Responsible Genes for Neuronal Migration in the Chromosome 17p13.3: Beyond Pafah1b1(Lis1), Crk and Ywhae(14-3-3ε)
Abstract
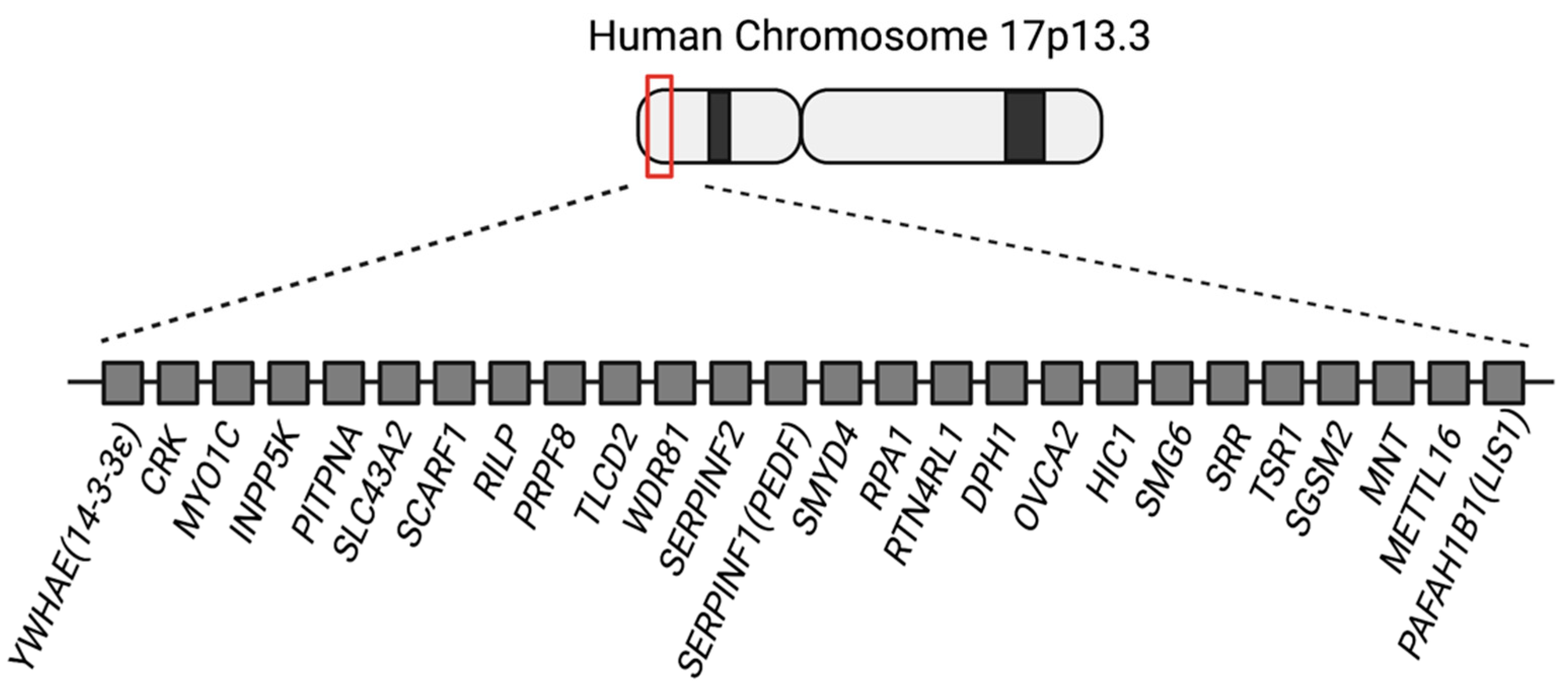
1. Introduction
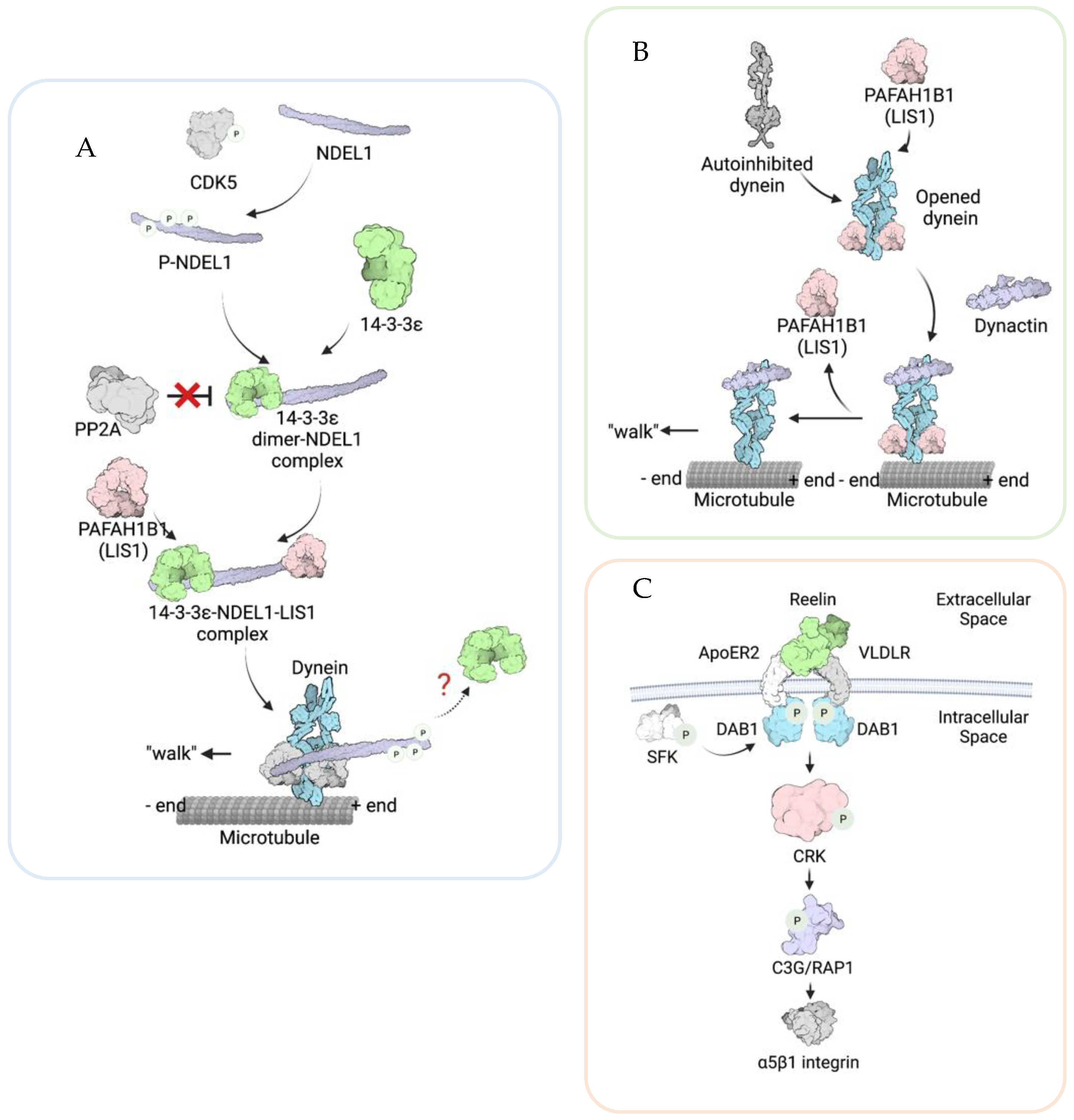
2. The Roles of YWHAE, PAFAH1B1, and CRK in Neuronal Migration
2.1. YWHAE/14-3-3ε
2.2. PAFAH1B1 (LIS1)
2.3. CRK
3. The Functions of Other Genes in the 17p13.3 Region and Their Potential Roles in Neuronal Migration
4. Conclusions
| Gene | Functions of the Protein | Involvement | References |
|---|---|---|---|
| MNT | Regulator of the MYC/MAX/MAD network |
| [124] Wu et al., 2012 [125] Toyo-oka et al., 2004 |
| SGSM2 | GTPase-activating protein involving in the modulation of the GTPases RAP and RAB |
| [126] Lin et al., 2019 |
| SRR | Production of D-serine from L-serine | Interacts with Disrupted-in-Schizophrenia-1 (DISC1), and DISC1 KD causes a defect in cortical neuron radial migration | [121] Jacobi et al., 2019 |
| HIC1 | Transcription repressor and tumor suppressor |
| [127] Ray et al., 2020 [128] Valenta et al., 2006 [129] Carter et al., 2000 |
| DPH1 (OVCA1) | Responsible for diphthamide biosynthesis | Dph1 KO causes craniofacial abnormalities in mice, but no observations indicate a defect in neuronal migration | [130] Yu et al., 2014 |
| RTN4RL1 (NGR3) |
| Regulates epithelial cell migration | [131] He et al., 2018 |
| RPA1 | Replication protein A | Overexpression causes 17p13.3 instability | [36] Outwin et al., 2011 |
| RILP |
| RILP inhibits cell migration in cancer cells | [132] Margiotta et al., 2017 [133] Wang et al., 2015 |
| SCARF1 | A member of the Scavenger receptor. Regulates endocytosis |
| [110] Patten et al., 2017 [111] Shibata et al. 2004 |
| SLC43A2 | Amino acid transporter for methionine uptake | Essential for mouse embryonic development | [134] Guetg et al., 2015 |
| MYO1C | Unconventional actin motor |
| [106] Fan et al., 2012 [108] Edimo et al., 2016 |
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Muraki, K.; Tanigaki, K. Neuronal migration abnormalities and its possible implications for schizophrenia. Front. Neurosci. 2015, 9, 74. [Google Scholar] [CrossRef]
- Catts, V.S.; Fung, S.J.; Long, L.E.; Joshi, D.; Vercammen, A.; Allen, K.M.; Fillman, S.G.; Rothmond, D.A.; Sinclair, D.; Tiwari, Y.; et al. Rethinking schizophrenia in the context of normal neurodevelopment. Front. Cell Neurosci. 2013, 7, 60. [Google Scholar] [CrossRef]
- Kato, M.; Dobyns, W.B. Lissencephaly and the molecular basis of neuronal migration. Hum. Mol. Genet. 2003, 12, R89–R96. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.S. Molecular genetics of neuronal migration disorders. Curr. Neurol. Neurosci. Rep. 2011, 11, 171–178. [Google Scholar] [CrossRef] [PubMed]
- Kuzniecky, R. Epilepsy and malformations of cortical development: New developments. Curr. Opin. Neurol. 2015, 28, 151–157. [Google Scholar] [CrossRef] [PubMed]
- Barkovich, A.J.; Dobyns, W.B.; Guerrini, R. Malformations of cortical development and epilepsy. Cold Spring Harb. Perspect. Med. 2015, 5, a022392. [Google Scholar] [CrossRef] [PubMed]
- Telias, M. Molecular Mechanisms of Synaptic Dysregulation in Fragile X Syndrome and Autism Spectrum Disorders. Front. Mol. Neurosci. 2019, 12, 51. [Google Scholar] [CrossRef] [PubMed]
- Almeida Montes, L.G.; Prado Alcántara, H.; Martínez García, R.B.; De La Torre, L.B.; Avila Acosta, D.; Duarte, M.G. Brain cortical thickness in ADHD: Age, sex, and clinical correlations. J. Atten. Disord. 2013, 17, 641–654. [Google Scholar] [CrossRef] [PubMed]
- Reiner, O.; Karzbrun, E.; Kshirsagar, A.; Kaibuchi, K. Regulation of neuronal migration, an emerging topic in autism spectrum disorders. J. Neurochem. 2016, 136, 440–456. [Google Scholar] [CrossRef] [PubMed]
- Haverfield, E.V.; Whited, A.J.; Petras, K.S.; Dobyns, W.B.; Das, S. Intragenic deletions and duplications of the LIS1 and DCX genes: A major disease-causing mechanism in lissencephaly and subcortical band heterotopia. Eur. J. Hum. Genet. 2009, 17, 911–918. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chang, B.S.; Duzcan, F.; Kim, S.; Cinbis, M.; Aggarwal, A.; Apse, K.A.; Ozdel, O.; Atmaca, M.; Zencir, S.; Bagci, H.; et al. The role of RELN in lissencephaly and neuropsychiatric disease. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2007, 144, 58–63. [Google Scholar] [CrossRef]
- Urquhart, J.E.; Beaman, G.; Byers, H.; Roberts, N.A.; Chervinsky, E.; O’Sullivan, J.; Pilz, D.; Fry, A.; Williams, S.G.; Bhaskar, S.S.; et al. DMRTA2 (DMRT5) is mutated in a novel cortical brain malformation. Clin. Genet. 2016, 89, 724–727. [Google Scholar] [CrossRef]
- Hebebrand, M.; Hüffmeier, U.; Trollmann, R.; Hehr, U.; Uebe, S.; Ekici, A.B.; Kraus, C.; Krumbiegel, M.; Reis, A.; Thiel, C.T.; et al. The mutational and phenotypic spectrum of TUBA1A-associated tubulinopathy. Orphanet J. Rare Dis. 2019, 14, 38. [Google Scholar] [CrossRef]
- Hikita, N.; Hattori, H.; Kato, M.; Sakuma, S.; Morotomi, Y.; Ishida, H.; Seto, T.; Tanaka, K.; Shimono, T.; Shintaku, H.; et al. A case of TUBA1A mutation presenting with lissencephaly and Hirschsprung disease. Brain Dev. 2014, 36, 159–162. [Google Scholar] [CrossRef]
- Marcorelles, P.; Laquerrière, A.; Adde-Michel, C.; Marret, S.; Saugier-Veber, P.; Beldjord, C.; Friocourt, G. Evidence for tangential migration disturbances in human lissencephaly resulting from a defect in LIS1, DCX and ARX genes. Acta Neuropathol. 2010, 120, 503–515. [Google Scholar] [CrossRef]
- Buchberg, A.M.; Brownell, E.; Nagata, S.; Jenkins, N.A.; Copeland, N.G. A comprehensive genetic map of murine chromosome 11 reveals extensive linkage conservation between mouse and human. Genetics 1989, 122, 153–161. [Google Scholar] [CrossRef]
- Hirotsune, S.; Pack, S.D.; Chong, S.S.; Robbins, C.M.; Pavan, W.J.; Ledbetter, D.H.; Wynshaw-Boris, A. Genomic organization of the murine Miller-Dieker/lissencephaly region: Conservation of linkage with the human region. Genome Res. 1997, 7, 625–634. [Google Scholar] [CrossRef]
- Cardoso, C.; Leventer, R.J.; Ward, H.L.; Toyo-Oka, K.; Chung, J.; Gross, A.; Martin, C.L.; Allanson, J.; Pilz, D.T.; Olney, A.H.; et al. Refinement of a 400-kb critical region allows genotypic differentiation between isolated lissencephaly, Miller-Dieker syndrome, and other phenotypes secondary to deletions of 17p13.3. Am. J. Hum. Genet. 2003, 72, 918–930. [Google Scholar] [CrossRef]
- Hatanaka, Y.; Zhu, Y.; Torigoe, M.; Kita, Y.; Murakami, F. From migration to settlement: The pathways, migration modes and dynamics of neurons in the developing brain. Proc. Jpn. Acad. Ser. B 2016, 92, 1–19. [Google Scholar] [CrossRef]
- Rivas, R.J.; Hatten, M.E. Motility and cytoskeletal organization of migrating cerebellar granule neurons. J. Neurosci. 1995, 15, 981–989. [Google Scholar] [CrossRef]
- Kopf, A.; Renkawitz, J.; Hauschild, R.; Girkontaite, I.; Tedford, K.; Merrin, J.; Thorn-Seshold, O.; Trauner, D.; Häcker, H.; Fischer, K.-D.; et al. Microtubules control cellular shape and coherence in amoeboid migrating cells. J. Cell Biol. 2020, 219. [Google Scholar] [CrossRef]
- Park, T.J.; Curran, T. Crk and Crk-like play essential overlapping roles downstream of disabled-1 in the Reelin pathway. J. Neurosci. 2008, 28, 13551–13562. [Google Scholar] [CrossRef]
- Dobyns, W.B.; Reiner, O.; Carrozzo, R.; Ledbetter, D.H. Lissencephaly. A human brain malformation associated with deletion of the LIS1 gene located at chromosome 17p13. JAMA 1993, 270, 2838–2842. [Google Scholar] [CrossRef]
- Bridges, D.; Moorhead, G.B.G. 14-3-3 Proteins: A Number of Functions for a Numbered Protein. Sci. STKE 2005, 2005, re10. [Google Scholar] [CrossRef]
- Toyo-oka, K.; Shionoya, A.; Gambello, M.J.; Cardoso, C.; Leventer, R.; Ward, H.L.; Ayala, R.; Tsai, L.H.; Dobyns, W.; Ledbetter, D.; et al. 14-3-3epsilon is important for neuronal migration by binding to NUDEL: A molecular explanation for Miller-Dieker syndrome. Nat. Genet. 2003, 34, 274–285. [Google Scholar] [CrossRef]
- Toyo-oka, K.; Wachi, T.; Hunt, R.F.; Baraban, S.C.; Taya, S.; Ramshaw, H.; Kaibuchi, K.; Schwarz, Q.P.; Lopez, A.F.; Wynshaw-Boris, A. 14-3-3ε and ζ regulate neurogenesis and differentiation of neuronal progenitor cells in the developing brain. J. Neurosci. 2014, 34, 12168–12181. [Google Scholar] [CrossRef] [PubMed]
- Blazejewski, S.M.; Bennison, S.A.; Smith, T.H.; Toyo-Oka, K. Neurodevelopmental Genetic Diseases Associated With Microdeletions and Microduplications of Chromosome 17p13.3. Front. Genet. 2018, 9, 80. [Google Scholar] [CrossRef] [PubMed]
- Tucker, M.E.; Escobar, L.F. Cleft lip/palate associated with 17p13.3 duplication involving a single candidate gene (YWHAE). Clin. Genet. 2014, 85, 600–601. [Google Scholar] [CrossRef] [PubMed]
- Al Kaissi, A.; Ganger, R.; Rötzer, K.M.; Klaushofer, K.; Grill, F. A child with split-hand/foot associated with tibial hemimelia (SHFLD syndrome) and thrombocytopenia maps to chromosome region 17p13.3. Am. J. Med. Genet. 2014, 164, 2338–2343. [Google Scholar] [CrossRef] [PubMed]
- Curry, C.J.; Rosenfeld, J.A.; Grant, E.; Gripp, K.W.; Anderson, C.; Aylsworth, A.S.; Saad, T.B.; Chizhikov, V.V.; Dybose, G.; Fagerberg, C.; et al. The duplication 17p13.3 phenotype: Analysis of 21 families delineates developmental, behavioral and brain abnormalities, and rare variant phenotypes. Am. J. Med. Genet. A 2013, 161, 1833–1852. [Google Scholar] [CrossRef] [PubMed]
- Capra, V.; Mirabelli-Badenier, M.; Stagnaro, M.; Rossi, A.; Tassano, E.; Gimelli, S.; Gimelli, G. Identification of a rare 17p13.3 duplication including the BHLHA9 and YWHAE genes in a family with developmental delay and behavioural problems. BMC Med. Genet. 2012, 13, 93. [Google Scholar] [CrossRef]
- Avela, K.; Aktan-Collan, K.; Horelli-Kuitunen, N.; Knuutila, S.; Somer, M. A microduplication on chromosome 17p13.1p13.3 including the PAFAH1B1 (LIS1) gene. Am. J. Med. Genet. 2011, 155, 875–879. [Google Scholar] [CrossRef]
- Bruno, D.L.; Anderlid, B.M.; Lindstrand, A.; van Ravenswaaij-Arts, C.; Ganesamoorthy, D.; Lundin, J.; Martin, C.L.; Douglas, J.; Nowak, C.; Adam, M.P.; et al. Further molecular and clinical delineation of co-locating 17p13.3 microdeletions and microduplications that show distinctive phenotypes. J. Med. Genet. 2010, 47, 299–311. [Google Scholar] [CrossRef]
- Hyon, C.; Marlin, S.; Chantot-Bastaraud, S.; Mabboux, P.; Beaujard, M.P.; Al Ageeli, E.; Vazquez, M.P.; Picard, A.; Siffroi, J.P.; Portnoi, M.F. A new 17p13.3 microduplication including the PAFAH1B1 and YWHAE genes resulting from an unbalanced X;17 translocation. Eur. J. Med. Genet. 2011, 54, 287–291. [Google Scholar] [CrossRef]
- Roos, L.; Jonch, A.E.; Kjaergaard, S.; Taudorf, K.; Simonsen, H.; Hamborg-Petersen, B.; Brondum-Nielsen, K.; Kirchhoff, M. A new microduplication syndrome encompassing the region of the Miller-Dieker (17p13 deletion) syndrome. J. Med. Genet. 2009, 46, 703–710. [Google Scholar] [CrossRef]
- Outwin, E.; Carpenter, G.; Bi, W.; Withers, M.A.; Lupski, J.R.; O’Driscoll, M. Increased RPA1 Gene Dosage Affects Genomic Stability Potentially Contributing to 17p13.3 Duplication Syndrome. PLoS Genet. 2011, 7, e1002247. [Google Scholar] [CrossRef]
- Hashimoto, R.; Nakazawa, T.; Tsurusaki, Y.; Yasuda, Y.; Nagayasu, K.; Matsumura, K.; Kawashima, H.; Yamamori, H.; Fujimoto, M.; Ohi, K.; et al. Whole-exome sequencing and neurite outgrowth analysis in autism spectrum disorder. J. Hum. Genet. 2015, 61, 199–206. [Google Scholar] [CrossRef]
- Doers, M.E.; Musser, M.T.; Nichol, R.; Berndt, E.R.; Baker, M.; Gomez, T.M.; Zhang, S.-C.; Abbeduto, L.; Bhattacharyya, A. iPSC-derived forebrain neurons from FXS individuals show defects in initial neurite outgrowth. Stem Cells Dev. 2014, 23, 1777–1787. [Google Scholar] [CrossRef] [PubMed]
- Phillips, M.; Pozzo-Miller, L. Dendritic spine dysgenesis in autism related disorders. Neurosci. Lett. 2015, 601, 30–40. [Google Scholar] [CrossRef] [PubMed]
- Cornell, B.; Wachi, T.; Zhukarev, V.; Toyo-Oka, K. Regulation of neuronal morphogenesis by 14-3-3epsilon (Ywhae) via the microtubule binding protein, doublecortin. Hum. Mol. Genet. 2016, 25, 4405–4418. [Google Scholar] [CrossRef] [PubMed]
- Torrico, B.; Antón-Galindo, E.; Fernàndez-Castillo, N.; Rojo-Francàs, E.; Ghorbani, S.; Pineda-Cirera, L.; Hervás, A.; Rueda, I.; Moreno, E.; Fullerton, J.M.; et al. Involvement of the 14-3-3 Gene Family in Autism Spectrum Disorder and Schizophrenia: Genetics, Transcriptomics and Functional Analyses. J. Clin. Med. 2020, 9, 1851. [Google Scholar] [CrossRef]
- Cheah, P.S.; Ramshaw, H.S.; Thomas, P.Q.; Toyo-Oka, K.; Xu, X.; Martin, S.; Coyle, P.; Guthridge, M.A.; Stomski, F.; van den Buuse, M.; et al. Neurodevelopmental and neuropsychiatric behaviour defects arise from 14-3-3ζ deficiency. Mol. Psychiatry 2012, 17, 451–466. [Google Scholar] [CrossRef]
- Blazejewski, S.M.; Bennison, S.A.; Ha, N.T.; Liu, X.; Smith, T.H.; Dougherty, K.J.; Toyo-Oka, K. Rpsa Signaling Regulates Cortical Neuronal Morphogenesis via Its Ligand, PEDF, and Plasma Membrane Interaction Partner, Itga6. Cereb. Cortex 2021. accepted. [Google Scholar] [CrossRef]
- Wynshaw-Boris, A. Lissencephaly and LIS1: Insights into the molecular mechanisms of neuronal migration and development. Clin. Genet. 2007, 72, 296–304. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Tsai, L.H.; Wynshaw-Boris, A. Life is a journey: A genetic look at neocortical development. Nat. Rev. Genet. 2002, 3, 342–355. [Google Scholar] [CrossRef] [PubMed]
- Vallee, R.B.; Tsai, J.W. The cellular roles of the lissencephaly gene LIS1, and what they tell us about brain development. Genes Dev. 2006, 20, 1384–1393. [Google Scholar] [CrossRef] [PubMed]
- Markus, S.M.; Marzo, M.G.; McKenney, R.J. New insights into the mechanism of dynein motor regulation by lissencephaly-1. ELife. 2020, 9, e59737. [Google Scholar] [CrossRef] [PubMed]
- Reiner, O.; Carrozzo, R.; Shen, Y.; Wehnert, M.; Faustinella, F.; Dobyns, W.B.; Caskey, C.T.; Ledbetter, D.H. Isolation of a Miller-Dieker lissencephaly gene containing G protein beta-subunit-like repeats. Nature 1993, 364, 717–721. [Google Scholar] [CrossRef] [PubMed]
- Hirotsune, S.; Fleck, M.W.; Gambello, M.J.; Bix, G.J.; Chen, A.; Clark, G.D.; Ledbetter, D.H.; McBain, C.J.; Wynshaw-Boris, A. Graded reduction of Pafah1b1 (Lis1) activity results in neuronal migration defects and early embryonic lethality. Nat. Genet. 1998, 19, 333–339. [Google Scholar] [CrossRef] [PubMed]
- Wynshaw-Boris, A.; Pramparo, T.; Youn, Y.H.; Hirotsune, S. Lissencephaly: Mechanistic insights from animal models and potential therapeutic strategies. Semin. Cell Dev. Biol. 2010, 21, 823–830. [Google Scholar] [CrossRef] [PubMed]
- Hattori, M.; Adachi, H.; Tsujimoto, M.; Arai, H.; Inoue, K. Miller-Dieker lissencephaly gene encodes a subunit of brain platelet-activating factor acetylhydrolase. Nature 1994, 370, 216–218. [Google Scholar] [CrossRef]
- Sapir, T.; Elbaum, M.; Reiner, O. Reduction of microtubule catastrophe events by LIS1, platelet-activating factor acetylhydrolase subunit. EMBO J. 1997, 16, 6977–6984. [Google Scholar] [CrossRef]
- Lo Nigro, C.; Chong, C.S.; Smith, A.C.; Dobyns, W.B.; Carrozzo, R.; Ledbetter, D.H. Point mutations and an intragenic deletion in LIS1, the lissencephaly causative gene in isolated lissencephaly sequence and Miller-Dieker syndrome. Hum. Mol. Genet. 1997, 6, 157–164. [Google Scholar] [CrossRef]
- Pilz, D.T.; Macha, M.E.; Precht, K.S.; Smith, A.C.; Dobyns, W.B.; Ledbetter, D.H. Fluorescence in situ hybridization analysis with LIS1 specific probes reveals a high deletion mutation rate in isolated lissencephaly sequence. Genet. Med. 1998, 1, 29–33. [Google Scholar] [CrossRef][Green Version]
- Hunt, R.F.; Dinday, M.T.; Hindle-Katel, W.; Baraban, S.C. LIS1 deficiency promotes dysfunctional synaptic integration of granule cells generated in the developing and adult dentate gyrus. J. Neurosci. 2012, 32, 12862–12875. [Google Scholar] [CrossRef]
- Shimojima, K.; Sugiura, C.; Takahashi, H.; Ikegami, M.; Takahashi, Y.; Ohno, K.; Matsuo, M.; Saito, K.; Yamamoto, T. Genomic copy number variations at 17p13.3 and epileptogenesis. Epilepsy Res. 2010, 89, 303–309. [Google Scholar] [CrossRef]
- Greenwood, J.S.; Wang, Y.; Estrada, R.C.; Ackerman, L.; Ohara, P.T.; Baraban, S.C. Seizures, enhanced excitation, and increased vesicle number in Lis1 mutant mice. Ann. Neurol. 2009, 66, 644–653. [Google Scholar] [CrossRef]
- Jones, D.L.; Baraban, S.C. Characterization of inhibitory circuits in the malformed hippocampus of Lis1 mutant mice. J. Neurophysiol. 2007, 98, 2737–2746. [Google Scholar] [CrossRef] [PubMed]
- Sudarov, A.; Gooden, F.; Tseng, D.; Gan, W.B.; Ross, M.E. Lis1 controls dynamics of neuronal filopodia and spines to impact synaptogenesis and social behaviour. EMBO Mol. Med. 2013, 5, 591–607. [Google Scholar] [CrossRef] [PubMed]
- Canty, J.T.; Yildiz, A. Activation and Regulation of Cytoplasmic Dynein. Trends Biochem. Sci. 2020, 45, 440–453. [Google Scholar] [CrossRef] [PubMed]
- Faulkner, N.E.; Dujardin, D.L.; Tai, C.Y.; Vaughan, K.T.; O’Connell, C.B.; Wang, Y.; Vallee, R.B. A role for the lissencephaly gene LIS1 in mitosis and cytoplasmic dynein function. Nat. Cell Biol. 2000, 2, 784–791. [Google Scholar] [CrossRef]
- Smith, D.S.; Niethammer, M.; Ayala, R.; Zhou, Y.; Gambello, M.J.; Wynshaw-Boris, A.; Tsai, L.H. Regulation of cytoplasmic dynein behaviour and microtubule organization by mammalian Lis1. Nat. Cell. Biol. 2000, 2, 767–775. [Google Scholar] [CrossRef]
- Toropova, K.; Zou, S.; Roberts, A.J.; Redwine, W.B.; Goodman, B.S.; Reck-Peterson, S.L.; Leschziner, A.E. Lis1 regulates dynein by sterically blocking its mechanochemical cycle. Elife 2014, 3, e03372. [Google Scholar] [CrossRef]
- Htet, Z.M.; Gillies, J.P.; Baker, R.W.; Leschziner, A.E.; DeSantis, M.E.; Reck-Peterson, S.L. LIS1 promotes the formation of activated cytoplasmic dynein-1 complexes. Nat. Cell Biol. 2020, 22, 518–525. [Google Scholar] [CrossRef]
- Chhatre, A.; Sanghavi, P.; Mallik, R. Lis1 co-localizes with actin in the phagocytic cup and regulates phagocytosis. Cytoskeleton 2020, 77, 249–260. [Google Scholar] [CrossRef]
- Chen, J.; Cai, Z.; Zhang, L.; Yin, Y.; Chen, X.; Chen, C.; Zhang, Y.; Zhai, S.; Long, X.; Liu, X.; et al. Lis1 Regulates Germinal Center B Cell Antigen Acquisition and Affinity Maturation. J. Immunol. 2021, 198, 4304–4311. [Google Scholar] [CrossRef] [PubMed]
- Jheng, G.-W.; Hur, S.S.; Chang, C.-M.; Wu, C.-C.; Cheng, J.-S.; Lee, H.-H.; Chung, B.-C.; Wang, Y.-K.; Lin, K.-H.; Del Álamo, J.C.; et al. Lis1 dysfunction leads to traction force reduction and cytoskeletal disorganization during cell migration. Biochem. Biophys. Res. Commun. 2018, 497, 869–875. [Google Scholar] [CrossRef]
- Chen, K.; Ochalski, P.G.; Tran, T.S.; Sahir, N.; Schubert, M.; Pramatarova, A.; Howell, B.W. Interaction between Dab1 and CrkII is promoted by Reelin signaling. J. Cell Sci. 2004, 117, 4527–4536. [Google Scholar] [CrossRef] [PubMed]
- Falconer, D.S. Two new mutants, ‘trembler’ and ‘reeler’, with neurological actions in the house mouse (Mus musculus L.). J. Genet. 1951, 50, 192–201. [Google Scholar] [CrossRef] [PubMed]
- Miao, G.G.; Smeyne, R.J.; D’Arcangelo, G.; Copeland, N.G.; Jenkins, N.A.; Morgan, J.I.; Curran, T. Isolation of an allele of reeler by insertional mutagenesis. Proc. Natl. Acad. Sci. USA 1994, 91, 11050–11054. [Google Scholar] [CrossRef] [PubMed]
- D’Arcangelo, G.; Miao, G.G.; Chen, S.C.; Soares, H.D.; Morgan, J.I.; Curran, T. A protein related to extracellular matrix proteins deleted in the mouse mutant reeler. Nature 1995, 374, 719–723. [Google Scholar] [CrossRef]
- Keshvara, L.; Benhayon, D.; Magdaleno, S.; Curran, T. Identification of reelin-induced sites of tyrosyl phosphorylation on disabled 1. J. Biol. Chem. 2001, 276, 16008–16014. [Google Scholar] [CrossRef]
- Kuo, G.; Arnaud, L.; Kronstad-O’Brien, P.; Cooper, J.A. Absence of Fyn and Src causes a reeler-like phenotype. J. Neurosci. 2005, 25, 8578–8586. [Google Scholar] [CrossRef]
- Bock, H.H.; Herz, J. Reelin activates SRC family tyrosine kinases in neurons. Curr. Biol. 2003, 13, 18–26. [Google Scholar] [CrossRef]
- Ballif, B.A.; Arnaud, L.; Arthur, W.T.; Guris, D.; Imamoto, A.; Cooper, J.A. Activation of a Dab1/CrkL/C3G/Rap1 pathway in Reelin-stimulated neurons. Curr. Biol. 2004, 14, 606–610. [Google Scholar] [CrossRef] [PubMed]
- Guris, D.L.; Fantes, J.; Tara, D.; Druker, B.J.; Imamoto, A. Mice lacking the homologue of the human 22q11.2 gene CRKL phenocopy neurocristopathies of DiGeorge syndrome. Nat. Genet. 2001, 27, 293–298. [Google Scholar] [CrossRef]
- Park, T.-J.; Boyd, K.; Curran, T. Cardiovascular and craniofacial defects in Crk-null mice. Mol. Cell. Biol. 2006, 26, 6272–6282. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sekine, K.; Kawauchi, T.; Kubo, K.-I.; Honda, T.; Herz, J.; Hattori, M.; Kinashi, T.; Nakajima, K. Reelin controls neuronal positioning by promoting cell-matrix adhesion via inside-out activation of integrin α5β1. Neuron 2012, 76, 353–369. [Google Scholar] [CrossRef] [PubMed]
- O’Dell, R.S.; Cameron, D.A.; Zipfel, W.R.; Olson, E.C. Reelin Prevents Apical Neurite Retraction during Terminal Translocation and Dendrite Initiation. J. Neurosci. 2015, 35, 10659. [Google Scholar] [CrossRef]
- Tenney, J.R.; Hopkin, R.J.; Schapiro, M.B. Deletion of 14-3-3ε and CRK: A clinical syndrome with macrocephaly, developmental delay, and generalized epilepsy. J. Child Neurol. 2011, 26, 223–227. [Google Scholar] [CrossRef]
- Romano, C.; Ferranti, S.; Mencarelli, M.A.; Longo, I.; Renieri, A.; Grosso, S. 17p13.3 microdeletion including YWHAE and CRK genes: Towards a clinical characterization. Neurol. Sci. 2020, 41, 2259–2262. [Google Scholar] [CrossRef]
- Fatemi, S.H.; Stary, J.M.; Halt, A.R.; Realmuto, G.R. Dysregulation of Reelin and Bcl-2 proteins in autistic cerebellum. J. Autism Dev. Disord. 2001, 31, 529–535. [Google Scholar] [CrossRef] [PubMed]
- Fatemi, S.H.; Stary, J.M.; Egan, E.A. Reduced blood levels of reelin as a vulnerability factor in pathophysiology of autistic disorder. Cell Mol. Neurobiol. 2002, 22, 139–152. [Google Scholar] [CrossRef] [PubMed]
- Darnell, J.C.; Van Driesche, S.J.; Zhang, C.; Hung, K.Y.; Mele, A.; Fraser, C.E.; Stone, E.F.; Chen, C.; Fak, J.J.; Chi, S.W.; et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell 2011, 146, 247–261. [Google Scholar] [CrossRef]
- Budimirovic, D.B.; Subramanian, M. Neurobiology of Disease. In Neurobiology of Autism and Intellectual Disability Fragile X Syndrome: Fragile X Syndrome; Oxford University Press: Oxford, UK, 2017. [Google Scholar]
- Pan, L.; Zhang, Y.Q.; Woodruff, E.; Broadie, K. The Drosophila fragile X gene negatively regulates neuronal elaboration and synaptic differentiation. Curr. Biol. 2004, 14, 1863–1870. [Google Scholar] [CrossRef] [PubMed]
- Budimirovic, D.B.; Schlageter, A.; Filipovic-Sadic, S.; Protic, D.D.; Bram, E.; Mahone, E.M.; Nicholson, K.; Culp, K.; Javanmardi, K.; Kemppainen, J.; et al. A Genotype-Phenotype Study of High-Resolution FMR1 Nucleic Acid and Protein Analyses in Fragile X Patients with Neurobehavioral Assessments. Brain Sci. 2020, 10, 694. [Google Scholar] [CrossRef]
- Wu, C.; Zhang, X.; Chen, P.; Ruan, X.; Liu, W.; Li, Y.; Sun, C.; Hou, L.; Yin, B.; Qiang, B.; et al. MicroRNA-129 modulates neuronal migration by targeting Fmr1 in the developing mouse cortex. Cell Death Dis. 2019, 10, 287. [Google Scholar] [CrossRef] [PubMed]
- Sidorov, M.S.; Auerbach, B.D.; Bear, M.F. Fragile X mental retardation protein and synaptic plasticity. Mol. Brain 2013, 6, 15. [Google Scholar] [CrossRef]
- Kazdoba, T.M.; Leach, P.T.; Silverman, J.L.; Crawley, J.N. Modeling fragile X syndrome in the Fmr1 knockout mouse. Intractable Rare Dis. Res. 2014, 3, 118–133. [Google Scholar] [CrossRef] [PubMed]
- Podhorna, J.; Didriksen, M. The heterozygous reeler mouse: Behavioural phenotype. Behav. Brain Res. 2004, 153, 43–54. [Google Scholar] [CrossRef]
- Sakai, K.; Shoji, H.; Kohno, T.; Miyakawa, T.; Hattori, M. Mice that lack the C-terminal region of Reelin exhibit behavioral abnormalities related to neuropsychiatric disorders. Sci. Rep. 2016, 6, 28636. [Google Scholar] [CrossRef]
- Kawaguchi, T.; Yamagishi, S.I.; Sata, M. Structure-function relationships of PEDF. Curr. Mol. Med. 2010, 10, 302–311. [Google Scholar] [CrossRef] [PubMed]
- Tombran-Tink, J.; Johnson, L.V. Neuronal differentiation of retinoblastoma cells induced by medium conditioned by human RPE cells. Investig. Ophthalmol. Vis. Sci. 1989, 30, 1700–1707. [Google Scholar]
- Sugita, Y.; Becerra, S.P.; Chader, G.J.; Schwartz, J.P. Pigment epithelium-derived factor (PEDF) has direct effects on the metabolism and proliferation of microglia and indirect effects on astrocytes. J. Neurosci. Res. 1997, 49, 710–718. [Google Scholar] [CrossRef]
- Cao, W.; Tombran-Tink, J.; Chen, W.; Mrazek, D.; Elias, R.; McGinnis, J.F. Pigment epithelium-derived factor protects cultured retinal neurons against hydrogen peroxide-induced cell death. J. Neurosci. Res. 1999, 57, 789–800. [Google Scholar] [CrossRef]
- Dawson, D.W.; Volpert, O.V.; Gillis, P.; Crawford, S.E.; Xu, H.; Benedict, W.; Bouck, N.P. Pigment epithelium-derived factor: A potent inhibitor of angiogenesis. Science 1999, 285, 245–248. [Google Scholar] [CrossRef] [PubMed]
- Steele, F.R.; Chader, G.J.; Johnson, L.V.; Tombran-Tink, J. Pigment epithelium-derived factor: Neurotrophic activity and identification as a member of the serine protease inhibitor gene family. Proc. Natl. Acad. Sci. USA 1993, 90, 1526–1530. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, P.; Notario, P.M.; Becerra, S.P. Pigment epithelium-derived factor receptor (PEDF-R): A plasma membrane-linked phospholipase with PEDF binding affinity. Adv. Exp. Med. Biol. 2010, 664, 29–37. [Google Scholar] [CrossRef]
- Bernard, A.; Gao-Li, J.; Franco, C.-A.; Bouceba, T.; Huet, A.; Li, Z. Laminin receptor involvement in the anti-angiogenic activity of pigment epithelium-derived factor. J. Biol. Chem. 2009, 284, 10480–10490. [Google Scholar] [CrossRef]
- Cheng, G.; Zhong, M.; Kawaguchi, R.; Kassai, M.; Al-Ubaidi, M.; Deng, J.; Ter-Stepanian, M.; Sun, H. Identification of PLXDC1 and PLXDC2 as the transmembrane receptors for the multifunctional factor PEDF. Elife 2014, 3, e05401. [Google Scholar] [CrossRef]
- He, T.; Hu, J.; Yan, G.; Li, L.; Zhang, D.; Zhang, Q.; Chen, B.; Huang, Y. Pigment epithelium-derived factor regulates microvascular permeability through adipose triglyceride lipase in sepsis. Clin. Sci. 2015, 129, 49–61. [Google Scholar] [CrossRef]
- Yao, S.; Zhang, Y.; Wang, X.; Zhao, F.; Sun, M.; Zheng, X.; Dong, H.; Guo, K. Pigment Epithelium-Derived Factor (PEDF) Protects Osteoblastic Cell Line from Glucocorticoid-Induced Apoptosis via PEDF-R. Int. J. Mol. Sci. 2016, 17, 730. [Google Scholar] [CrossRef]
- Beaty, R.M.; Edwards, J.B.; Boon, K.; Siu, I.M.; Conway, J.E.; Riggins, G.J. PLXDC1 (TEM7) is identified in a genome-wide expression screen of glioblastoma endothelium. J. Neurooncol. 2007, 81, 241–248. [Google Scholar] [CrossRef]
- Miller-Delaney, S.F.C.; Lieberam, I.; Murphy, P.; Mitchell, K.J. Plxdc2 Is a Mitogen for Neural Progenitors. PLoS ONE 2011, 6, e14565. [Google Scholar] [CrossRef]
- Fan, Y.; Eswarappa, S.M.; Hitomi, M.; Fox, P.L. Myo1c facilitates G-actin transport to the leading edge of migrating endothelial cells. J. Cell Biol. 2012, 198, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Brandstaetter, H.; Kendrick-Jones, J.; Buss, F. Myo1c regulates lipid raft recycling to control cell spreading, migration and Salmonella invasion. J. Cell Sci. 2012, 125, 1991–2003. [Google Scholar] [CrossRef] [PubMed]
- Edimo, W.E.; Ramos, A.R.; Ghosh, S.; Vanderwinden, J.M.; Erneux, C. The SHIP2 interactor Myo1c is required for cell migration in 1321 N1 glioblastoma cells. Biochem. Biophys. Res. Commun. 2016, 476, 508–514. [Google Scholar] [CrossRef] [PubMed]
- Visuttijai, K.; Pettersson, J.; Mehrbani Azar, Y.; van den Bout, I.; Örndal, C.; Marcickiewicz, J.; Nilsson, S.; Hörnquist, M.; Olsson, B.; Ejeskär, K.; et al. Lowered Expression of Tumor Suppressor Candidate MYO1C Stimulates Cell Proliferation, Suppresses Cell Adhesion and Activates AKT. PLoS ONE 2016, 11, e0164063. [Google Scholar] [CrossRef] [PubMed]
- Patten, D.A.; Kamarajah, S.K.; Rose, J.M.; Tickle, J.; Shepherd, E.L.; Adams, D.H.; Weston, C.J.; Shetty, S. SCARF-1 promotes adhesion of CD4(+) T cells to human hepatic sinusoidal endothelium under conditions of shear stress. Sci. Rep. 2017, 7, 17600. [Google Scholar] [CrossRef] [PubMed]
- Shibata, M.; Ishii, J.; Koizumi, H.; Shibata, N.; Dohmae, N.; Takio, K.; Adachi, H.; Tsujimoto, M.; Arai, H. Type F scavenger receptor SREC-I interacts with advillin, a member of the gelsolin/villin family, and induces neurite-like outgrowth. J. Biol. Chem. 2004, 279, 40084–40090. [Google Scholar] [CrossRef] [PubMed]
- Wolosker, H.; Sheth, K.N.; Takahashi, M.; Mothet, J.-P.; Brady, R.O.; Ferris, C.D.; Snyder, S.H. Purification of serine racemase: Biosynthesis of the neuromodulator D-serine. Proc. Natl. Acad. Sci. USA 1999, 96, 721. [Google Scholar] [CrossRef]
- Foltyn, V.N.; Bendikov, I.; De Miranda, J.; Panizzutti, R.; Dumin, E.; Shleper, M.; Li, P.; Toney, M.D.; Kartvelishvily, E.; Wolosker, H. Serine racemase modulates intracellular D-serine levels through an alpha,beta-elimination activity. J. Biol. Chem. 2005, 280, 1754–1763. [Google Scholar] [CrossRef]
- Lau, C.G.; Takeuchi, K.; Rodenas-Ruano, A.; Takayasu, Y.; Murphy, J.; Bennett, M.V.L.; Zukin, R.S. Regulation of NMDA receptor Ca2+ signalling and synaptic plasticity. Biochem. Soc. Trans. 2009, 37, 1369–1374. [Google Scholar] [CrossRef]
- Xing, L.; Larsen, R.S.; Bjorklund, G.R.; Li, X.; Wu, Y.; Philpot, B.D.; Snider, W.D.; Newbern, J.M. Layer specific and general requirements for ERK/MAPK signaling in the developing neocortex. Elife 2016, 5, e11123. [Google Scholar] [CrossRef] [PubMed]
- Millar, J.K.; Christie, S.; Semple, C.A.M.; Porteous, D.J. Chromosomal Location and Genomic Structure of the Human Translin-Associated Factor X Gene (TRAX; TSNAX) Revealed by Intergenic Splicing to DISC1, a Gene Disrupted by a Translocation Segregating with Schizophrenia. Genomics 2000, 67, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Brandon, N.J.; Sawa, A. Linking neurodevelopmental and synaptic theories of mental illness through DISC1. Nat. Rev. Neurosci. 2011, 12, 707–722. [Google Scholar] [CrossRef] [PubMed]
- Tomita, K.; Kubo, K.-I.; Ishii, K.; Nakajima, K. Disrupted-in-Schizophrenia-1 (Disc1) is necessary for migration of the pyramidal neurons during mouse hippocampal development. Hum. Mol. Genet. 2011, 20, 2834–2845. [Google Scholar] [CrossRef] [PubMed]
- Steinecke, A.; Gampe, C.; Nitzsche, F.; Bolz, J. DISC1 knockdown impairs the tangential migration of cortical interneurons by affecting the actin cytoskeleton. Front. Cell. Neurosci. 2014, 8, 190. [Google Scholar] [CrossRef]
- Fukuda, T.; Sugita, S.; Inatome, R.; Yanagi, S. CAMDI, a novel disrupted in schizophrenia 1 (DISC1)-binding protein, is required for radial migration. J. Biol. Chem. 2010, 285, 40554–40561. [Google Scholar] [CrossRef]
- Jacobi, A.A.; Halawani, S.; Lynch, D.R.; Lin, H. Neuronal serine racemase associates with Disrupted-In-Schizophrenia-1 and DISC1 agglomerates: Implications for schizophrenia. Neurosci. Lett. 2019, 692, 107–114. [Google Scholar] [CrossRef]
- Bershteyn, M.; Nowakowski, T.J.; Pollen, A.A.; Di Lullo, E.; Nene, A.; Wynshaw-Boris, A.; Kriegstein, A.R. Human iPSC-Derived Cerebral Organoids Model Cellular Features of Lissencephaly and Reveal Prolonged Mitosis of Outer Radial Glia. Cell Stem Cell 2017, 20, 435–449.e434. [Google Scholar] [CrossRef]
- Bershteyn, M.; Hayashi, Y.; Desachy, G.; Hsiao, E.C.; Sami, S.; Tsang, K.M.; Weiss, L.A.; Kriegstein, A.R.; Yamanaka, S.; Wynshaw-Boris, A. Cell-autonomous correction of ring chromosomes in human induced pluripotent stem cells. Nature 2014, 507, 99–103. [Google Scholar] [CrossRef]
- Wu, J.; Zhou, Q.; Wang, Y.; Zhou, X.; Li, J. MNT inhibits the migration of human hepatocellular carcinoma SMMC7721 cells. Biochem. Biophys. Res. Commun. 2012, 418, 93–97. [Google Scholar] [CrossRef]
- Toyo-oka, K.; Hirotsune, S.; Gambello, M.J.; Zhou, Z.-Q.; Olson, L.; Rosenfeld, M.G.; Eisenman, R.; Hurlin, P.; Wynshaw-Boris, A. Loss of the Max-interacting protein Mnt in mice results in decreased viability, defective embryonic growth and craniofacial defects: Relevance to Miller–Dieker syndrome. Hum. Mol. Genet. 2004, 13, 1057–1067. [Google Scholar] [CrossRef]
- Lin, J.-H.; Lee, W.-J.; Wu, H.-C.; Wu, C.-H.; Chen, L.-C.; Huang, C.-C.; Chang, H.-L.; Cheng, T.-C.; Chang, H.-W.; Ho, C.-T.; et al. Small G protein signalling modulator 2 (SGSM2) is involved in oestrogen receptor-positive breast cancer metastasis through enhancement of migratory cell adhesion via interaction with E-cadherin. Cell Adhes. Migr. 2019, 13, 120–137. [Google Scholar] [CrossRef] [PubMed]
- Ray, H.; Chang, C. The transcription factor Hypermethylated in Cancer 1 (Hic1) regulates neural crest migration via interaction with Wnt signaling. Dev. Biol. 2020, 463, 169–181. [Google Scholar] [CrossRef]
- Valenta, T.; Lukas, J.; Doubravska, L.; Fafilek, B.; Korinek, V. HIC1 attenuates Wnt signaling by recruitment of TCF-4 and beta-catenin to the nuclear bodies. EMBO J. 2006, 25, 2326–2337. [Google Scholar] [CrossRef]
- Carter, M.G.; Johns, M.A.; Zeng, X.; Zhou, L.; Zink, M.C.; Mankowski, J.L.; Donovan, D.M.; Baylin, S.B. Mice deficient in the candidate tumor suppressor gene Hic1 exhibit developmental defects of structures affected in the Miller–Dieker syndrome. Hum. Mol. Genet. 2000, 9, 413–419. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.-R.; You, L.-R.; Yan, Y.-T.; Chen, C.-M. Role of OVCA1/DPH1 in craniofacial abnormalities of Miller–Dieker syndrome. Hum. Mol. Genet. 2014, 23, 5579–5596. [Google Scholar] [CrossRef]
- He, J.Y.; Han, P.; Zhang, Y.; Liu, Y.D.; Song, S.J.; Feng, G.K.; An, Y.; Zhou, A.J.; Wang, H.B.; Yuan, L.; et al. Overexpression of Nogo receptor 3 (NgR3) correlates with poor prognosis and contributes to the migration of epithelial cells of nasopharyngeal carcinoma patients. J. Mol. Med. 2018, 96, 265–279. [Google Scholar] [CrossRef] [PubMed]
- Margiotta, A.; Progida, C.; Bakke, O.; Bucci, C. Characterization of the role of RILP in cell migration. EJH 2017, 61, 2783. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhou, Y.; Hu, X.; Chen, W.; Lin, X.; Sun, L.; Xu, X.; Hong, W.; Wang, T. RILP suppresses invasion of breast cancer cells by modulating the activity of RalA through interaction with RalGDS. Cell Death Dis. 2015, 6, e1923. [Google Scholar] [CrossRef] [PubMed]
- Guetg, A.; Mariotta, L.; Bock, L.; Herzog, B.; Fingerhut, R.; Camargo, S.M.; Verrey, F. Essential amino acid transporter Lat4 (Slc43a2) is required for mouse development. J. Physiol. 2015, 593, 1273–1289. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, X.; Bennison, S.A.; Robinson, L.; Toyo-oka, K. Responsible Genes for Neuronal Migration in the Chromosome 17p13.3: Beyond Pafah1b1(Lis1), Crk and Ywhae(14-3-3ε). Brain Sci. 2022, 12, 56. https://doi.org/10.3390/brainsci12010056
Liu X, Bennison SA, Robinson L, Toyo-oka K. Responsible Genes for Neuronal Migration in the Chromosome 17p13.3: Beyond Pafah1b1(Lis1), Crk and Ywhae(14-3-3ε). Brain Sciences. 2022; 12(1):56. https://doi.org/10.3390/brainsci12010056
Chicago/Turabian StyleLiu, Xiaonan, Sarah A. Bennison, Lozen Robinson, and Kazuhito Toyo-oka. 2022. "Responsible Genes for Neuronal Migration in the Chromosome 17p13.3: Beyond Pafah1b1(Lis1), Crk and Ywhae(14-3-3ε)" Brain Sciences 12, no. 1: 56. https://doi.org/10.3390/brainsci12010056
APA StyleLiu, X., Bennison, S. A., Robinson, L., & Toyo-oka, K. (2022). Responsible Genes for Neuronal Migration in the Chromosome 17p13.3: Beyond Pafah1b1(Lis1), Crk and Ywhae(14-3-3ε). Brain Sciences, 12(1), 56. https://doi.org/10.3390/brainsci12010056







