Detection of Resting-State Functional Connectivity from High-Density Electroencephalography Data: Impact of Head Modeling Strategies
Abstract
:1. Introduction
2. Materials and Methods
2.1. EEG Experiment
2.2. Standard EEG Data Analysis
2.2.1. EEG Signal Pre-Processing
2.2.2. Realistic Head Model Creation
2.2.3. Source Activity Reconstruction
2.2.4. Functional Connectivity
2.3. Impact of Head Modeling Strategies
Generation of Test Models
2.4. Statistical Analysis
3. Results
4. Discussion
4.1. Impact of Electrode Localization Error
4.2. Impact of Head Tissue Segmentation
4.3. Differential Impact of Electrode Localization and Head Tissue Segmentation
4.4. Analysis of Robustness for Different Frequency Bands and Networks
4.5. Study Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Michel, C.M.; Murray, M.M. Towards the utilization of EEG as a brain imaging tool. Neuroimage 2012, 61, 371–385. [Google Scholar] [CrossRef] [PubMed]
- Pfurtscheller, G.; Aranibar, A. Event-related cortical desynchronization detected by power measurements of scalp EEG. Electroencephalogr. Clin. Neurophysiol. 1977, 42, 817–826. [Google Scholar] [CrossRef]
- Pfurtscheller, G. Event-related synchronization (ERS): An electrophysiological correlate of cortical areas at rest. Electroencephalogr. Clin. Neurophysiol. 1992, 83, 62–69. [Google Scholar] [CrossRef]
- Frund, I.; Schadow, J.; Busch, N.A.; Korner, U.; Herrmann, C.S. Evoked gamma oscillations in human scalp EEG are test-retest reliable. Clin. Neurophysiol. 2007, 118, 221–227. [Google Scholar] [CrossRef] [PubMed]
- Morash, V.; Bai, O.; Furlani, S.; Lin, P.; Hallett, M. Classifying EEG signals preceding right hand, left hand, tongue, and right foot movements and motor imageries. Clin. Neurophysiol. 2008, 119, 2570–2578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pfurtscheller, G.; Lopes da Silva, F.H. Event-related EEG/MEG synchronization and desynchronization: Basic principles. Clin. Neurophysiol. 1999, 110, 1842–1857. [Google Scholar] [CrossRef]
- Ruiz, Y.; Pockett, S.; Freeman, W.J.; Gonzalez, E.; Li, G. A method to study global spatial patterns related to sensory perception in scalp EEG. J. Neurosci. Methods 2010, 191, 110–118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brookes, M.J.; Woolrich, M.; Luckhoo, H.; Price, D.; Hale, J.R.; Stephenson, M.C.; Barnes, G.R.; Smith, S.M.; Morris, P.G. Investigating the electrophysiological basis of resting state networks using magnetoencephalography. Proc. Natl. Acad. Sci. USA 2011, 108, 16783–16788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Pasquale, F.; Della Penna, S.; Snyder, A.Z.; Lewis, C.; Mantini, D.; Marzetti, L.; Belardinelli, P.; Ciancetta, L.; Pizzella, V.; Romani, G.L.; et al. Temporal dynamics of spontaneous MEG activity in brain networks. Proc. Natl. Acad. Sci. USA 2010, 107, 6040–6045. [Google Scholar] [CrossRef] [Green Version]
- Liu, Q.; Farahibozorg, S.; Porcaro, C.; Wenderoth, N.; Mantini, D. Detecting large-scale networks in the human brain using high-density electroencephalography. Hum. Brain Mapp. 2017, 38, 4631–4643. [Google Scholar] [CrossRef] [Green Version]
- Marino, M.; Arcara, G.; Porcaro, C.; Mantini, D. Hemodynamic Correlates of Electrophysiological Activity in the Default Mode Network. Front. Neurosci. 2019, 13, 1060. [Google Scholar] [CrossRef]
- Samogin, J.; Liu, Q.; Marino, M.; Wenderoth, N.; Mantini, D. Shared and connection-specific intrinsic interactions in the default mode network. Neuroimage 2019, 200, 474–481. [Google Scholar] [CrossRef]
- Sockeel, S.; Schwartz, D.; Pelegrini-Issac, M.; Benali, H. Large-Scale Functional Networks Identified from Resting-State EEG Using Spatial ICA. PLoS ONE 2016, 11, e0146845. [Google Scholar] [CrossRef] [PubMed]
- Hassan, M.; Dufor, O.; Merlet, I.; Berrou, C.; Wendling, F. EEG source connectivity analysis: From dense array recordings to brain networks. PLoS ONE 2014, 9, e105041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klamer, S.; Elshahabi, A.; Lerche, H.; Braun, C.; Erb, M.; Scheffler, K.; Focke, N.K. Differences between MEG and high-density EEG source localizations using a distributed source model in comparison to fMRI. Brain Topogr. 2015, 28, 87–94. [Google Scholar] [CrossRef]
- Liu, Q.; Ganzetti, M.; Wenderoth, N.; Mantini, D. Detecting Large-Scale Brain Networks Using EEG: Impact of Electrode Density, Head Modeling and Source Localization. Front. Neuroinform. 2018, 12, 4. [Google Scholar] [CrossRef]
- Michel, C.M.; Murray, M.M.; Lantz, G.; Gonzalez, S.; Spinelli, L.; Grave de Peralta, R. EEG source imaging. Clin. Neurophysiol. 2004, 115, 2195–2222. [Google Scholar] [CrossRef] [PubMed]
- Bola, M.; Sabel, B.A. Dynamic reorganization of brain functional networks during cognition. Neuroimage 2015, 114, 398–413. [Google Scholar] [CrossRef] [PubMed]
- Brodbeck, V.; Spinelli, L.; Lascano, A.M.; Wissmeier, M.; Vargas, M.I.; Vulliemoz, S.; Pollo, C.; Schaller, K.; Michel, C.M.; Seeck, M. Electroencephalographic source imaging: A prospective study of 152 operated epileptic patients. Brain 2011, 134, 2887–2897. [Google Scholar] [CrossRef]
- Canuet, L.; Ishii, R.; Pascual-Marqui, R.D.; Iwase, M.; Kurimoto, R.; Aoki, Y.; Ikeda, S.; Takahashi, H.; Nakahachi, T.; Takeda, M. Resting-state EEG source localization and functional connectivity in schizophrenia-like psychosis of epilepsy. PLoS ONE 2011, 6, e27863. [Google Scholar] [CrossRef] [Green Version]
- Zhao, M.; Marino, M.; Samogin, J.; Swinnen, S.P.; Mantini, D. Hand, foot and lip representations in primary sensorimotor cortex: A high-density electroencephalography study. Sci. Rep. 2019, 9, 19464. [Google Scholar] [CrossRef]
- Li, K.; Papademetris, X.; Tucker, D.M. BrainK for Structural Image Processing: Creating Electrical Models of the Human Head. Comput. Intell. Neurosci. 2016, 2016, 1349851. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montes-Restrepo, V.; van Mierlo, P.; Strobbe, G.; Staelens, S.; Vandenberghe, S.; Hallez, H. Influence of skull modeling approaches on EEG source localization. Brain Topogr. 2014, 27, 95–111. [Google Scholar] [CrossRef] [Green Version]
- Ramon, C.; Haueisen, J.; Schimpf, P.H. Influence of head models on neuromagnetic fields and inverse source localizations. Biomed. Eng. Online 2006, 5, 55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taberna, G.A.; Samogin, J.; Mantini, D. Automated Head Tissue Modelling Based on Structural Magnetic Resonance Images for Electroencephalographic Source Reconstruction. Neuroinformatics 2021. [Google Scholar] [CrossRef]
- Hallez, H.; Vanrumste, B.; Grech, R.; Muscat, J.; De Clercq, W.; Vergult, A.; D’Asseler, Y.; Camilleri, K.P.; Fabri, S.G.; Van Huffel, S.; et al. Review on solving the forward problem in EEG source analysis. J. Neuroeng. Rehabil. 2007, 4, 46. [Google Scholar] [CrossRef]
- Akalin Acar, Z.; Makeig, S. Effects of forward model errors on EEG source localization. Brain Topogr. 2013, 26, 378–396. [Google Scholar] [CrossRef] [Green Version]
- Cespedes-Villar, Y.; Martinez-Vargas, J.D.; Castellanos-Dominguez, G. Influence of Patient-Specific Head Modeling on EEG Source Imaging. Comput. Math. Methods Med. 2020, 2020, 5076865. [Google Scholar] [CrossRef]
- Von Ellenrieder, N.; Muravchik, C.H.; Wagner, M.; Nehorai, A. Effect of head shape variations among individuals on the EEG/MEG forward and inverse problems. IEEE Trans. Biomed. Eng. 2009, 56, 587–597. [Google Scholar] [CrossRef] [PubMed]
- Wagner, S.; Rampersad, S.M.; Aydin, U.; Vorwerk, J.; Oostendorp, T.F.; Neuling, T.; Herrmann, C.S.; Stegeman, D.F.; Wolters, C.H. Investigation of tDCS volume conduction effects in a highly realistic head model. J. Neural. Eng. 2014, 11, 016002. [Google Scholar] [CrossRef] [PubMed]
- Collins, D.L.; Zijdenbos, A.P.; Kollokian, V.; Sled, J.G.; Kabani, N.J.; Holmes, C.J.; Evans, A.C. Design and construction of a realistic digital brain phantom. IEEE Trans. Med. Imaging 1998, 17, 463–468. [Google Scholar] [CrossRef]
- Huang, Y.; Parra, L.C.; Haufe, S. The New York Head-A precise standardized volume conductor model for EEG source localization and tES targeting. Neuroimage 2016, 140, 150–162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mazziotta, J.; Toga, A.; Evans, A.; Fox, P.; Lancaster, J.; Zilles, K.; Woods, R.; Paus, T.; Simpson, G.; Pike, B.; et al. A probabilistic atlas and reference system for the human brain: International Consortium for Brain Mapping (ICBM). Philos. Trans. R. Soc. Lond. B Biol. Sci. 2001, 356, 1293–1322. [Google Scholar] [CrossRef] [PubMed]
- Valdes-Hernandez, P.A.; von Ellenrieder, N.; Ojeda-Gonzalez, A.; Kochen, S.; Aleman-Gomez, Y.; Muravchik, C.; Valdes-Sosa, P.A. Approximate average head models for EEG source imaging. J. Neurosci. Methods 2009, 185, 125–132. [Google Scholar] [CrossRef]
- Fischl, B. FreeSurfer. Neuroimage 2012, 62, 774–781. [Google Scholar] [CrossRef] [Green Version]
- Holdefer, R.N.; Sadleir, R.; Russell, M.J. Predicted current densities in the brain during transcranial electrical stimulation. Clin. Neurophysiol. 2006, 117, 1388–1397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Homma, S.; Musha, T.; Nakajima, Y.; Okamoto, Y.; Blom, S.; Flink, R.; Hagbarth, K.E.; Mostrom, U. Location of electric current sources in the human brain estimated by the dipole tracing method of the scalp-skull-brain (SSB) head model. Electroencephalogr. Clin. Neurophysiol. 1994, 91, 374–382. [Google Scholar] [CrossRef]
- Oostenveld, R.; Fries, P.; Maris, E.; Schoffelen, J.M. FieldTrip: Open source software for advanced analysis of MEG, EEG, and invasive electrophysiological data. Comput. Intell. Neurosci. 2011, 2011, 156869. [Google Scholar] [CrossRef]
- Le, J.; Lu, M.; Pellouchoud, E.; Gevins, A. A rapid method for determining standard 10/10 electrode positions for high resolution EEG studies. Electroencephalogr. Clin. Neurophysiol. 1998, 106, 554–558. [Google Scholar] [CrossRef]
- Whalen, C.; Maclin, E.L.; Fabiani, M.; Gratton, G. Validation of a method for coregistering scalp recording locations with 3D structural MR images. Hum. Brain Mapp. 2008, 29, 1288–1301. [Google Scholar] [CrossRef]
- Clausner, T.; Dalal, S.S.; Crespo-Garcia, M. Photogrammetry-Based Head Digitization for Rapid and Accurate Localization of EEG Electrodes and MEG Fiducial Markers Using a Single Digital SLR Camera. Front. Neurosci. 2017, 11, 264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Russell, G.S.; Jeffrey Eriksen, K.; Poolman, P.; Luu, P.; Tucker, D.M. Geodesic photogrammetry for localizing sensor positions in dense-array EEG. Clin. Neurophysiol. 2005, 116, 1130–1140. [Google Scholar] [CrossRef]
- Homolle, S.; Oostenveld, R. Using a structured-light 3D scanner to improve EEG source modeling with more accurate electrode positions. J. Neurosci. Methods 2019, 326, 108378. [Google Scholar] [CrossRef] [PubMed]
- Taberna, G.A.; Guarnieri, R.; Mantini, D. SPOT3D: Spatial positioning toolbox for head markers using 3D scans. Sci. Rep. 2019, 9, 12813. [Google Scholar] [CrossRef] [Green Version]
- Taberna, G.A.; Marino, M.; Ganzetti, M.; Mantini, D. Spatial localization of EEG electrodes using 3D scanning. J. Neural. Eng. 2019, 16, 026020. [Google Scholar] [CrossRef] [PubMed]
- Koessler, L.; Cecchin, T.; Caspary, O.; Benhadid, A.; Vespignani, H.; Maillard, L. EEG-MRI co-registration and sensor labeling using a 3D laser scanner. Ann. Biomed. Eng. 2011, 39, 983–995. [Google Scholar] [CrossRef]
- Koessler, L.; Maillard, L.; Benhadid, A.; Vignal, J.P.; Braun, M.; Vespignani, H. Spatial localization of EEG electrodes. Neurophysiol. Clin. 2007, 37, 97–102. [Google Scholar] [CrossRef]
- Shirazi, S.Y.; Huang, H.J. More Reliable EEG Electrode Digitizing Methods Can Reduce Source Estimation Uncertainty, but Current Methods Already Accurately Identify Brodmann Areas. Front. Neurosci. 2019, 13, 1159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Samogin, J.; Marino, M.; Porcaro, C.; Wenderoth, N.; Dupont, P.; Swinnen, S.P.; Mantini, D. Frequency-dependent functional connectivity in resting state networks. Hum. Brain Mapp. 2020, 41, 5187–5198. [Google Scholar] [CrossRef]
- Cuartas Morales, E.; Acosta-Medina, C.D.; Castellanos-Dominguez, G.; Mantini, D. A Finite-Difference Solution for the EEG Forward Problem in Inhomogeneous Anisotropic Media. Brain Topogr. 2019, 32, 229–239. [Google Scholar] [CrossRef]
- Mantini, D.; Franciotti, R.; Romani, G.L.; Pizzella, V. Improving MEG source localizations: An automated method for complete artifact removal based on independent component analysis. Neuroimage 2008, 40, 160–173. [Google Scholar] [CrossRef]
- Liu, Q.; Balsters, J.H.; Baechinger, M.; van der Groen, O.; Wenderoth, N.; Mantini, D. Estimating a neutral reference for electroencephalographic recordings: The importance of using a high-density montage and a realistic head model. J. Neural. Eng. 2015, 12, 056012. [Google Scholar] [CrossRef] [PubMed]
- Yao, D. A method to standardize a reference of scalp EEG recordings to a point at infinity. Physiol. Meas. 2001, 22, 693–711. [Google Scholar] [CrossRef]
- Yao, D.; Wang, L.; Oostenveld, R.; Nielsen, K.D.; Arendt-Nielsen, L.; Chen, A.C. A comparative study of different references for EEG spectral mapping: The issue of the neutral reference and the use of the infinity reference. Physiol. Meas. 2005, 26, 173–184. [Google Scholar] [CrossRef]
- Haueisen, J.; Ramon, C.; Eiselt, M.; Brauer, H.; Nowak, H. Influence of tissue resistivities on neuromagnetic fields and electric potentials studied with a finite element model of the head. IEEE Trans. Biomed. Eng. 1997, 44, 727–735. [Google Scholar] [CrossRef]
- Pascual-Marqui, R.D.; Lehmann, D.; Koukkou, M.; Kochi, K.; Anderer, P.; Saletu, B.; Tanaka, H.; Hirata, K.; John, E.R.; Prichep, L.; et al. Assessing interactions in the brain with exact low-resolution electromagnetic tomography. Philos. Trans. A Math. Phys. Eng. Sci. 2011, 369, 3768–3784. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Shi, L.; Lei, Y.; Liang, P.; Li, K.; Chu, W.C.; Wang, D.; Alzheimer’s Disease Neuroimaging, I. Mapping the “What” and “Where” Visual Cortices and Their Atrophy in Alzheimer’s Disease: Combined Activation Likelihood Estimation with Voxel-Based Morphometry. Front. Hum. Neurosci. 2016, 10, 333. [Google Scholar] [CrossRef]
- Fishbein, D.H.; Eldreth, D.L.; Hyde, C.; Matochik, J.A.; London, E.D.; Contoreggi, C.; Kurian, V.; Kimes, A.S.; Breeden, A.; Grant, S. Risky decision making and the anterior cingulate cortex in abstinent drug abusers and nonusers. Brain Res. Cogn. Brain Res. 2005, 23, 119–136. [Google Scholar] [CrossRef]
- Kaufmann, C.; Wehrle, R.; Wetter, T.C.; Holsboer, F.; Auer, D.P.; Pollmacher, T.; Czisch, M. Brain activation and hypothalamic functional connectivity during human non-rapid eye movement sleep: An EEG/fMRI study. Brain 2006, 129, 655–667. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Buuren, M.; Vink, M.; Kahn, R.S. Default-mode network dysfunction and self-referential processing in healthy siblings of schizophrenia patients. Schizophr. Res. 2012, 142, 237–243. [Google Scholar] [CrossRef] [Green Version]
- Hipp, J.F.; Hawellek, D.J.; Corbetta, M.; Siegel, M.; Engel, A.K. Large-scale cortical correlation structure of spontaneous oscillatory activity. Nat. Neurosci. 2012, 15, 884–890. [Google Scholar] [CrossRef] [Green Version]
- de Pasquale, F.; Della Penna, S.; Snyder, A.Z.; Marzetti, L.; Pizzella, V.; Romani, G.L.; Corbetta, M. A cortical core for dynamic integration of functional networks in the resting human brain. Neuron 2012, 74, 753–764. [Google Scholar] [CrossRef] [Green Version]
- King, B.R.; van Ruitenbeek, P.; Leunissen, I.; Cuypers, K.; Heise, K.F.; Santos Monteiro, T.; Hermans, L.; Levin, O.; Albouy, G.; Mantini, D.; et al. Age-Related Declines in Motor Performance are Associated With Decreased Segregation of Large-Scale Resting State Brain Networks. Cereb. Cortex 2018, 28, 4390–4402. [Google Scholar] [CrossRef]
- Newton, A.T.; Morgan, V.L.; Rogers, B.P.; Gore, J.C. Modulation of steady state functional connectivity in the default mode and working memory networks by cognitive load. Hum. Brain Mapp. 2011, 32, 1649–1659. [Google Scholar] [CrossRef] [Green Version]
- Vema Krishna Murthy, S.; MacLellan, M.; Beyea, S.; Bardouille, T. Faster and improved 3-D head digitization in MEG using Kinect. Front. Neurosci. 2014, 8, 326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iacono, M.I.; Neufeld, E.; Akinnagbe, E.; Bower, K.; Wolf, J.; Vogiatzis Oikonomidis, I.; Sharma, D.; Lloyd, B.; Wilm, B.J.; Wyss, M.; et al. MIDA: A Multimodal Imaging-Based Detailed Anatomical Model of the Human Head and Neck. PLoS ONE 2015, 10, e0124126. [Google Scholar] [CrossRef]
- Searle, S.R.; Speed, F.M.; Milliken, G.A. Population Marginal Means in the Linear Model: An Alternative to Least Squares Means. Am. Stat. 1980, 34, 216–221. [Google Scholar] [CrossRef]
- Coito, A.; Michel, C.M.; van Mierlo, P.; Vulliemoz, S.; Plomp, G. Directed Functional Brain Connectivity Based on EEG Source Imaging: Methodology and Application to Temporal Lobe Epilepsy. IEEE Trans. Biomed. Eng. 2016, 63, 2619–2628. [Google Scholar] [CrossRef]
- He, B.; Dai, Y.; Astolfi, L.; Babiloni, F.; Yuan, H.; Yang, L. eConnectome: A MATLAB toolbox for mapping and imaging of brain functional connectivity. J. Neurosci. Methods 2011, 195, 261–269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michel, C.M.; Brunet, D. EEG Source Imaging: A Practical Review of the Analysis Steps. Front. Neurol. 2019, 10, 325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beltrachini, L.; von Ellenrieder, N.; Muravchik, C.H. General bounds for electrode mislocation on the EEG inverse problem. Comput. Methods Programs Biomed. 2011, 103, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Gotman, J. The influence of electrode location errors on EEG dipole source localization with a realistic head model. Clin. Neurophysiol. 2001, 112, 1777–1780. [Google Scholar] [CrossRef]
- Marino, M.; Liu, Q.; Brem, S.; Wenderoth, N.; Mantini, D. Automated detection and labeling of high-density EEG electrodes from structural MR images. J. Neural. Eng. 2016, 13, 056003. [Google Scholar] [CrossRef]
- De Munck, J.C.; Goncalves, S.I.; Huijboom, L.; Kuijer, J.P.; Pouwels, P.J.; Heethaar, R.M.; Lopes da Silva, F.H. The hemodynamic response of the alpha rhythm: An EEG/fMRI study. Neuroimage 2007, 35, 1142–1151. [Google Scholar] [CrossRef]
- Scarff, C.J.; Reynolds, A.; Goodyear, B.G.; Ponton, C.W.; Dort, J.C.; Eggermont, J.J. Simultaneous 3-T fMRI and high-density recording of human auditory evoked potentials. Neuroimage 2004, 23, 1129–1142. [Google Scholar] [CrossRef] [PubMed]


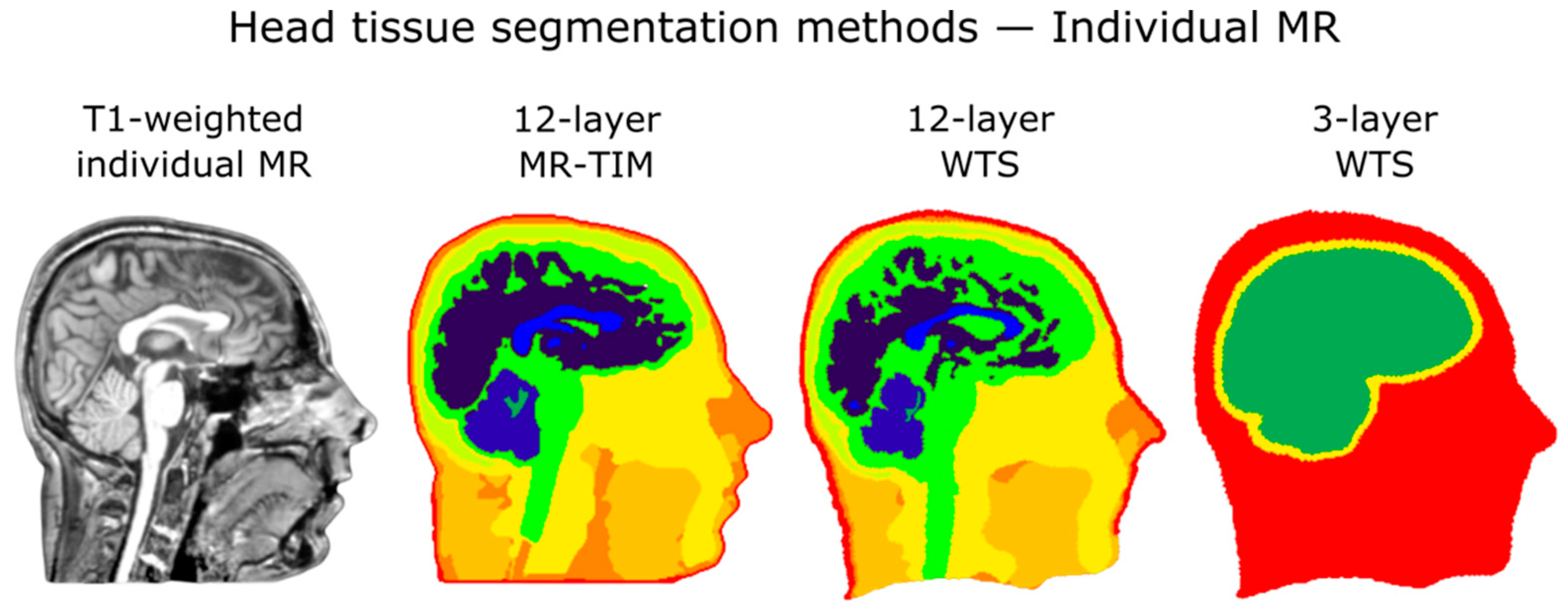
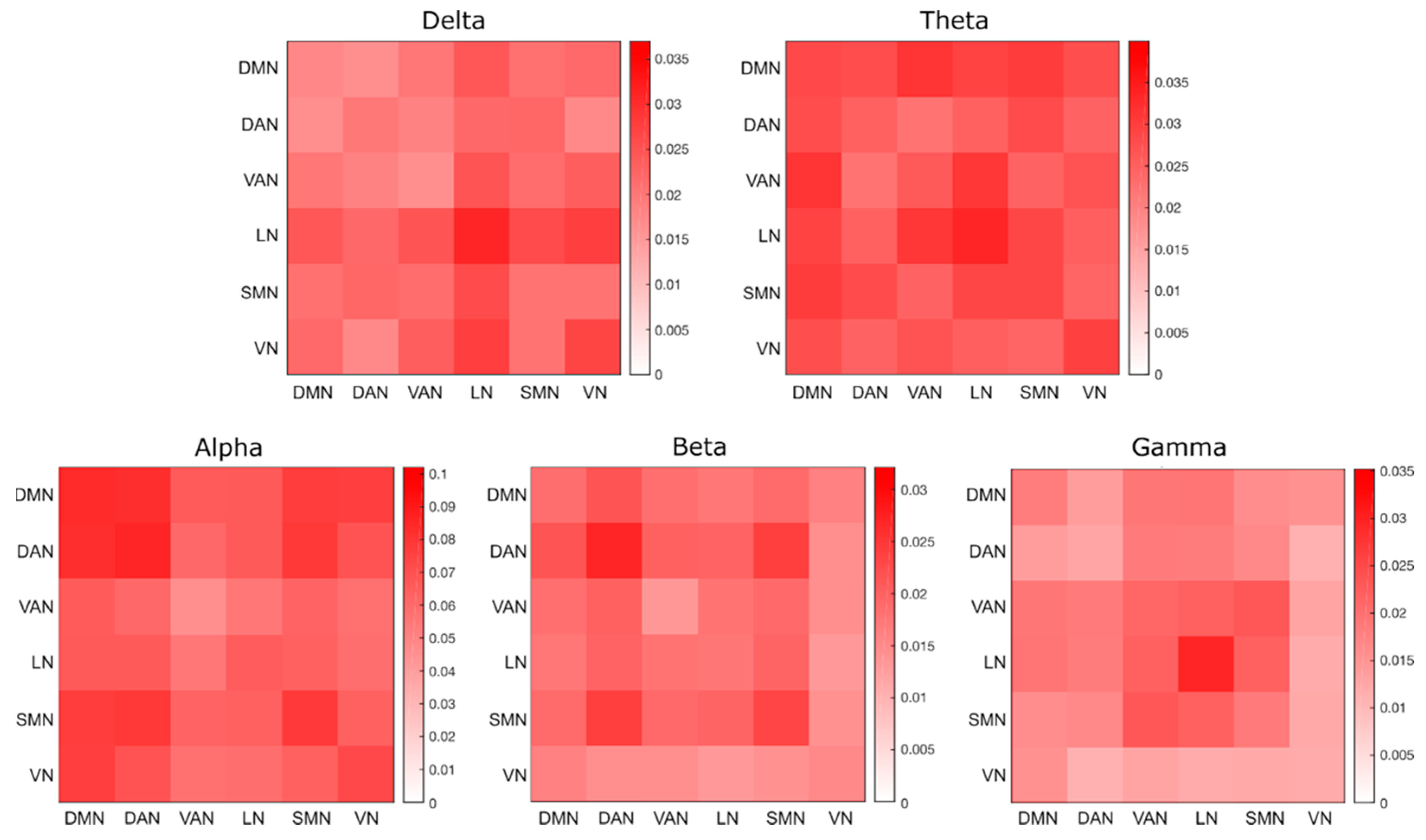
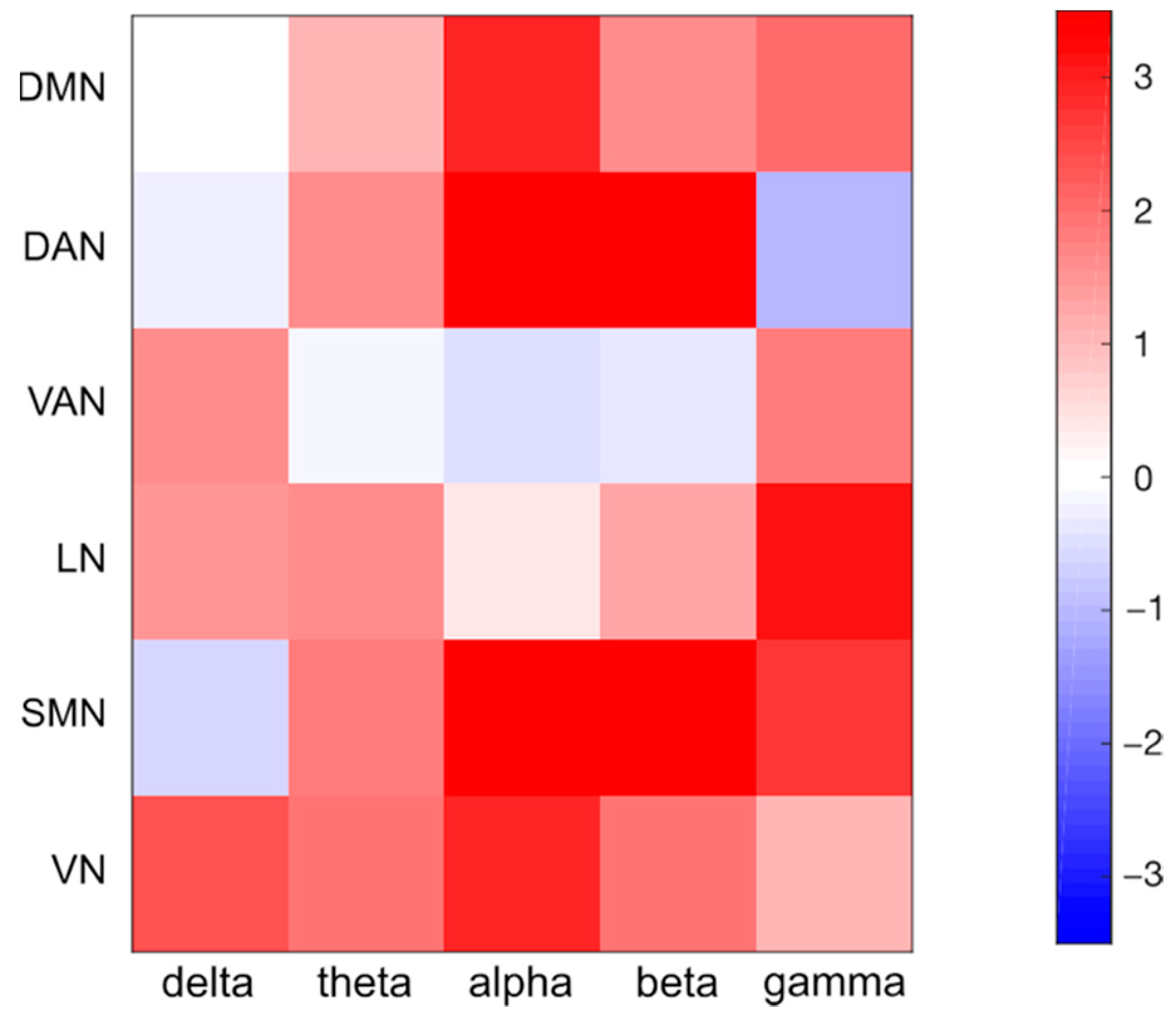
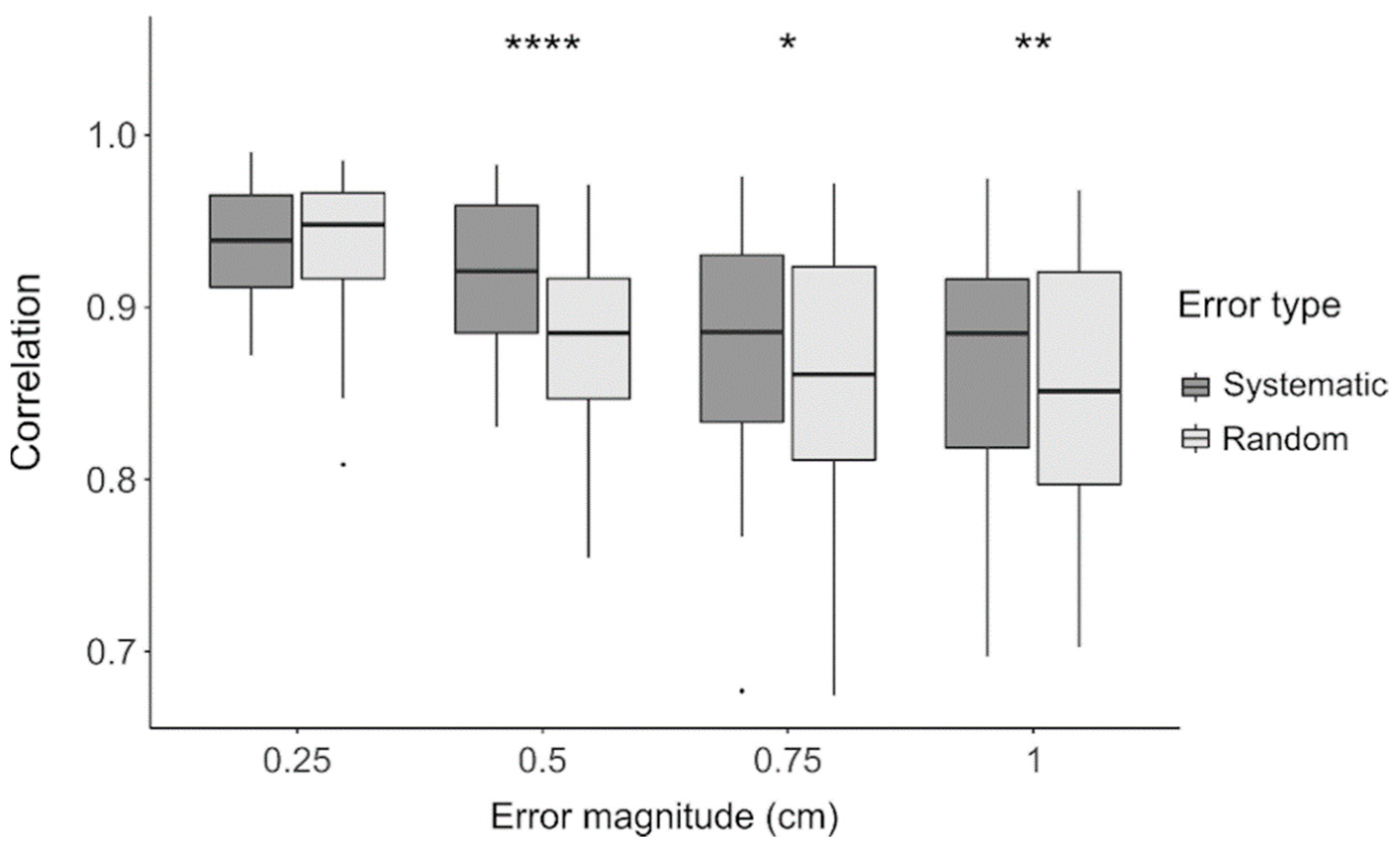


| Electrode Localization Errors | df | Sum Squares | Mean Square | F | p-Value |
|---|---|---|---|---|---|
| Network | 5 | 6.58 | 1.32 | 79.39 | <0.001 |
| Band | 4 | 18.10 | 4.53 | 273.19 | <0.001 |
| Error magnitude | 3 | 7.28 | 2.43 | 146.54 | <0.001 |
| Error type | 1 | 0.46 | 0.46 | 27.72 | <0.001 |
| Residuals | 226 | 3.74 | 0.02 | ||
| Total | 239 | 36.16 |
| Head Tissue Segmentation Methods | df | Sum Squares | Mean Square | F | p-Value |
|---|---|---|---|---|---|
| Network | 5 | 1.18 | 0.24 | 19.52 | <0.001 |
| Band | 4 | 5.83 | 1.46 | 120.42 | <0.001 |
| Segmentation method | 2 | 0.29 | 0.15 | 12.17 | <0.001 |
| Residuals | 78 | 0.94 | 0.01 | ||
| Total | 89 | 8.24 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Taberna, G.A.; Samogin, J.; Marino, M.; Mantini, D. Detection of Resting-State Functional Connectivity from High-Density Electroencephalography Data: Impact of Head Modeling Strategies. Brain Sci. 2021, 11, 741. https://doi.org/10.3390/brainsci11060741
Taberna GA, Samogin J, Marino M, Mantini D. Detection of Resting-State Functional Connectivity from High-Density Electroencephalography Data: Impact of Head Modeling Strategies. Brain Sciences. 2021; 11(6):741. https://doi.org/10.3390/brainsci11060741
Chicago/Turabian StyleTaberna, Gaia Amaranta, Jessica Samogin, Marco Marino, and Dante Mantini. 2021. "Detection of Resting-State Functional Connectivity from High-Density Electroencephalography Data: Impact of Head Modeling Strategies" Brain Sciences 11, no. 6: 741. https://doi.org/10.3390/brainsci11060741






