Interleukin-11/IL-11 Receptor Promotes Glioblastoma Cell Proliferation, Epithelial–Mesenchymal Transition, and Invasion
Abstract
:1. Introduction
2. Materials and Methods
3. Results
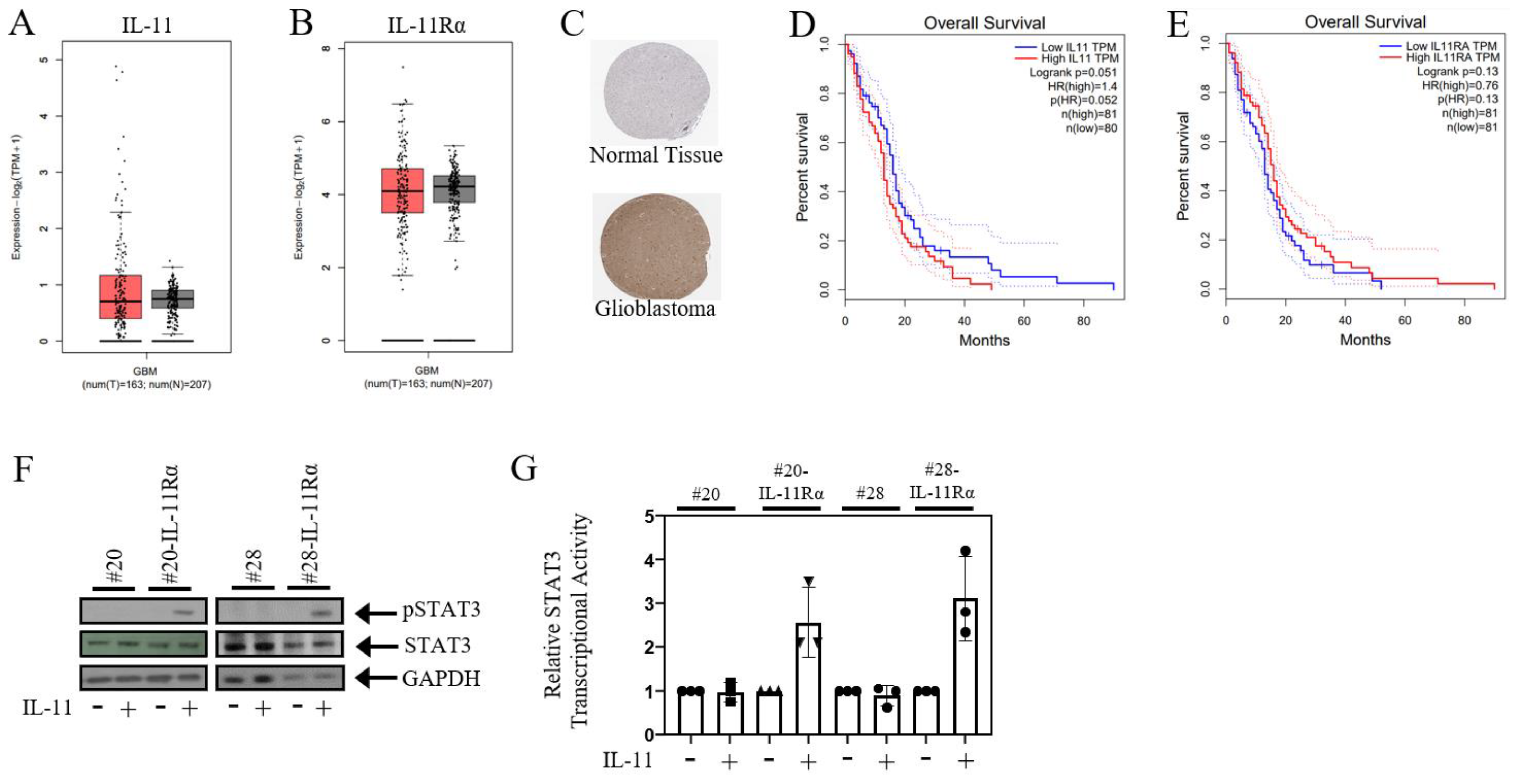
3.1. IL-11Rα Promotes a Pro-Tumourigenic Protein Signature
3.2. IL-11Rα Promotes Glioblastoma Cell Proliferation
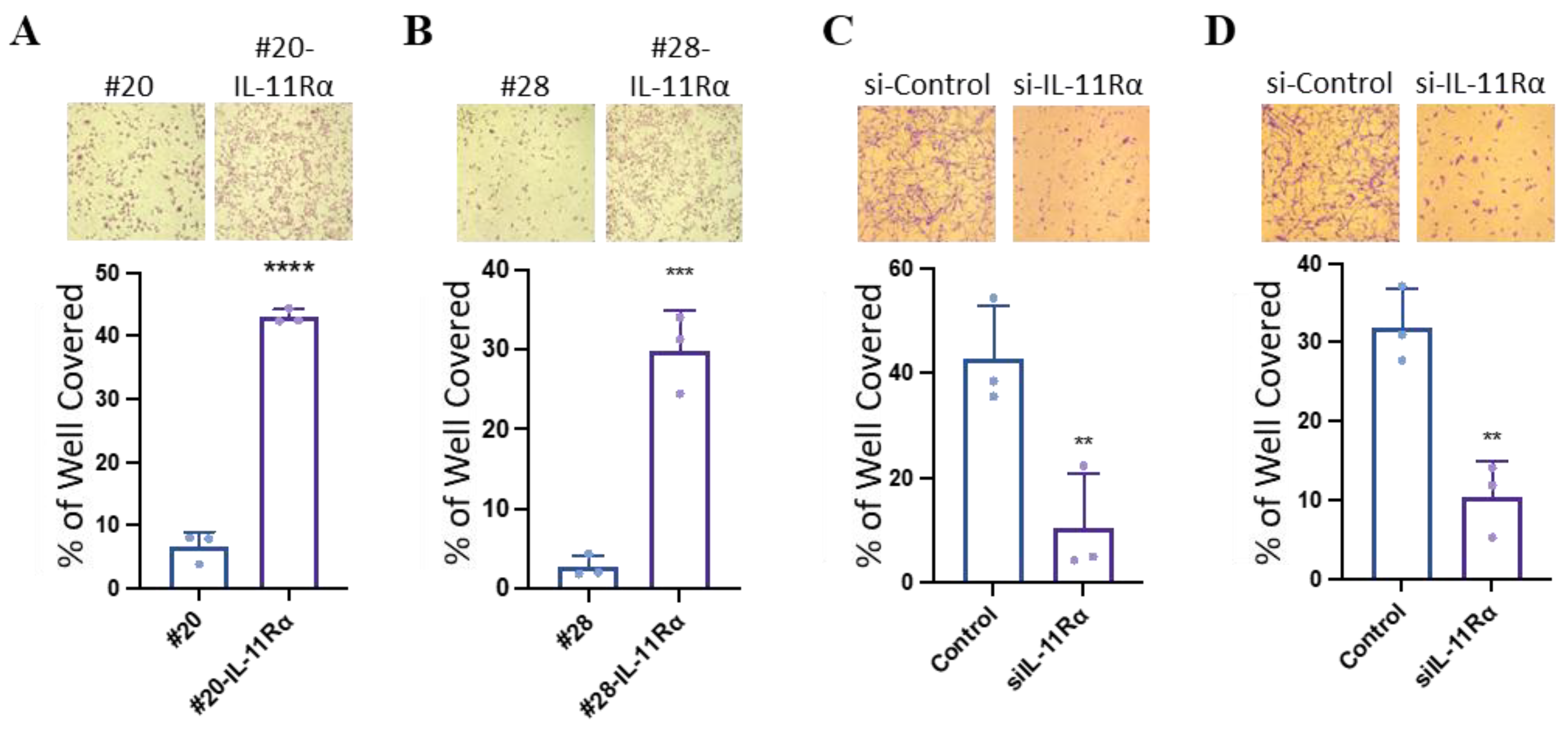
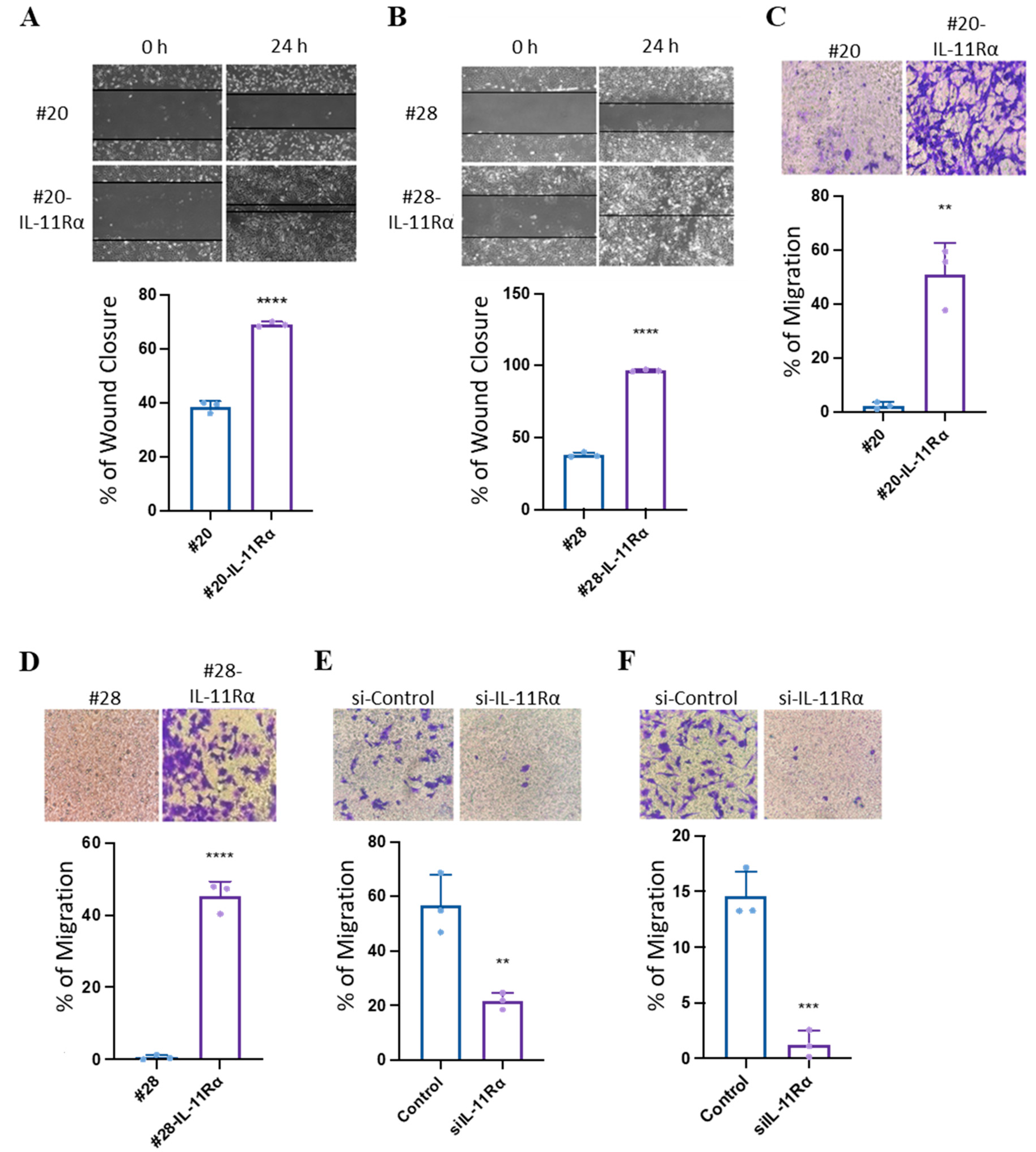
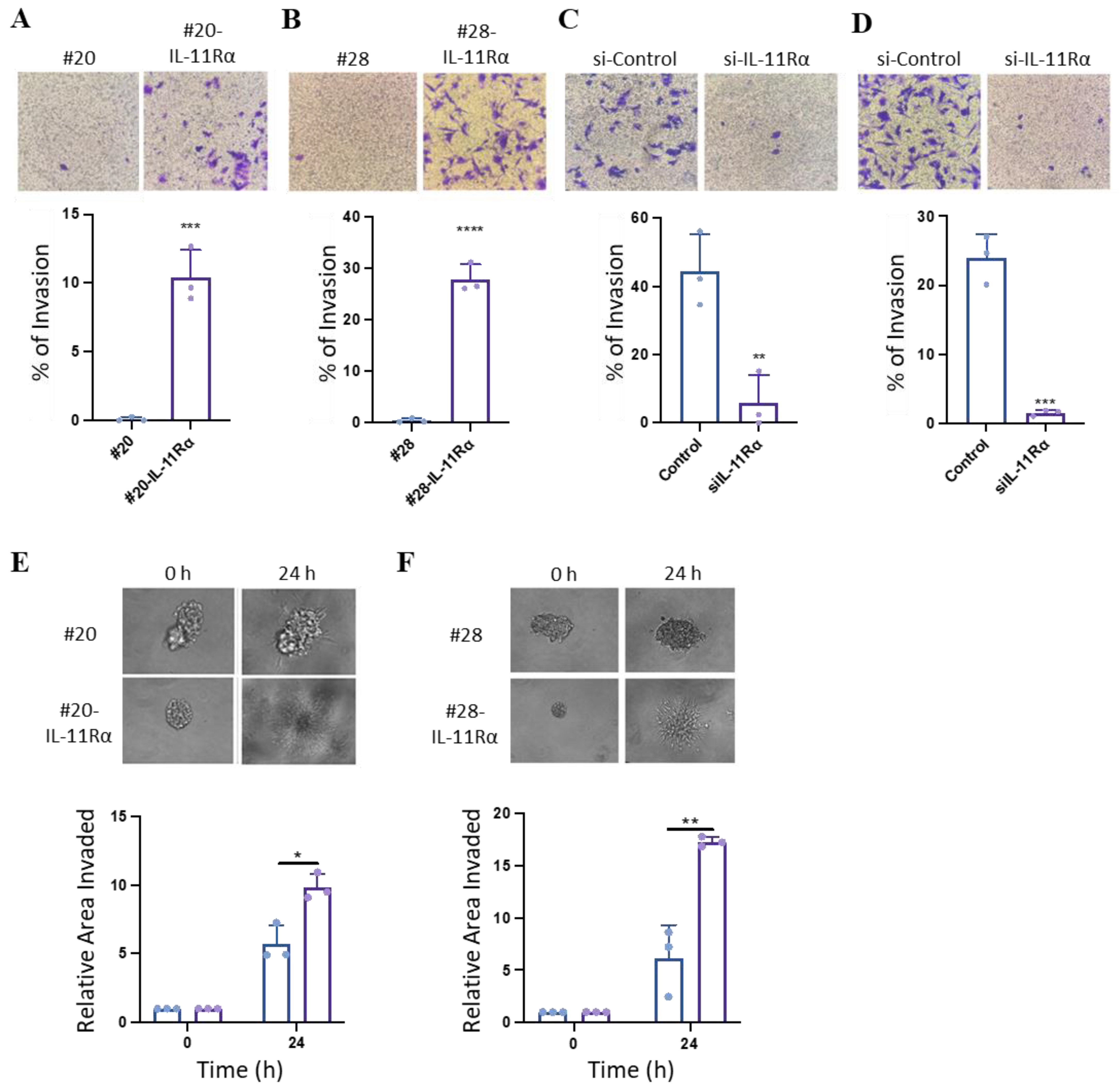
3.3. IL-11Rα Promotes Glioblastoma Cell Migration and Invasion
3.4. IL-11Rα Promotes an EMT-like Phenotype
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, C.; Li, Y.; Chen, H.; Huang, K.; Liu, X.; Qiu, M.; Liu, Y.; Yang, Y.; Yang, J. CYP4X1 Inhibition by Flavonoid CH625 Normalizes Glioma Vasculature through Reprogramming TAMs via CB2 and EGFR-STAT3 Axis. J. Pharmacol. Exp. Ther. 2018, 365, 72–83. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Clark, P.A.; Kuo, J.S.; Kalejta, R.F. Human Cytomegalovirus-Infected Glioblastoma Cells Display Stem Cell-Like Phenotypes. mSphere 2017, 2, e00137-17. [Google Scholar] [CrossRef] [PubMed]
- Mohme, M.; Schliffke, S.; Maire, C.L.; Runger, A.; Glau, L.; Mende, K.C.; Matschke, J.; Gehbauer, C.; Akyuz, N.; Zapf, S.; et al. Immunophenotyping of Newly Diagnosed and Recurrent Glioblastoma Defines Distinct Immune Exhaustion Profiles in Peripheral and Tumor-infiltrating Lymphocytes. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2018, 24, 4187–4200. [Google Scholar] [CrossRef] [PubMed]
- Nakada, M.; Furuta, T.; Hayashi, Y.; Minamoto, T.; Hamada, J. The strategy for enhancing temozolomide against malignant glioma. Front. Oncol. 2012, 2, 98. [Google Scholar] [CrossRef] [PubMed]
- McFarland, B.C.; Hong, S.W.; Rajbhandari, R.; Twitty, G.B., Jr.; Gray, G.K.; Yu, H.; Benveniste, E.N.; Nozell, S.E. NF-kappaB-induced IL-6 ensures STAT3 activation and tumor aggressiveness in glioblastoma. PLoS ONE 2013, 8, e78728. [Google Scholar] [CrossRef]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A summary. Neuro-Oncol. 2021, 23, 1231–1251. [Google Scholar] [CrossRef]
- Nørøxe, D.S.; Poulsen, H.S.; Lassen, U. Hallmarks of glioblastoma: A systematic review. ESMO Open 2016, 1, e000144. [Google Scholar] [CrossRef]
- Alexander, B.M.; Cloughesy, T.F. Adult Glioblastoma. J. Clin. Oncol. 2017, 35, 2402–2409. [Google Scholar] [CrossRef]
- Li, J.; Liang, R.; Song, C.; Xiang, Y.; Liu, Y. Prognostic significance of epidermal growth factor receptor expression in glioma patients. Onco Targets Ther. 2018, 11, 731–742. [Google Scholar] [CrossRef]
- Soomro, S.H.; Ting, L.R.; Qing, Y.Y.; Ren, M. Molecular biology of glioblastoma: Classification and mutational locations. J. Pak. Med. Assoc. 2017, 67, 1410–1414. [Google Scholar]
- Leibetseder, A.; Ackerl, M.; Flechl, B.; Wohrer, A.; Widhalm, G.; Dieckmann, K.; Kreinecker, S.S.; Pichler, J.; Hainfellner, J.; Preusser, M.; et al. Outcome and molecular characteristics of adolescent and young adult patients with newly diagnosed primary glioblastoma: A study of the Society of Austrian Neurooncology (SANO). Neuro Oncol. 2013, 15, 112–121. [Google Scholar] [CrossRef] [PubMed]
- Alphandéry, E. Glioblastoma Treatments: An Account of Recent Industrial Developments. Front. Pharmacol. 2018, 9, 879. [Google Scholar] [CrossRef] [PubMed]
- Perrin, S.L.; Samuel, M.S.; Koszyca, B.; Brown, M.P.; Ebert, L.M.; Oksdath, M.; Gomez, G.A. Glioblastoma heterogeneity and the tumour microenvironment: Implications for preclinical research and development of new treatments. Biochem. Soc. Trans. 2019, 47, 625–638. [Google Scholar] [CrossRef] [PubMed]
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef] [PubMed]
- Jensen, K.V.; Cseh, O.; Aman, A.; Weiss, S.; Luchman, H.A. The JAK2/STAT3 inhibitor pacritinib effectively inhibits patient-derived GBM brain tumor initiating cells in vitro and when used in combination with temozolomide increases survival in an orthotopic xenograft model. PLoS ONE 2017, 12, e0189670. [Google Scholar] [CrossRef] [PubMed]
- Cattaneo, M.G.; Vanetti, C.; Samarani, M.; Aureli, M.; Bassi, R.; Sonnino, S.; Giussani, P. Cross-talk between sphingosine-1-phosphate and EGFR signaling pathways enhances human glioblastoma cell invasiveness. FEBS Lett. 2018, 592, 949–961. [Google Scholar] [CrossRef]
- Lamano, J.B.; Lamano, J.B.; Li, Y.D.; DiDomenico, J.D.; Choy, W.; Veliceasa, D.; Oyon, D.E.; Fakurnejad, S.; Ampie, L.; Kesavabhotla, K.; et al. Glioblastoma-Derived IL-6 Induces Immunosuppressive Peripheral Myeloid Cell PD-L1 and Promotes Tumor Growth. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2019, 25, 3643–3657. [Google Scholar] [CrossRef]
- Combs, S.E.; Schulz-Ertner, D.; Roth, W.; Herold-Mende, C.; Debus, J.; Weber, K.J. In vitro responsiveness of glioma cell lines to multimodality treatment with radiotherapy, temozolomide, and epidermal growth factor receptor inhibition with cetuximab. Int. J. Radiat. Oncol. Biol. Phys. 2007, 68, 873–882. [Google Scholar] [CrossRef]
- Sun, L.; Yu, S.; Xu, H.; Zheng, Y.; Lin, J.; Wu, M.; Wang, J.; Wang, A.; Lan, Q.; Furnari, F.; et al. FHL2 interacts with EGFR to promote glioblastoma growth. Oncogene 2018, 37, 1386–1398. [Google Scholar] [CrossRef]
- da Hora, C.C.; Schweiger, M.W.; Wurdinger, T.; Tannous, B.A. Patient-Derived Glioma Models: From Patients to Dish to Animals. Cells 2019, 8, 1177. [Google Scholar] [CrossRef]
- Pearson, J.R.D.; Regad, T. Targeting cellular pathways in glioblastoma multiforme. Signal Transduct. Target. Ther. 2017, 2, 17040. [Google Scholar] [CrossRef] [PubMed]
- Czapski, B.; Baluszek, S.; Herold-Mende, C.; Kaminska, B. Clinical and immunological correlates of long term survival in glioblastoma. Contemp. Oncol. 2018, 22, 81–85. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Fried, A.; Hussaini, R.; White, R.; Baidoo, J.; Yalamanchi, S.; Banerjee, P. Phytosomal curcumin causes natural killer cell-dependent repolarization of glioblastoma (GBM) tumor-associated microglia/macrophages and elimination of GBM and GBM stem cells. J. Exp. Clin. Cancer Res. CR 2018, 37, 168. [Google Scholar] [CrossRef] [PubMed]
- Mofatteh, M.; Mashayekhi, M.S.; Arfaie, S.; Chen, Y.; Malhotra, A.K.; Alvi, M.A.; Sader, N.; Antonick, V.; Fatehi Hassanabad, M.; Mansouri, A.; et al. Suicidal ideation and attempts in brain tumor patients and survivors: A systematic review. Neurooncol. Adv. 2023, 5, vdad058. [Google Scholar] [CrossRef]
- Lokau, J.; Agthe, M.; Flynn, C.M.; Garbers, C. Proteolytic control of Interleukin-11 and Interleukin-6 biology. Biochim. Biophys. Acta Mol. Cell Res. 2017, 1864, 2105–2117. [Google Scholar] [CrossRef]
- Metcalfe, R.D.; Aizel, K.; Zlatic, C.O.; Nguyen, P.M.; Morton, C.J.; Lio, D.S.; Cheng, H.C.; Dobson, R.C.J.; Parker, M.W.; Gooley, P.R.; et al. The structure of the extracellular domains of human interleukin 11alpha receptor reveals mechanisms of cytokine engagement. J. Biol. Chem. 2020, 295, 8285–8301. [Google Scholar] [CrossRef]
- Wei, J.; Ma, L.; Lai, Y.H.; Zhang, R.; Li, H.; Li, C.; Lin, J. Bazedoxifene as a novel GP130 inhibitor for Colon Cancer therapy. J. Exp. Clin. Cancer Res. 2019, 38, 63. [Google Scholar] [CrossRef]
- Dams-Kozlowska, H.; Gryska, K.; Kwiatkowska-Borowczyk, E.; Izycki, D.; Rose-John, S.; Mackiewicz, A. A designer hyper interleukin 11 (H11) is a biologically active cytokine. BMC Biotechnol. 2012, 12, 8. [Google Scholar] [CrossRef]
- Putoczki, T.L.; Thiem, S.; Loving, A.; Busuttil, R.A.; Wilson, N.J.; Ziegler, P.K.; Nguyen, P.M.; Preaudet, A.; Farid, R.; Edwards, K.M.; et al. Interleukin-11 is the dominant IL-6 family cytokine during gastrointestinal tumorigenesis and can be targeted therapeutically. Cancer Cell 2013, 24, 257–271. [Google Scholar] [CrossRef]
- Wang, X.; Che, X.; Liu, C.; Fan, Y.; Bai, M.; Hou, K.; Shi, X.; Zhang, X.; Liu, B.; Zheng, C.; et al. Cancer-associated fibroblasts-stimulated interleukin-11 promotes metastasis of gastric cancer cells mediated by upregulation of MUC1. Exp. Cell Res. 2018, 368, 184–193. [Google Scholar] [CrossRef]
- Ernst, M.; Putoczki, T.L. Molecular pathways: IL11 as a tumor-promoting cytokine-translational implications for cancers. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2014, 20, 5579–5588. [Google Scholar] [CrossRef] [PubMed]
- Johnstone, C.N.; Chand, A.; Putoczki, T.L.; Ernst, M. Emerging roles for IL-11 signaling in cancer development and progression: Focus on breast cancer. Cytokine Growth Factor. Rev. 2015, 26, 489–498. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Cao, Y.; Xiao, H.; Li, C.; Lin, J. Bazedoxifene as a Novel GP130 Inhibitor for Pancreatic Cancer Therapy. Mol. Cancer Ther. 2016, 15, 2609–2619. [Google Scholar] [CrossRef] [PubMed]
- Howlett, M.; Menheniott, T.R.; Judd, L.M.; Giraud, A.S. Cytokine signalling via gp130 in gastric cancer. Biochim. Biophys. Acta 2009, 1793, 1623–1633. [Google Scholar] [CrossRef]
- Nguyen, P.M.; Abdirahman, S.M.; Putoczki, T.L. Emerging roles for Interleukin-11 in disease. Growth Factors 2019, 37, 1–11. [Google Scholar] [CrossRef]
- Stuart, S.F.; Bezawork-Geleta, A.; Areeb, Z.; Gomez, J.; Tsui, V.; Zulkifli, A.; Paradiso, L.; Jones, J.; Nguyen, H.P.T.; Putoczki, T.L.; et al. The Interleukin-11/IL-11 Receptor Promotes Glioblastoma Survival and Invasion under Glucose-Starved Conditions through Enhanced Glutaminolysis. Int. J. Mol. Sci. 2023, 24, 3356. [Google Scholar] [CrossRef]
- Murphy, G.M., Jr.; Bitting, L.; Majewska, A.; Schmidt, K.; Song, Y.; Wood, C.R. Expression of interleukin-11 and its encoding mRNA by glioblastoma cells. Neurosci. Lett. 1995, 196, 153–156. [Google Scholar] [CrossRef]
- Li, J.; Kaneda, M.M.; Ma, J.; Li, M.; Shepard, R.M.; Patel, K.; Koga, T.; Sarver, A.; Furnari, F.; Xu, B.; et al. PI3Kγ inhibition suppresses microglia/TAM accumulation in glioblastoma microenvironment to promote exceptional temozolomide response. Proc. Natl. Acad. Sci. USA 2021, 118, e2009290118. [Google Scholar] [CrossRef]
- Guzman, C.; Bagga, M.; Kaur, A.; Westermarck, J.; Abankwa, D. ColonyArea: An ImageJ plugin to automatically quantify colony formation in clonogenic assays. PLoS ONE 2014, 9, e92444. [Google Scholar] [CrossRef]
- Tan, F.H.; Putoczki, T.L.; Lou, J.; Hinde, E.; Hollande, F.; Giraud, J.; Stylli, S.S.; Paradiso, L.; Zhu, H.J.; Sieber, O.M.; et al. Ponatinib Inhibits Multiple Signaling Pathways Involved in STAT3 Signaling and Attenuates Colorectal Tumor Growth. Cancers 2018, 10, 526. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Grivennikov, S.I. IL-11: A prominent pro-tumorigenic member of the IL-6 family. Cancer Cell 2013, 24, 145–147. [Google Scholar] [CrossRef]
- Zhao, M.; Liu, Y.; Liu, R.; Qi, J.; Hou, Y.; Chang, J.; Ren, L. Upregulation of IL-11, an IL-6 Family Cytokine, Promotes Tumor Progression and Correlates with Poor Prognosis in Non-Small Cell Lung Cancer. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2018, 45, 2213–2224. [Google Scholar] [CrossRef]
- Ren, C.; Chen, Y.; Han, C.; Fu, D.; Chen, H. Plasma interleukin-11 (IL-11) levels have diagnostic and prognostic roles in patients with pancreatic cancer. Tumour Biol. 2014, 35, 11467–11472. [Google Scholar] [CrossRef]
- Hanavadi, S.; Martin, T.A.; Watkins, G.; Mansel, R.E.; Jiang, W.G. Expression of interleukin 11 and its receptor and their prognostic value in human breast cancer. Ann. Surg. Oncol. 2006, 13, 802–808. [Google Scholar] [CrossRef]
- Pan, D.; Xu, L.; Liu, H.; Zhang, W.; Liu, W.; Liu, Y.; Fu, Q.; Xu, J. High expression of interleukin-11 is an independent indicator of poor prognosis in clear-cell renal cell carcinoma. Cancer Sci. 2015, 106, 592–597. [Google Scholar] [CrossRef]
- Lewis, V.O.; Devarajan, E.; Cardó-Vila, M.; Thomas, D.G.; Kleinerman, E.S.; Marchiò, S.; Sidman, R.L.; Pasqualini, R.; Arap, W. BMTP-11 is active in preclinical models of human osteosarcoma and a candidate targeted drug for clinical translation. Proc. Natl. Acad. Sci. USA 2017, 114, 8065–8070. [Google Scholar] [CrossRef] [PubMed]
- Lewis, V.O.; Ozawa, M.G.; Deavers, M.T.; Wang, G.; Shintani, T.; Arap, W.; Pasqualini, R. The interleukin-11 receptor alpha as a candidate ligand-directed target in osteosarcoma: Consistent data from cell lines, orthotopic models, and human tumor samples. Cancer Res. 2009, 69, 1995–1999. [Google Scholar] [CrossRef] [PubMed]
- Lokau, J.; Schoeder, V.; Garbers, C. Die Rolle von Interleukin-11 beim Osteosarkom. Der Pathol. 2020, 41, 163–167. [Google Scholar] [CrossRef]
- Lauko, A.; Bayik, D.; Lathia, J.D. IL-11 drives postsurgical hepatocellular carcinoma recurrence. EBioMedicine 2019, 47, 18–19. [Google Scholar] [CrossRef]
- Yang, W.; Zhang, S.; Ou, T.; Jiang, H.; Jia, D.; Qi, Z.; Zou, Y.; Qian, J.; Sun, A.; Ge, J. Interleukin-11 regulates the fate of adipose-derived mesenchymal stem cells via STAT3 signalling pathways. Cell Prolif. 2020, 53, e12771. [Google Scholar] [CrossRef]
- Lay, V.; Yap, J.; Sonderegger, S.; Dimitriadis, E. Interleukin 11 regulates endometrial cancer cell adhesion and migration via STAT3. Int. J. Oncol. 2012, 41, 759–764. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, K.; Karin, M. IL-6 and related cytokines as the critical lynchpins between inflammation and cancer. Semin. Immunol. 2014, 26, 54–74. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.H. Inhibition of the Interleukin-11-STAT3 Axis Attenuates Hypoxia-Induced Migration and Invasion in MDA-MB-231 Breast Cancer Cells. Korean J. Physiol. Pharmacol. 2014, 18, 391–396. [Google Scholar] [CrossRef]
- Yamazumi, K.; Nakayama, T.; Kusaba, T.; Wen, C.Y.; Yoshizaki, A.; Yakata, Y.; Nagayasu, T.; Sekine, I. Expression of interleukin-11 and interleukin-11 receptor alpha in human colorectal adenocarcinoma; immunohistochemical analyses and correlation with clinicopathological factors. World J. Gastroenterol. 2006, 12, 317–321. [Google Scholar] [CrossRef] [PubMed]
- Pan, D.; Xu, L.; Liu, H.; Zhang, W.; Zhu, Y.; Xu, J.; Gu, J. Interleukin-11 receptor predicts post-operative clinical outcome in patients with early-stage clear-cell renal cell carcinoma. Jpn. J. Clin. Oncol. 2015, 45, 202–209. [Google Scholar] [CrossRef]
- Briukhovetska, D.; Dörr, J.; Endres, S.; Libby, P.; Dinarello, C.A.; Kobold, S. Interleukins in cancer: From biology to therapy. Nat. Rev. Cancer 2021, 21, 481–499. [Google Scholar] [CrossRef]
- Zhang, X.; Putoczki, T.; Markovic-Plese, S. IL-11 in multiple sclerosis. Oncotarget 2015, 6, 32297–32298. [Google Scholar] [CrossRef]
- Li, T.M.; Wu, C.M.; Huang, H.C.; Chou, P.C.; Fong, Y.C.; Tang, C.H. Interleukin-11 increases cell motility and up-regulates intercellular adhesion molecule-1 expression in human chondrosarcoma cells. J. Cell Biochem. 2012, 113, 3353–3362. [Google Scholar] [CrossRef]
- Karjalainen, K.; Jaalouk, D.E.; Bueso-Ramos, C.; Bover, L.; Sun, Y.; Kuniyasu, A.; Driessen, W.H.; Cardó-Vila, M.; Rietz, C.; Zurita, A.J.; et al. Targeting IL11 Receptor in Leukemia and Lymphoma: A Functional Ligand-Directed Study and Hematopathology Analysis of Patient-Derived Specimens. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2015, 21, 3041–3051. [Google Scholar] [CrossRef]
- Lewis, V.O. IL-11Rα: A Novel Target for the Treatment of Osteosarcoma. In Current Advances in Osteosarcoma; Kleinerman, M.D.E.S., Ed.; Springer International Publishing: Cham, Swizterland, 2014; pp. 285–289. [Google Scholar] [CrossRef]
- Liu, T.; Ma, Q.; Zhang, Y.; Ke, S.; Yan, K.; Chen, X.; Wen, Y.; Fan, Q.; Qiu, X. Interleukin-11 receptor α is overexpressed in human osteosarcoma, and near-infrared-labeled IL-11Rα imaging agent could detect osteosarcoma in mouse tumor xenografts. Tumour Biol. 2015, 36, 2369–2375. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stuart, S.F.; Curpen, P.; Gomes, A.J.; Lan, M.C.; Nie, S.; Williamson, N.A.; Kannourakis, G.; Morokoff, A.P.; Achuthan, A.A.; Luwor, R.B. Interleukin-11/IL-11 Receptor Promotes Glioblastoma Cell Proliferation, Epithelial–Mesenchymal Transition, and Invasion. Brain Sci. 2024, 14, 89. https://doi.org/10.3390/brainsci14010089
Stuart SF, Curpen P, Gomes AJ, Lan MC, Nie S, Williamson NA, Kannourakis G, Morokoff AP, Achuthan AA, Luwor RB. Interleukin-11/IL-11 Receptor Promotes Glioblastoma Cell Proliferation, Epithelial–Mesenchymal Transition, and Invasion. Brain Sciences. 2024; 14(1):89. https://doi.org/10.3390/brainsci14010089
Chicago/Turabian StyleStuart, Sarah F., Peter Curpen, Adele J. Gomes, Michelle C. Lan, Shuai Nie, Nicholas A. Williamson, George Kannourakis, Andrew P. Morokoff, Adrian A. Achuthan, and Rodney B. Luwor. 2024. "Interleukin-11/IL-11 Receptor Promotes Glioblastoma Cell Proliferation, Epithelial–Mesenchymal Transition, and Invasion" Brain Sciences 14, no. 1: 89. https://doi.org/10.3390/brainsci14010089
APA StyleStuart, S. F., Curpen, P., Gomes, A. J., Lan, M. C., Nie, S., Williamson, N. A., Kannourakis, G., Morokoff, A. P., Achuthan, A. A., & Luwor, R. B. (2024). Interleukin-11/IL-11 Receptor Promotes Glioblastoma Cell Proliferation, Epithelial–Mesenchymal Transition, and Invasion. Brain Sciences, 14(1), 89. https://doi.org/10.3390/brainsci14010089








