The Crosstalk between the EGFR and IFN-γ Pathways and Synergistic Roles in Survival Prediction and Immune Escape in Gliomas
Abstract
:1. Introduction
2. Methods
2.1. Samples and Datasets
2.2. Construction of EGFR-Related and IFN-γ-Related Prognostic Gene Signatures
2.3. Biological Function and Signaling Pathway Analysis
2.4. Comprehensive Analysis of Immune and Molecular Characteristics
2.5. Cell Culture for Western Blot (WB) and Flow Cytometry (FCM)
2.6. Statistical Analysis
3. Results
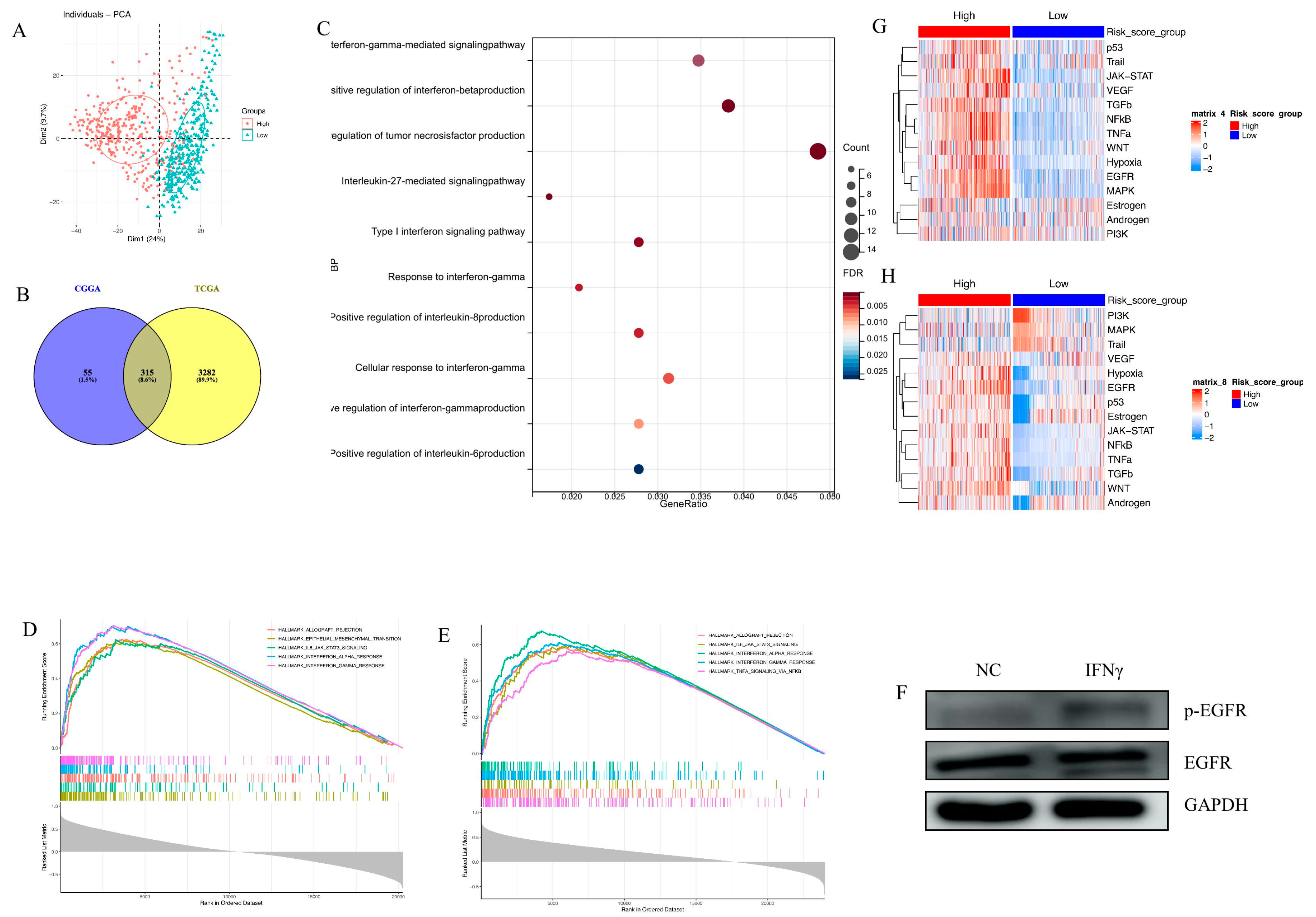
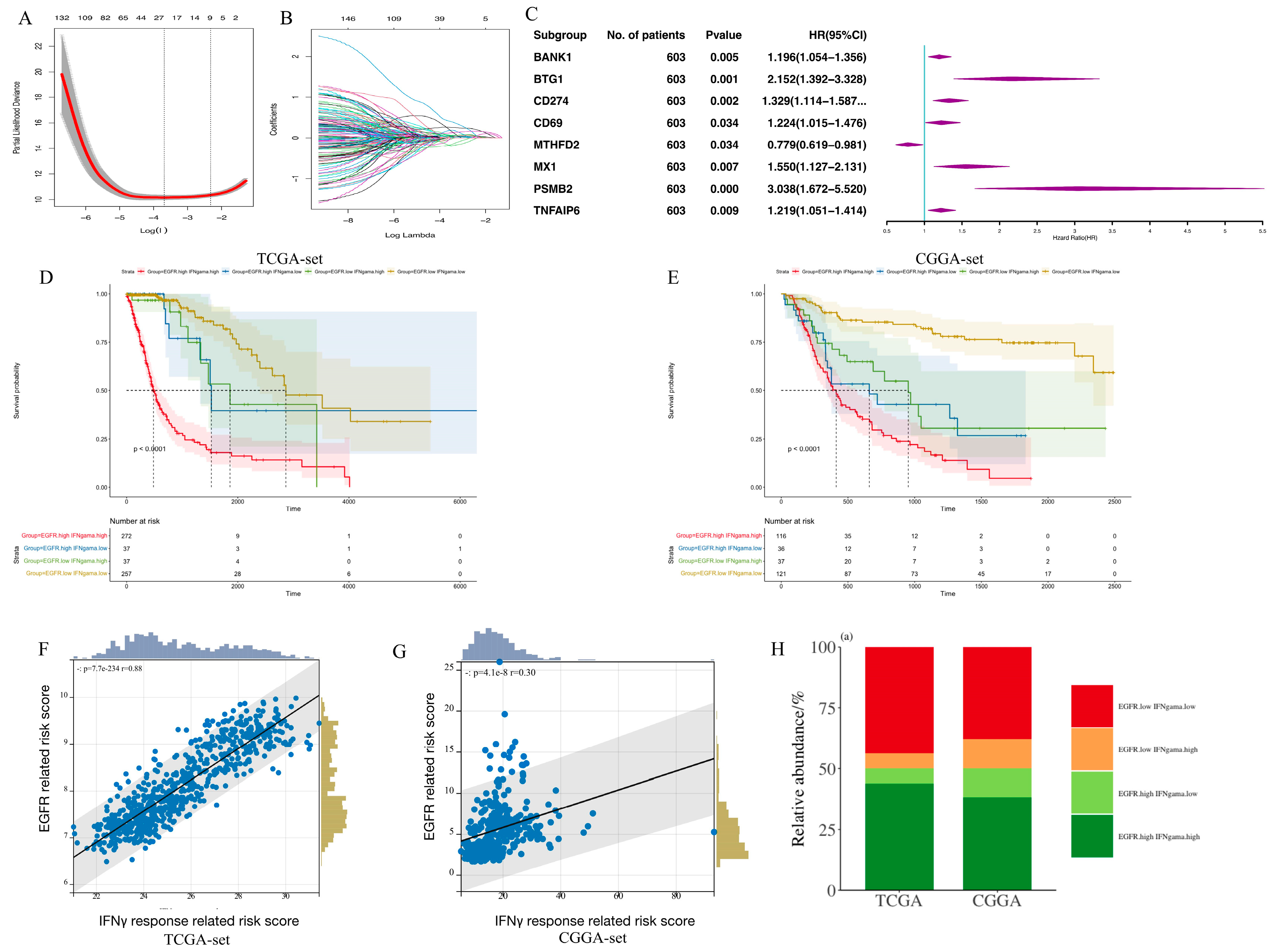
3.1. Establishment of an EGFR Pathway-Related Prognostic Gene Signature
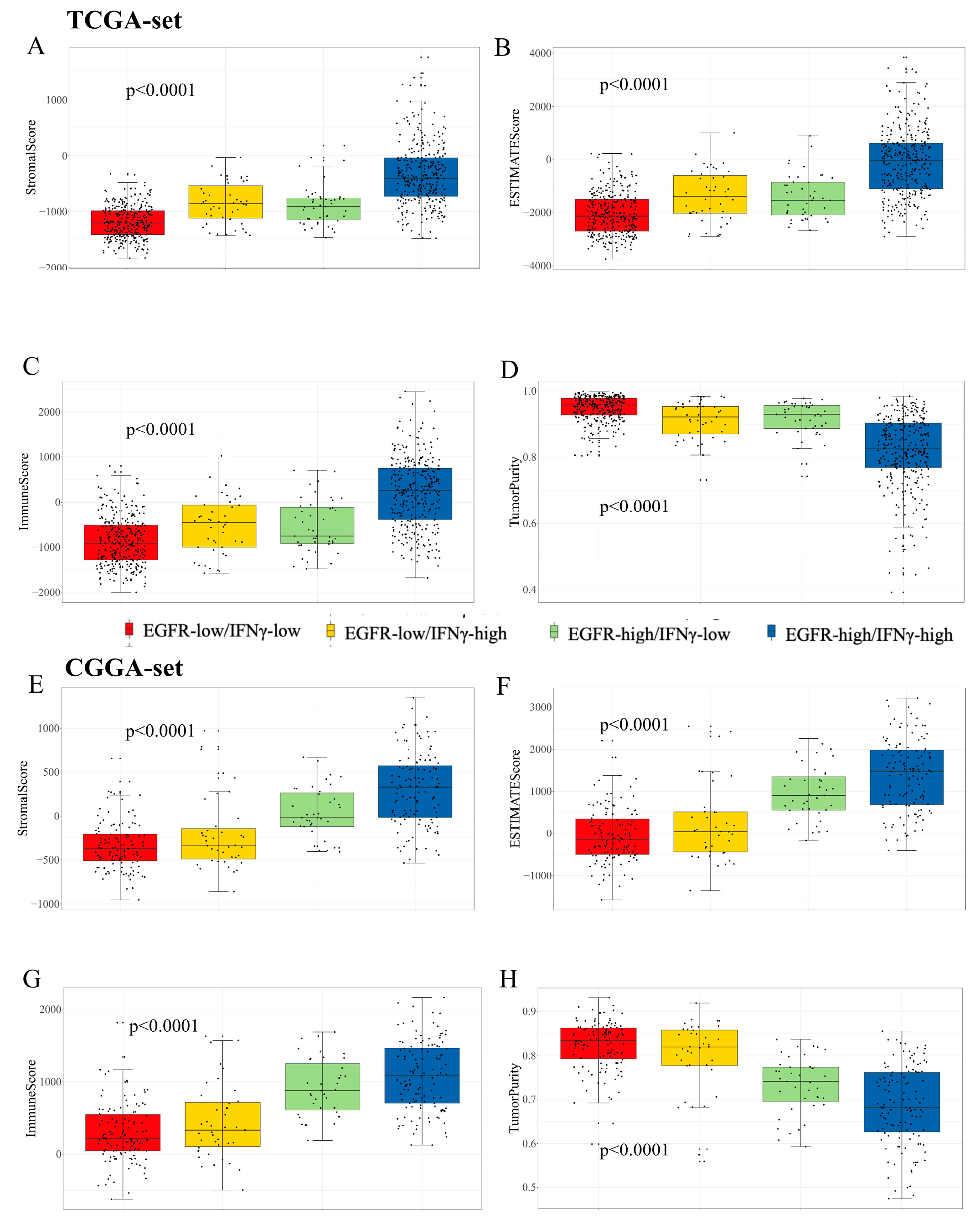
3.2. Clinic Pathological Features Related to the EGFR Pathway-Related Prognostic Gene Signature in Gliomas
3.3. Biological Processes and Signaling Pathway Analysis
3.4. Establishment of an IFN-γ-Related Prognostic Gene Signature
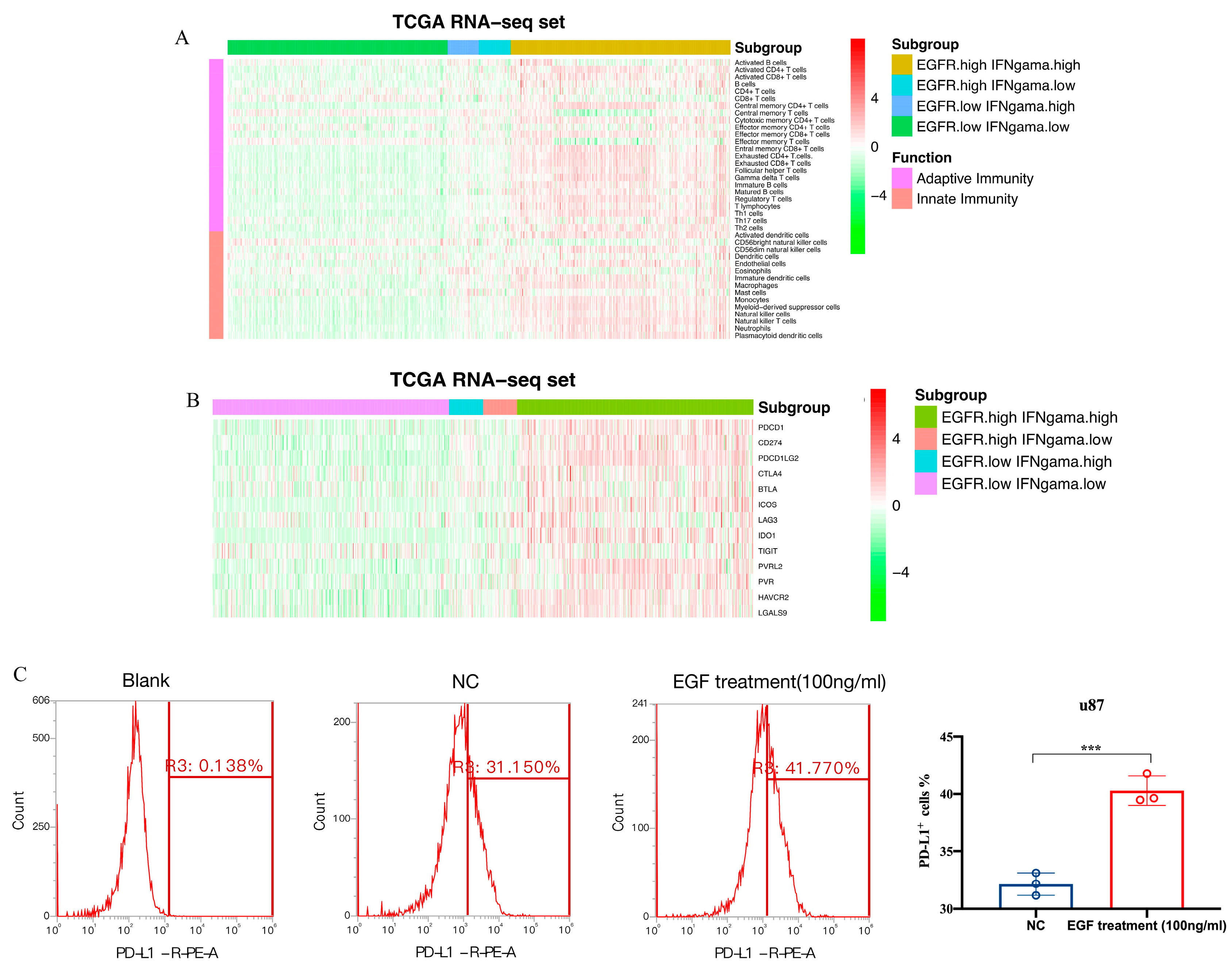
3.5. Immune Cell Infiltration and Inflammatory Profiles Related to the Gene Signature
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ostrom, Q.T.; Cioffi, G.; Waite, K.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2014–2018. Neuro Oncol. 2021, 23, iii1–iii105. [Google Scholar] [CrossRef]
- Yu, M.W.; Quail, D.F. Immunotherapy for Glioblastoma: Current Progress and Challenges. Front. Immunol. 2021, 12, 676301. [Google Scholar] [CrossRef]
- Patel, A.P.; Tirosh, I.; Trombetta, J.J.; Shalek, A.K.; Gillespie, S.M.; Wakimoto, H.; Cahill, D.P.; Nahed, B.V.; Curry, W.T.; Martuza, R.L.; et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 2014, 344, 1396–1401. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A summary. Neuro Oncol. 2021, 23, 1231–1251. [Google Scholar] [CrossRef] [PubMed]
- Jiang, T.; Nam, D.H.; Ram, Z.; Poon, W.S.; Wang, J.; Boldbaatar, D.; Mao, Y.; Ma, W.; Mao, Q.; You, Y.; et al. Clinical practice guidelines for the management of adult diffuse gliomas. Cancer Lett. 2021, 499, 60–72. [Google Scholar] [CrossRef]
- Järvelä, S.; Helin, H.; Haapasalo, J.; Järvelä, T.; Junttila, T.T.; Elenius, K.; Tanner, M.; Haapasalo, H.; Isola, J. Amplification of the epidermal growth factor receptor in astrocytic tumours by chromogenic in situ hybridization: Association with clinicopathological features and patient survival. Neuropathol. Appl. Neurobiol. 2006, 32, 441–450. [Google Scholar] [CrossRef] [PubMed]
- Houillier, C.; Lejeune, J.; Benouaich-Amiel, A.; Laigle-Donadey, F.; Criniere, E.; Mokhtari, K.; Thillet, J.; Delattre, J.Y.; Hoang-Xuan, K.; Sanson, M. Prognostic impact of molecular markers in a series of 220 primary glioblastomas. Cancer 2006, 106, 2218–2223. [Google Scholar] [CrossRef]
- Saadeh, F.S.; Mahfouz, R.; Assi, H.I. EGFR as a clinical marker in glioblastomas and other gliomas. Int. J. Biol. Markers 2018, 33, 22–32. [Google Scholar] [CrossRef]
- Stichel, D.; Ebrahimi, A.; Reuss, D.; Schrimpf, D.; Ono, T.; Shirahata, M.; Reifenberger, G.; Weller, M.; Hanggi, D.; Wick, W.; et al. Distribution of EGFR amplification, combined chromosome 7 gain and chromosome 10 loss, and TERT promoter mutation in brain tumors and their potential for the reclassification of IDHwt astrocytoma to glioblastoma. Acta Neuropathol. 2018, 136, 793–803. [Google Scholar] [CrossRef]
- Oprita, A.; Baloi, S.C.; Staicu, G.A.; Alexandru, O.; Tache, D.E.; Danoiu, S.; Micu, E.S.; Sevastre, A.S. Updated Insights on EGFR Signaling Pathways in Glioma. Int. J. Mol. Sci. 2021, 22, 587. [Google Scholar] [CrossRef]
- Oda, K.; Matsuoka, Y.; Funahashi, A.; Kitano, H. A comprehensive pathway map of epidermal growth factor receptor signaling. Mol. Syst. Biol. 2005, 1, 2005.0010. [Google Scholar] [CrossRef] [PubMed]
- An, Z.; Knobbe-Thomsen, C.B.; Wan, X.; Fan, Q.W.; Reifenberger, G.; Weiss, W.A. EGFR Cooperates with EGFRvIII to Recruit Macrophages in Glioblastoma. Cancer Res. 2018, 78, 6785–6794. [Google Scholar] [CrossRef] [PubMed]
- Litak, J.; Mazurek, M.; Grochowski, C.; Kamieniak, P.; Roliński, J. PD-L1/PD-1 Axis in Glioblastoma Multiforme. Int. J. Mol. Sci. 2019, 20, 5347. [Google Scholar] [CrossRef]
- Barthel, L.; Hadamitzky, M.; Dammann, P.; Schedlowski, M.; Sure, U.; Thakur, B.K.; Hetze, S. Glioma: Molecular signature and crossroads with tumor microenvironment. Cancer Metastasis Rev. 2022, 41, 53–75. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.M.; Xing, B.; Li, J.C.; Cheung, B.K.; Lau, A.S. Interferon-gamma regulation of TNFalpha-induced matrix metalloproteinase 3 expression and migration of human glioma T98G cells. Int. J. Cancer 2007, 121, 1190–1196. [Google Scholar] [CrossRef] [PubMed]
- George, J.; Banik, N.L.; Ray, S.K. Knockdown of hTERT and concurrent treatment with interferon-gamma inhibited proliferation and invasion of human glioblastoma cell lines. Int. J. Biochem. Cell Biol. 2010, 42, 1164–1173. [Google Scholar] [CrossRef]
- Alizadeh, D.; Wong, R.A.; Gholamin, S.; Maker, M.; Aftabizadeh, M.; Yang, X.; Pecoraro, J.R.; Jeppson, J.D.; Wang, D.; Aguilar, B.; et al. IFNγ Is Critical for CAR T Cell-Mediated Myeloid Activation and Induction of Endogenous Immunity. Cancer Discov. 2021, 11, 2248–2265. [Google Scholar] [CrossRef]
- Larson, R.C.; Kann, M.C.; Bailey, S.R.; Haradhvala, N.J.; Llopis, P.M.; Bouffard, A.A.; Scarfó, I.; Leick, M.B.; Grauwet, K.; Berger, T.R.; et al. CAR T cell killing requires the IFNγR pathway in solid but not liquid tumours. Nature 2022, 604, 563–570. [Google Scholar] [CrossRef]
- Cloughesy, T.F.; Mochizuki, A.Y.; Orpilla, J.R.; Hugo, W.; Lee, A.H.; Davidson, T.B.; Wang, A.C.; Ellingson, B.M.; Rytlewski, J.A.; Sanders, C.M.; et al. Neoadjuvant anti-PD-1 immunotherapy promotes a survival benefit with intratumoral and systemic immune responses in recurrent glioblastoma. Nat. Med. 2019, 25, 477–486. [Google Scholar] [CrossRef]
- Qian, J.; Wang, C.; Wang, B.; Yang, J.; Wang, Y.; Luo, F.; Xu, J.; Zhao, C.; Liu, R.; Chu, Y. The IFN-γ/PD-L1 axis between T cells and tumor microenvironment: Hints for glioma anti-PD-1/PD-L1 therapy. J. Neuroinflamm. 2018, 15, 290. [Google Scholar] [CrossRef]
- Burova, E.; Vassilenko, K.; Dorosh, V.; Gonchar, I.; Nikolsky, N. Interferon gamma-dependent transactivation of epidermal growth factor receptor. FEBS Lett. 2007, 581, 1475–1480. [Google Scholar] [CrossRef] [PubMed]
- Boente, M.P.; Berchuck, A.; Rodriguez, G.C.; Davidoff, A.; Whitaker, R.; Xu, F.J.; Marks, J.; Clarke-Pearson, D.L.; Bast, R.C., Jr. The effect of interferon gamma on epidermal growth factor receptor expression in normal and malignant ovarian epithelial cells. Am. J. Obstet. Gynecol. 1992, 167, 1877–1882. [Google Scholar] [CrossRef]
- Chakravarthy, A.; Chen, L.C.; Mehta, D.; Hamburger, A.W. Modulation of epidermal growth factor receptors by gamma interferon in a breast cancer cell line. Anticancer Res. 1991, 11, 347–351. [Google Scholar]
- Safran, M.; Dalah, I.; Alexander, J.; Rosen, N.; Iny Stein, T.; Shmoish, M.; Nativ, N.; Bahir, I.; Doniger, T.; Krug, H.; et al. GeneCards Version 3: The human gene integrator. Database 2010, 2010, baq020. [Google Scholar] [CrossRef] [PubMed]
- Tibshirani, R. The lasso method for variable selection in the Cox model. Stat. Med. 1997, 16, 385–395. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Schubert, M.; Klinger, B.; Klünemann, M.; Sieber, A.; Uhlitz, F.; Sauer, S.; Garnett, M.J.; Blüthgen, N.; Saez-Rodriguez, J. Perturbation-response genes reveal signaling footprints in cancer gene expression. Nat. Commun. 2018, 9, 20. [Google Scholar] [CrossRef] [PubMed]
- Hänzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013, 14, 7. [Google Scholar] [CrossRef] [PubMed]
- Yoshihara, K.; Shahmoradgoli, M.; Martínez, E.; Vegesna, R.; Kim, H.; Torres-Garcia, W.; Treviño, V.; Shen, H.; Laird, P.W.; Levine, D.A.; et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013, 4, 2612. [Google Scholar] [CrossRef] [PubMed]
- Therneau, T.M. A Package for Survival Analysis in R. 2020. Available online: https://CRAN.R-project.org/package=survival (accessed on 1 May 2023).
- Zhang, C.; Cheng, W.; Ren, X.; Wang, Z.; Liu, X.; Li, G.; Han, S.; Jiang, T.; Wu, A. Tumor Purity as an Underlying Key Factor in Glioma. Clin. Cancer Res. 2017, 23, 6279–6291. [Google Scholar] [CrossRef]
- Feng, E.; Liang, T.; Wang, X.; Du, J.; Tang, K.; Wang, X.; Wang, F.; You, G. Correlation of alteration of HLA-F expression and clinical characterization in 593 brain glioma samples. J. Neuroinflamm. 2019, 16, 33. [Google Scholar] [CrossRef]
- Zhao, B.; Wang, Y.; Wang, Y.; Dai, C.; Wang, Y.; Ma, W. Investigation of Genetic Determinants of Glioma Immune Phenotype by Integrative Immunogenomic Scale Analysis. Front. Immunol. 2021, 12, 557994. [Google Scholar] [CrossRef]
- Qian, Y.; Ding, P.; Xu, J.; Nie, X.; Lu, B. CCL2 activates AKT signaling to promote glycolysis and chemoresistance in glioma cells. Cell Biol. Int. 2022, 46, 819–828. [Google Scholar] [CrossRef]
- Gurgis, F.M.; Yeung, Y.T.; Tang, M.X.; Heng, B.; Buckland, M.; Ammit, A.J.; Haapasalo, J.; Haapasalo, H.; Guillemin, G.J.; Grewal, T.; et al. The p38-MK2-HuR pathway potentiates EGFRvIII-IL-1beta-driven IL-6 secretion in glioblastoma cells. Oncogene 2015, 34, 2934–2942. [Google Scholar] [CrossRef]
- Miao, R.; Huang, D.; Zhao, K.; Li, Y.; Zhang, X.; Cheng, Y.; Guo, N. VAV3 regulates glioblastoma cell proliferation, migration, invasion and cancer stem-like cell self-renewal. Mol. Med. Rep. 2023, 27, 94. [Google Scholar] [CrossRef]
- Nicolas, S.; Abdellatef, S.; Haddad, M.A.; Fakhoury, I.; El-Sibai, M. Hypoxia and EGF Stimulation Regulate VEGF Expression in Human Glioblastoma Multiforme (GBM) Cells by Differential Regulation of the PI3K/Rho-GTPase and MAPK Pathways. Cells 2019, 8, 1397. [Google Scholar] [CrossRef]
- Zhao, L.; Li, X.; Su, J.; Wang Gong, F.; Lu, J.; Wei, Y. STAT1 determines aggressiveness of glioblastoma both in vivo and in vitro through wnt/beta-catenin signalling pathway. Cell Biochem. Funct. 2020, 38, 630–641. [Google Scholar] [CrossRef]
- Jung, J.U.; Ravi, S.; Lee, D.W.; McFadden, K.; Kamradt, M.L.; Toussaint, L.G.; Sitcheran, R. NIK/MAP3K14 Regulates Mitochondrial Dynamics and Trafficking to Promote Cell Invasion. Curr. Biol. 2016, 26, 3288–3302. [Google Scholar] [CrossRef] [PubMed]
- Xing, S.; Chen, S.; Yang, X.; Huang, W. Role of MAPK activity in PD-L1 expression in hepatocellular carcinoma cells. J. Buon 2020, 25, 1875–1882. [Google Scholar]
- Garcia-Diaz, A.; Shin, D.S.; Moreno, B.H.; Saco, J.; Escuin-Ordinas, H.; Rodriguez, G.A.; Zaretsky, J.M.; Sun, L.; Hugo, W.; Wang, X.; et al. Interferon Receptor Signaling Pathways Regulating PD-L1 and PD-L2 Expression. Cell Rep. 2017, 19, 1189–1201. [Google Scholar] [CrossRef]
- Cheng, C.C.; Lin, H.C.; Tsai, K.J.; Chiang, Y.W.; Lim, K.H.; Chen, C.G.; Su, Y.W.; Peng, C.L.; Ho, A.S.; Huang, L.; et al. Epidermal growth factor induces STAT1 expression to exacerbate the IFNr-mediated PD-L1 axis in epidermal growth factor receptor-positive cancers. Mol. Carcinog. 2018, 57, 1588–1598. [Google Scholar] [CrossRef] [PubMed]
- Sidorov, M.; Dighe, P.; Woo, R.W.L.; Rodriguez-Brotons, A.; Chen, M.; Ice, R.J.; Vaquero, E.; Jian, D.; Desprez, P.Y.; Nosrati, M.; et al. Dual Targeting of EGFR and MTOR Pathways Inhibits Glioblastoma Growth by Modulating the Tumor Microenvironment. Cells 2023, 12, 547. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, X.; Liang, T.; Ge, Y.; Wang, Y.; Ma, W. The Crosstalk between the EGFR and IFN-γ Pathways and Synergistic Roles in Survival Prediction and Immune Escape in Gliomas. Brain Sci. 2023, 13, 1349. https://doi.org/10.3390/brainsci13091349
Zhou X, Liang T, Ge Y, Wang Y, Ma W. The Crosstalk between the EGFR and IFN-γ Pathways and Synergistic Roles in Survival Prediction and Immune Escape in Gliomas. Brain Sciences. 2023; 13(9):1349. https://doi.org/10.3390/brainsci13091349
Chicago/Turabian StyleZhou, Xingang, Tingyu Liang, Yulu Ge, Yu Wang, and Wenbin Ma. 2023. "The Crosstalk between the EGFR and IFN-γ Pathways and Synergistic Roles in Survival Prediction and Immune Escape in Gliomas" Brain Sciences 13, no. 9: 1349. https://doi.org/10.3390/brainsci13091349
APA StyleZhou, X., Liang, T., Ge, Y., Wang, Y., & Ma, W. (2023). The Crosstalk between the EGFR and IFN-γ Pathways and Synergistic Roles in Survival Prediction and Immune Escape in Gliomas. Brain Sciences, 13(9), 1349. https://doi.org/10.3390/brainsci13091349






