on Pediatric Brain Tumors Imaging: Narrative Review of Techniques and Challenges. Brain Sciences. 2021; 11(6):716. https://doi.org/10.3390/brainsci11060716
Chicago/Turabian StyleShaari, Hala, Jasmin Kevrić, Samed Jukić, Larisa Bešić, Dejan Jokić, Nuredin Ahmed, and Vladimir Rajs. 2021. "Deep Learning-Based Studies on Pediatric Brain Tumors Imaging: Narrative Review of Techniques and Challenges" Brain Sciences 11, no. 6: 716. https://doi.org/10.3390/brainsci11060716
APA StyleShaari, H., Kevrić, J., Jukić, S., Bešić, L., Jokić, D., Ahmed, N., & Rajs, V. (2021). Deep Learning-Based Studies on Pediatric Brain Tumors Imaging: Narrative Review of Techniques and Challenges. Brain Sciences, 11(6), 716. https://doi.org/10.3390/brainsci11060716








