Developmental Dyslexia: Insights from EEG-Based Findings and Molecular Signatures—A Pilot Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Participants
2.2. DASS21 Scale
2.3. Peripheral Blood Sample Collection, DNA and Total RNA Extraction
2.3.1. Peripheral Blood Sample Collection
2.3.2. DNA Extraction
2.3.3. Total RNA Extraction
2.4. Stress-Associated SNV Detection
2.5. Evaluation of mRNA Expression Levels of Stress-Related Genes
2.6. NR3C1 Gene Methylation Profile
2.6.1. Bioinformatic Analysis
2.6.2. Methylation Analysis
2.7. Mitochondrial DNA Copy Number (mtDNAcn) Estimation in Blood
2.8. EEG Recordings
2.8.1. EEG Data Acquisition
2.8.2. Preprocessing
2.8.3. Feature Extraction
2.9. Gene Network Analysis
2.10. Statistical Analysis
3. Results
3.1. Demographic Data, DASS21 Scores and SNVs
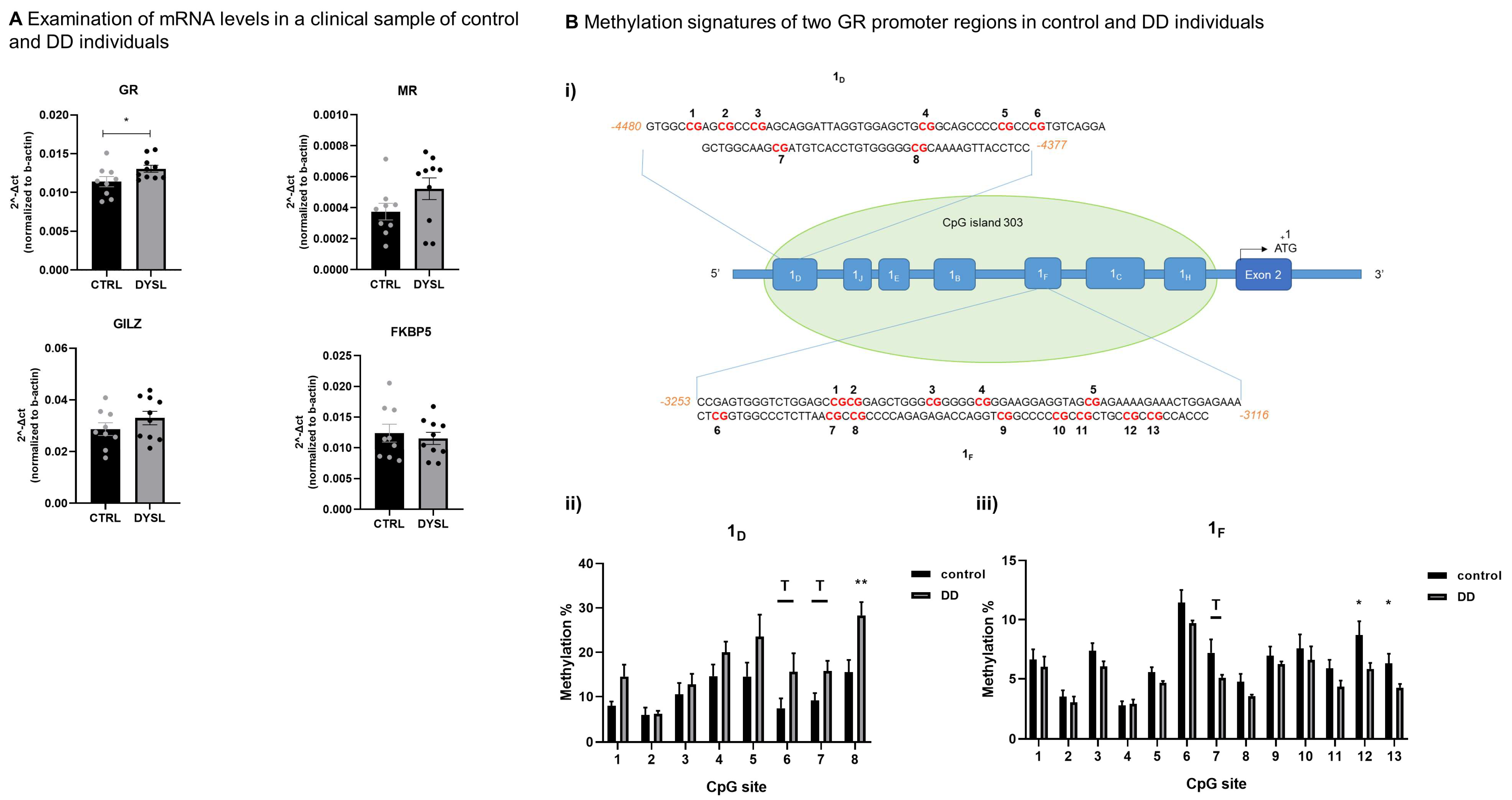
3.2. Altered mRNA and Methylation Levels in NR3C1 Gene in DD
3.3. Correlation of Stress-Related Molecular Changes and Behavioral Characteristics
3.4. Correlation of mRNA Levels with EEG Recordings
3.5. NR3C1 Changes Do Not Affect Mitochondrial DNA Copy Number (mtDNAcn)
3.6. Gene Network Analysis
4. Discussion
5. Limitations
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Snowling, M.J.; Hulme, C.; Nation, K. Defining and understanding dyslexia: Past, present and future. Oxf. Rev. Educ. 2020, 46, 501–513. [Google Scholar] [CrossRef] [PubMed]
- Elliott, J.G.; Grigorenko, E.L. The Dyslexia Debate; Cambridge University Press: Cambridge, UK, 2014. [Google Scholar]
- D’Mello, A.M.; Gabrieli, J.D.E. Cognitive Neuroscience of Dyslexia. Lang. Speech Hear. Serv. Sch. 2018, 49, 798–809. [Google Scholar] [CrossRef] [PubMed]
- Wolf, M.; Bowers, P.G. Naming-speed processes and developmental reading disabilities: An introduction to the special issue on the double-deficit hypothesis. J. Learn. Disabil. 2000, 33, 322–324. [Google Scholar] [CrossRef]
- Paulesu, E.; Demonet, J.F.; Fazio, F.; McCrory, E.; Chanoine, V.; Brunswick, N.; Cappa, S.F.; Cossu, G.; Habib, M.; Frith, C.D.; et al. Dyslexia: Cultural diversity and biological unity. Science 2001, 291, 2165–2167. [Google Scholar] [CrossRef] [PubMed]
- Gathercole, S.E.; Baddeley, A.D. Working Memory and Language; Psychology Press: London, UK, 2014. [Google Scholar]
- Goswami, U. A temporal sampling framework for developmental dyslexia. Trends Cogn. Sci. 2011, 15, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Nkomo, D.; Dube, B. The magnocellular theory of dyslexia: An overview, sub-categories and application in teaching. Eur. J. Spec. Educ. Res. 2019, 5. [Google Scholar] [CrossRef]
- Dubois, M.; Kyllingsbaek, S.; Prado, C.; Musca, S.C.; Peiffer, E.; Lassus-Sangosse, D.; Valdois, S. Fractionating the multi-character processing deficit in developmental dyslexia: Evidence from two case studies. Cortex 2010, 46, 717–738. [Google Scholar] [CrossRef]
- Snowling, M.J. Specific disorders and broader phenotypes: The case of dyslexia. Q. J. Exp. Psychol. 2008, 61, 142–156. [Google Scholar] [CrossRef]
- Livingston, E.M.; Siegel, L.S.; Ribary, U. Developmental dyslexia: Emotional impact and consequences. Aust. J. Learn. Difficulties 2018, 23, 107–135. [Google Scholar] [CrossRef]
- Menghini, D.; Finzi, A.; Benassi, M.; Bolzani, R.; Facoetti, A.; Giovagnoli, S.; Ruffino, M.; Vicari, S. Different underlying neurocognitive deficits in developmental dyslexia: A comparative study. Neuropsychologia 2010, 48, 863–872. [Google Scholar] [CrossRef]
- Peterson, R.L.; Pennington, B.F. Developmental dyslexia. Annu. Rev. Clin. Psychol. 2015, 11, 283–307. [Google Scholar] [CrossRef] [PubMed]
- Ramus, F.; Marshall, C.R.; Rosen, S.; van der Lely, H.K. Phonological deficits in specific language impairment and developmental dyslexia: Towards a multidimensional model. Brain 2013, 136, 630–645. [Google Scholar] [CrossRef] [PubMed]
- Cavalli, E.; Cole, P.; Leloup, G.; Poracchia-George, F.; Sprenger-Charolles, L.; El Ahmadi, A. Screening for Dyslexia in French-Speaking University Students: An Evaluation of the Detection Accuracy of the Alouette Test. J. Learn. Disabil. 2018, 51, 268–282. [Google Scholar] [CrossRef] [PubMed]
- Kershner, J.R. Dyslexia as an adaptation to cortico-limbic stress system reactivity. Neurobiol. Stress. 2020, 12, 100223. [Google Scholar] [CrossRef]
- Kershner, J.R. An Evolutionary Perspective of Dyslexia, Stress, and Brain Network Homeostasis. Front. Hum. Neurosci. 2020, 14, 575546. [Google Scholar] [CrossRef]
- Erbeli, F.; Rice, M.; Paracchini, S. Insights into Dyslexia Genetics Research from the Last Two Decades. Brain Sci. 2021, 12, 27. [Google Scholar] [CrossRef]
- Berretz, G.; Wolf, O.T.; Gunturkun, O.; Ocklenburg, S. Atypical lateralization in neurodevelopmental and psychiatric disorders: What is the role of stress? Cortex 2020, 125, 215–232. [Google Scholar] [CrossRef]
- Georgitsi, M.; Dermitzakis, I.; Soumelidou, E.; Bonti, E. The Polygenic Nature and Complex Genetic Architecture of Specific Learning Disorder. Brain Sci. 2021, 11, 631. [Google Scholar] [CrossRef]
- Skeide, M.A. Neurogenetic Insights into the Origins of Dyslexia and Dyscalculia. In The Cambridge Handbook of Dyslexia and Dyscalculia; Skeide, M.A., Ed.; Cambridge Handbooks in Psychology; Cambridge University Press: Cambridge, UK, 2022; pp. 155–161. [Google Scholar]
- Gialluisi, A.; Andlauer, T.F.M.; Mirza-Schreiber, N.; Moll, K.; Becker, J.; Hoffmann, P.; Ludwig, K.U.; Czamara, D.; Pourcain, B.S.; Honbolygo, F.; et al. Genome-wide association study reveals new insights into the heritability and genetic correlates of developmental dyslexia. Mol. Psychiatry 2021, 26, 3004–3017. [Google Scholar] [CrossRef]
- Hoeft, F.; Bouhali, F. Pre- and Postnatal Environmental Effects on Learning to Read and Mathematical Learning. In The Cambridge Handbook of Dyslexia and Dyscalculia; Skeide, M.A., Ed.; Cambridge Handbooks in Psychology; Cambridge University Press: Cambridge, UK, 2022; pp. 115–150. [Google Scholar]
- McEwen, B.S.; Gianaros, P.J. Stress- and allostasis-induced brain plasticity. Annu. Rev. Med. 2011, 62, 431–445. [Google Scholar] [CrossRef]
- Radley, J.; Morilak, D.; Viau, V.; Campeau, S. Chronic stress and brain plasticity: Mechanisms underlying adaptive and maladaptive changes and implications for stress-related CNS disorders. Neurosci. Biobehav. Rev. 2015, 58, 79–91. [Google Scholar] [CrossRef]
- Papadopoulou, Z.; Vlaikou, A.M.; Theodoridou, D.; Markopoulos, G.S.; Tsoni, K.; Agakidou, E.; Drosou-Agakidou, V.; Turck, C.W.; Filiou, M.D.; Syrrou, M. Stressful Newborn Memories: Pre-Conceptual, In Utero, and Postnatal Events. Front. Psychiatry 2019, 10, 220. [Google Scholar] [CrossRef]
- Mascheretti, S.; Bureau, A.; Battaglia, M.; Simone, D.; Quadrelli, E.; Croteau, J.; Cellino, M.R.; Giorda, R.; Beri, S.; Maziade, M.; et al. An assessment of gene-by-environment interactions in developmental dyslexia-related phenotypes. Genes. Brain Behav. 2013, 12, 47–55. [Google Scholar] [CrossRef]
- Sheerin, C.M.; Lind, M.J.; Bountress, K.E.; Marraccini, M.E.; Amstadter, A.B.; Bacanu, S.A.; Nugent, N.R. Meta-Analysis of Associations Between Hypothalamic-Pituitary-Adrenal Axis Genes and Risk of Posttraumatic Stress Disorder. J. Trauma. Stress. 2020, 33, 688–698. [Google Scholar] [CrossRef]
- Zakopoulou, V.; Vlaikou, A.M.; Darsinou, M.; Papadopoulou, Z.; Theodoridou, D.; Papageorgiou, K.; Alexiou, G.A.; Bougias, H.; Siafaka, V.; Zoccolotti, P.; et al. Linking Early Life Hypothalamic-Pituitary-Adrenal Axis Functioning, Brain Asymmetries, and Personality Traits in Dyslexia: An Informative Case Study. Front. Hum. Neurosci. 2019, 13, 327. [Google Scholar] [CrossRef]
- Bortoluzzi, A.; Blaya, C.; Rosa, E.D.d.; Paim, M.; Rosa, V.; Leistner-Segal, S.; Manfro, G.G. What can HPA axis-linked genes tell us about anxiety disorders in adolescents? Trends Psychiatry Psychother. 2015, 37, 232–237. [Google Scholar] [CrossRef] [PubMed]
- Oberlander, T.F.; Weinberg, J.; Papsdorf, M.; Grunau, R.; Misri, S.; Devlin, A.M. Prenatal exposure to maternal depression, neonatal methylation of human glucocorticoid receptor gene (NR3C1) and infant cortisol stress responses. Epigenetics 2008, 3, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Smart, C.; Strathdee, G.; Watson, S.; Murgatroyd, C.; McAllister-Williams, R.H. Early life trauma, depression and the glucocorticoid receptor gene--an epigenetic perspective. Psychol. Med. 2015, 45, 3393–3410. [Google Scholar] [CrossRef] [PubMed]
- Park, C.; Rosenblat, J.D.; Brietzke, E.; Pan, Z.; Lee, Y.; Cao, B.; Zuckerman, H.; Kalantarova, A.; McIntyre, R.S. Stress, epigenetics and depression: A systematic review. Neurosci. Biobehav. Rev. 2019, 102, 139–152. [Google Scholar] [CrossRef]
- Thiagarajah, A.S.; Eades, L.E.; Thomas, P.R.; Guymer, E.K.; Morand, E.F.; Clarke, D.M.; Leech, M. GILZ: Glitzing up our understanding of the glucocorticoid receptor in psychopathology. Brain Res. 2014, 1574, 60–69. [Google Scholar] [CrossRef] [PubMed]
- Zannas, A.S.; Wiechmann, T.; Gassen, N.C.; Binder, E.B. Gene-Stress-Epigenetic Regulation of FKBP5: Clinical and Translational Implications. Neuropsychopharmacology 2016, 41, 261–274. [Google Scholar] [CrossRef]
- Matosin, N.; Halldorsdottir, T.; Binder, E.B. Understanding the Molecular Mechanisms Underpinning Gene by Environment Interactions in Psychiatric Disorders: The FKBP5 Model. Biol. Psychiatry 2018, 83, 821–830. [Google Scholar] [CrossRef]
- Zannas, A.S.; Binder, E.B. Gene-environment interactions at the FKBP5 locus: Sensitive periods, mechanisms and pleiotropism. Genes. Brain Behav. 2014, 13, 25–37. [Google Scholar] [CrossRef] [PubMed]
- Grabe, H.J.; Schwahn, C.; Appel, K.; Mahler, J.; Schulz, A.; Spitzer, C.; Barnow, S.; John, U.; Freyberger, H.J.; Rosskopf, D.; et al. Update on the 2005 paper: Moderation of mental and physical distress by polymorphisms in the 5-HT transporter gene by interacting with social stressors and chronic disease burden. Mol. Psychiatry 2011, 16, 354–356. [Google Scholar] [CrossRef]
- Palma-Gudiel, H.; Fananas, L. An integrative review of methylation at the serotonin transporter gene and its dialogue with environmental risk factors, psychopathology and 5-HTTLPR. Neurosci. Biobehav. Rev. 2017, 72, 190–209. [Google Scholar] [CrossRef]
- Du, J.; Wang, Y.; Hunter, R.; Wei, Y.; Blumenthal, R.; Falke, C.; Khairova, R.; Zhou, R.; Yuan, P.; Machado-Vieira, R.; et al. Dynamic regulation of mitochondrial function by glucocorticoids. Proc. Natl. Acad. Sci. USA 2009, 106, 3543–3548. [Google Scholar] [CrossRef] [PubMed]
- Hunter, R.G.; Seligsohn, M.; Rubin, T.G.; Griffiths, B.B.; Ozdemir, Y.; Pfaff, D.W.; Datson, N.A.; McEwen, B.S. Stress and corticosteroids regulate rat hippocampal mitochondrial DNA gene expression via the glucocorticoid receptor. Proc. Natl. Acad. Sci. USA 2016, 113, 9099–9104. [Google Scholar] [CrossRef]
- Picard, M.; McEwen, B.S. Psychological Stress and Mitochondria: A Systematic Review. Psychosom. Med. 2018, 80, 141–153. [Google Scholar] [CrossRef]
- Lopes, S.; Teplytska, L.; Vaz-Silva, J.; Dioli, C.; Trindade, R.; Morais, M.; Webhofer, C.; Maccarrone, G.; Almeida, O.F.X.; Turck, C.W.; et al. Tau Deletion Prevents Stress-Induced Dendritic Atrophy in Prefrontal Cortex: Role of Synaptic Mitochondria. Cereb. Cortex 2017, 27, 2580–2591. [Google Scholar] [CrossRef] [PubMed]
- Filiou, M.D.; Sandi, C. Anxiety and Brain Mitochondria: A Bidirectional Crosstalk. Trends Neurosci. 2019, 42, 573–588. [Google Scholar] [CrossRef]
- Nussbaumer, M.; Asara, J.M.; Teplytska, L.; Murphy, M.P.; Logan, A.; Turck, C.W.; Filiou, M.D. Selective Mitochondrial Targeting Exerts Anxiolytic Effects In Vivo. Neuropsychopharmacology 2016, 41, 1751–1758. [Google Scholar] [CrossRef] [PubMed]
- Filiou, M.D.; Zhang, Y.; Teplytska, L.; Reckow, S.; Gormanns, P.; Maccarrone, G.; Frank, E.; Kessler, M.S.; Hambsch, B.; Nussbaumer, M.; et al. Proteomics and metabolomics analysis of a trait anxiety mouse model reveals divergent mitochondrial pathways. Biol. Psychiatry 2011, 70, 1074–1082. [Google Scholar] [CrossRef] [PubMed]
- Christodoulides, P.; Zakopoulou, V. The contribution of EEG rhythms’ changes to the audiovisual recognition of words in university students with dyslexia. In Psychology Applications & Developments VII Advances in Psychology and Psychological Trends Series; Pracana, C., Wang, M., Eds.; inScience Press: Lisbon, Portugal, 2021; p. 181. [Google Scholar]
- Filiou, M.D.; Asara, J.M.; Nussbaumer, M.; Teplytska, L.; Landgraf, R.; Turck, C.W. Behavioral extremes of trait anxiety in mice are characterized by distinct metabolic profiles. J. Psychiatr. Res. 2014, 58, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Petretto, D.R.; Masala, C. Dyslexia and specific learning disorders: New international diagnostic criteria. J. Child. Dev. Disord. 2017, 3, 4–19. [Google Scholar] [CrossRef]
- Hamilton, S.S.; Glascoe, F.P. Evaluation of children with reading difficulties. Am. Fam. Physician 2006, 74, 2079–2084. [Google Scholar]
- Francis, D.A.; Caruana, N.; Hudson, J.L.; McArthur, G.M. The association between poor reading and internalising problems: A systematic review and meta-analysis. Clin. Psychol. Rev. 2019, 67, 45–60. [Google Scholar] [CrossRef]
- Giovagnoli, S.; Mandolesi, L.; Magri, S.; Gualtieri, L.; Fabbri, D.; Tossani, E.; Benassi, M. Internalizing Symptoms in Developmental Dyslexia: A Comparison Between Primary and Secondary School. Front. Psychol. 2020, 11, 461. [Google Scholar] [CrossRef]
- Schoenberg, P.L. Linear and nonlinear EEG-based functional networks in anxiety disorders. Anxiety Disord. 2020, 1191, 35–59. [Google Scholar]
- Vajs, I.; Kovic, V.; Papic, T.; Savic, A.M.; Jankovic, M.M. Spatiotemporal Eye-Tracking Feature Set for Improved Recognition of Dyslexic Reading Patterns in Children. Sensors 2022, 22, 4900. [Google Scholar] [CrossRef]
- Jakovljevic, T.; Jankovic, M.M.; Savic, A.M.; Soldatovic, I.; Todorovic, P.; Jere Jakulin, T.; Papa, G.; Kovic, V. The Sensor Hub for Detecting the Developmental Characteristics in Reading in Children on a White vs. Colored Background/Colored Overlays. Sensors 2021, 21, 406. [Google Scholar] [CrossRef]
- Janković, M.M. Biomarker-based approaches for dyslexia screening: A review. In Proceedings of the 2022 IEEE Zooming Innovation in Consumer Technologies Conference (ZINC), Novi Sad, Serbia, 25–26 May 2022; pp. 28–33. [Google Scholar]
- Christoforou, C.; Fella, A.; Leppanen, P.H.T.; Georgiou, G.K.; Papadopoulos, T.C. Fixation-related potentials in naming speed: A combined EEG and eye-tracking study on children with dyslexia. Clin. Neurophysiol. 2021, 132, 2798–2807. [Google Scholar] [CrossRef]
- Evans, K.C.; Dougherty, D.D.; Pollack, M.H.; Rauch, S.L. Using neuroimaging to predict treatment response in mood and anxiety disorders. Ann. Clin. Psychiatry 2006, 18, 33–42. [Google Scholar] [CrossRef]
- Garcia Chimeno, Y.; Garcia Zapirain, B.; Saralegui Prieto, I.; Fernandez-Ruanova, B. Automatic classification of dyslexic children by applying machine learning to fMRI images. Biomed. Mater. Eng. 2014, 24, 2995–3002. [Google Scholar] [CrossRef]
- Ortiz, A.; Martinez-Murcia, F.J.; Luque, J.L.; Gimenez, A.; Morales-Ortega, R.; Ortega, J. Dyslexia Diagnosis by EEG Temporal and Spectral Descriptors: An Anomaly Detection Approach. Int. J. Neural Syst. 2020, 30, 2050029. [Google Scholar] [CrossRef]
- Thiede, A.; Glerean, E.; Kujala, T.; Parkkonen, L. Atypical MEG inter-subject correlation during listening to continuous natural speech in dyslexia. Neuroimage 2020, 216, 116799. [Google Scholar] [CrossRef] [PubMed]
- Xing, S.; McCardle, R.; Xie, S.S.Q. Reading the mind: The potential of electroencephalography in brain computer interfaces. In Proceedings of the 2012 19th International Conference on Mechatronics and Machine Vision in Practice (M2VIP), Auckland, New Zealand, 28–30 November 2012; pp. 275–280. [Google Scholar]
- Martínez-Murcia, F.; Ortiz, A.; Morales, R.; López, P.; Luque, J.; Castillo-Barnes, D.; Segovia, F.; Illan, I.; Ortega, J.; Ramírez, J.; et al. Periodogram Connectivity of EEG Signals for the Detection of Dyslexia. In Understanding the Brain Function and Emotions: 8th International Work-Conference on the Interplay Between Natural and Artificial Computation, IWINAC 2019, Almería, Spain, June 3–7, 2019, Proceedings; Springer International Publishing: Berlin/Heidelberg, Germany, 2019; pp. 350–359. [Google Scholar]
- Xue, H.; Wang, Z.; Tan, Y.; Yang, H.; Fu, W.; Xue, L.; Zhao, J. Resting-state EEG reveals global network deficiency in dyslexic children. Neuropsychologia 2020, 138, 107343. [Google Scholar] [CrossRef] [PubMed]
- Axmacher, N.; Mormann, F.; Fernandez, G.; Elger, C.E.; Fell, J. Memory formation by neuronal synchronization. Brain Res. Rev. 2006, 52, 170–182. [Google Scholar] [CrossRef] [PubMed]
- McCormick, D.A.; Contreras, D. On the cellular and network bases of epileptic seizures. Annu. Rev. Physiol. 2001, 63, 815–846. [Google Scholar] [CrossRef] [PubMed]
- Rippon, G.; Brunswick, N. Trait and state EEG indices of information processing in developmental dyslexia. Int. J. Psychophysiol. 2000, 36, 251–265. [Google Scholar] [CrossRef] [PubMed]
- Papagiannopoulou, E.A.; Lagopoulos, J. Resting State EEG Hemispheric Power Asymmetry in Children with Dyslexia. Front. Pediatr. 2016, 4, 11. [Google Scholar] [CrossRef]
- Kandel, S.; Lassus-Sangosse, D.; Grosjacques, G.; Perret, C. The impact of developmental dyslexia and dysgraphia on movement production during word writing. Cogn. Neuropsychol. 2017, 34, 219–251. [Google Scholar] [CrossRef]
- Perera, H.; Shiratuddin, M.F.; Wong, K.W. Review of EEG-based pattern classification frameworks for dyslexia. Brain Inform. 2018, 5, 4. [Google Scholar] [CrossRef] [PubMed]
- Peyrin, C.; Lallier, M.; Demonet, J.F.; Pernet, C.; Baciu, M.; Le Bas, J.F.; Valdois, S. Neural dissociation of phonological and visual attention span disorders in developmental dyslexia: FMRI evidence from two case reports. Brain Lang. 2012, 120, 381–394. [Google Scholar] [CrossRef] [PubMed]
- Steinbrink, C.; Groth, K.; Lachmann, T.; Riecker, A. Neural correlates of temporal auditory processing in developmental dyslexia during German vowel length discrimination: An fMRI study. Brain Lang. 2012, 121, 1–11. [Google Scholar] [CrossRef]
- Pecini, C.; Biagi, L.; Brizzolara, D.; Cipriani, P.; Di Lieto, M.C.; Guzzetta, A.; Tosetti, M.; Chilosi, A.M. How many functional brains in developmental dyslexia? When the history of language delay makes the difference. Cogn. Behav. Neurol. 2011, 24, 85–92. [Google Scholar] [CrossRef]
- Dufor, O.; Serniclaes, W.; Sprenger-Charolles, L.; Demonet, J.F. Left premotor cortex and allophonic speech perception in dyslexia: A PET study. Neuroimage 2009, 46, 241–248. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nuwer, M.R. EEG topographic mapping and frequency analysis: Techniques and studies in clinical setting. Semin. Neurol. 1990, 10, 166–177. [Google Scholar] [CrossRef]
- Kershner, J.R. Neuroscience and education: Cerebral lateralization of networks and oscillations in dyslexia. Laterality 2020, 25, 109–125. [Google Scholar] [CrossRef]
- Di Liberto, G.M.; Peter, V.; Kalashnikova, M.; Goswami, U.; Burnham, D.; Lalor, E.C. Atypical cortical entrainment to speech in the right hemisphere underpins phonemic deficits in dyslexia. Neuroimage 2018, 175, 70–79. [Google Scholar] [CrossRef]
- Christodoulides, P.; Miltiadous, A.; Tzimourta, K.D.; Peschos, D.; Ntritsos, G.; Zakopoulou, V.; Giannakeas, N.; Astrakas, L.G.; Tsipouras, M.G.; Tsamis, K.I. Classification of EEG signals from young adults with dyslexia combining a Brain Computer Interface device and an Interactive Linguistic Software Tool. Biomed. Signal Process. Control. 2022, 76, 103646. [Google Scholar] [CrossRef]
- Eroglu, G.; Köprülü, M.; Karabacak, B. Developmental dyslexia biomarker detection with Quantitative electroencephalography (QEEG) data in children: Feasibility, acceptability, economic impact. Qeios 2022. [Google Scholar] [CrossRef]
- Kaisar, S. Developmental dyslexia detection using machine learning techniques: A survey. ICT Express 2020, 6, 181–184. [Google Scholar] [CrossRef]
- Ahire, N.; Awale, R.; Patnaik, S.; Wagh, A. A comprehensive review of machine learning approaches for dyslexia diagnosis. Multimed. Tools Appl. 2022, 82, 13557–13577. [Google Scholar] [CrossRef]
- Rezvani, Z.; Zare, M.; Žarić, G.; Bonte, M.; Tijms, J.; Molen, M.W.V.d.; González, G.F. Machine learning Classification of Dyslexic Children based on EEG Local Network Features. bioRxiv 2019. [Google Scholar] [CrossRef]
- Karim, I.; Qayoom, A.; Wahab, A.; Kamaruddin, N. Early Identification of Dyslexic Preschoolers Based on Neurophysiological Signals. In Proceedings of the 2013 International Conference on Advanced Computer Science Applications and Technologies, Kuching, Malaysia, 23–24 December 2013; pp. 362–366. [Google Scholar]
- Zainuddin, A.Z.A.; Lee, K.Y.; Mansor, W.; Mahmoodin, Z. Optimized KNN classify rule for EEG based differentiation between capable dyslexic and normal children. In Proceedings of the 2016 IEEE EMBS Conference on Biomedical Engineering and Sciences (IECBES), Kuala Lumpur, Malaysia, 4–8 December 2016; pp. 685–688. [Google Scholar]
- Jakovljevic, T.; Jankovic, M.M.; Savic, A.M.; Soldatovic, I.; Colic, G.; Jakulin, T.J.; Papa, G.; Kovic, V. The Relation between Physiological Parameters and Colour Modifications in Text Background and Overlay during Reading in Children with and without Dyslexia. Brain Sci. 2021, 11, 539. [Google Scholar] [CrossRef] [PubMed]
- Mammarella, I.C.; Cardillo, R.; Zoccante, L. Differences in visuospatial processing in individuals with nonverbal learning disability or autism spectrum disorder without intellectual disability. Neuropsychology 2019, 33, 123–134. [Google Scholar] [CrossRef] [PubMed]
- Mahmoodin, Z.; Mansor, W.; Lee, K.Y.; Mohamad, N.B. An analysis of EEG signal power spectrum density generated during writing in children with dyslexia. In Proceedings of the 2015 IEEE 11th International Colloquium on Signal Processing & Its Applications (CSPA), Kuala Lumpur, Malaysia, 6–8 March 2015; pp. 156–160. [Google Scholar]
- Penolazzi, B.; Spironelli, C.; Vio, C.; Angrilli, A. Brain plasticity in developmental dyslexia after phonological treatment: A beta EEG band study. Behav. Brain Res. 2010, 209, 179–182. [Google Scholar] [CrossRef]
- Slikker, W., Jr. Biomarkers and their impact on precision medicine. Exp. Biol. Med. 2018, 243, 211–212. [Google Scholar] [CrossRef] [PubMed]
- Abi-Dargham, A.; Moeller, S.J.; Ali, F.; DeLorenzo, C.; Domschke, K.; Horga, G.; Jutla, A.; Kotov, R.; Paulus, M.P.; Rubio, J.M.; et al. Candidate biomarkers in psychiatric disorders: State of the field. World Psychiatry 2023, 22, 236–262. [Google Scholar] [CrossRef]
- Kuznetsova, I.L.; Ponomareva, N.V.; Alemastseva, E.A.; Manakhov, A.D.; Andreeva, T.V.; Gusev, F.E.; Rogaev, E.I. The Interactive Effect of Genetic and Epigenetic Variations in FKBP5 and ApoE Genes on Anxiety and Brain EEG Parameters. Genes 2022, 13, 164. [Google Scholar] [CrossRef]
- Wirz, L.; Reuter, M.; Wacker, J.; Felten, A.; Schwabe, L. A Haplotype Associated with Enhanced Mineralocorticoid Receptor Expression Facilitates the Stress-Induced Shift from “Cognitive” to “Habit” Learning. eNeuro 2017, 4, 6. [Google Scholar] [CrossRef]
- Theodoridou, D.; Christodoulides, P.; Zakopoulou, V.; Syrrou, M. Developmental Dyslexia: Environment Matters. Brain Sci. 2021, 11, 7821. [Google Scholar] [CrossRef]
- Becker, N.; Vasconcelos, M.; Oliveira, V.; Santos, F.C.D.; Bizarro, L.; Almeida, R.M.M.; Salles, J.F.; Carvalho, M.R.S. Genetic and environmental risk factors for developmental dyslexia in children: Systematic review of the last decade. Dev. Neuropsychol. 2017, 42, 423–445. [Google Scholar] [CrossRef]
- Christodoulides, P.; Zakopoulou, V.; Tzimourta, K.D.; Tzallas, A.T.; Peschos, D. The contribution of EEG recordings to the audiovisual recognition of words in university students with dyslexia. Psychol. Appl. Conf. Trends 2021. [Google Scholar] [CrossRef]
- Lovibond, P.F.; Lovibond, S.H. The structure of negative emotional states: Comparison of the Depression Anxiety Stress Scales (DASS) with the Beck Depression and Anxiety Inventories. Behav. Res. Ther. 1995, 33, 335–343. [Google Scholar] [CrossRef] [PubMed]
- Lan, H.T.Q.; Long, N.T.; Hanh, N.V. Validation of Depression, Anxiety and Stress Scales (DASS-21): Immediate Psychological Responses of Students in the E-Learning Environment. Int. J. High. Educ. 2020, 9, 125–133. [Google Scholar] [CrossRef]
- Lyrakos, G.; Arvaniti, C.; Smyrnioti, M.; Kostopanagiotou, G. Translation and validation study of the depression anxiety stress scale in the Greek general population and in a psychiatric patient’s sample. Eur. Psychiatry 2011, 26, 1731. [Google Scholar] [CrossRef]
- Xie, H. Chapter 5—Personalized Epigenetics: Analysis and Interpretation of DNA Methylation Variation. In Personalized Epigenetics; Tollefsbol, T.O., Ed.; Academic Press: Boston, MA, USA, 2015; pp. 123–150. [Google Scholar]
- World Medical Association Declaration of Helsinki: Ethical principles for medical research involving human subjects. JAMA 2013, 310, 2191–2194. [CrossRef]
- Kosek, E.; Jensen, K.B.; Lonsdorf, T.B.; Schalling, M.; Ingvar, M. Genetic variation in the serotonin transporter gene (5-HTTLPR, rs25531) influences the analgesic response to the short acting opioid Remifentanil in humans. Mol. Pain. 2009, 5, 37. [Google Scholar] [CrossRef] [PubMed]
- Al-Eitan, L.N.; Jaradat, S.A.; Qin, W.; Wildenauer, D.M.; Wildenauer, D.D.; Hulse, G.K.; Tay, G.K. Characterization of serotonin transporter gene (SLC6A4) polymorphisms and its association with drug dependence in a Jordanian Arab population. Toxicol. Ind. Health 2014, 30, 598–610. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Kent, W.J.; Sugnet, C.W.; Furey, T.S.; Roskin, K.M.; Pringle, T.H.; Zahler, A.M.; Haussler, D. The human genome browser at UCSC. Genome Res. 2002, 12, 996–1006. [Google Scholar] [CrossRef]
- Jiang, Z.; Qian, L.; Zou, H.; Jia, Y.; Ni, Y.; Yang, X.; Jiang, Z.; Zhao, R. Porcine glucocorticoid receptor (NR3C1) gene: Tissue-specificity of transcriptional strength and glucocorticoid responsiveness of alternative promoters. J. Steroid Biochem. Mol. Biol. 2014, 141, 87–93. [Google Scholar] [CrossRef]
- Chen, S.; Li, Z.; He, Y.; Zhang, F.; Li, H.; Liao, Y.; Wei, Z.; Wan, G.; Xiang, X.; Hu, M.; et al. Elevated mitochondrial DNA copy number in peripheral blood cells is associated with childhood autism. BMC Psychiatry 2015, 15, 50. [Google Scholar] [CrossRef]
- Turri, C.; Di Dona, G.; Santoni, A.; Zamfira, D.A.; Franchin, L.; Melcher, D.; Ronconi, L. Periodic and Aperiodic EEG Features as Potential Markers of Developmental Dyslexia. Biomedicines 2023, 11, 1607. [Google Scholar] [CrossRef]
- Chikhi, S.; Matton, N.; Blanchet, S. EEG power spectral measures of cognitive workload: A meta-analysis. Psychophysiology 2022, 59, e14009. [Google Scholar] [CrossRef] [PubMed]
- Basar, E.; Golbasi, B.T.; Tulay, E.; Aydin, S.; Basar-Eroglu, C. Best method for analysis of brain oscillations in healthy subjects and neuropsychiatric diseases. Int. J. Psychophysiol. 2016, 103, 22–42. [Google Scholar] [CrossRef] [PubMed]
- Katmah, R.; Al-Shargie, F.; Tariq, U.; Babiloni, F.; Al-Mughairbi, F.; Al-Nashash, H. A Review on Mental Stress Assessment Methods Using EEG Signals. Sensors 2021, 21, 5043. [Google Scholar] [CrossRef] [PubMed]
- Warde-Farley, D.; Donaldson, S.L.; Comes, O.; Zuberi, K.; Badrawi, R.; Chao, P.; Franz, M.; Grouios, C.; Kazi, F.; Lopes, C.T.; et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010, 38, W214–W220. [Google Scholar] [CrossRef] [PubMed]
- Menyhart, O.; Weltz, B.; Gyorffy, B. MultipleTesting.com: A tool for life science researchers for multiple hypothesis testing correction. PLoS ONE 2021, 16, e0245824. [Google Scholar] [CrossRef]
- Doust, C.; Gordon, S.D.; Garden, N.; Fisher, S.E.; Martin, N.G.; Bates, T.C.; Luciano, M. The Association of Dyslexia and Developmental Speech and Language Disorder Candidate Genes with Reading and Language Abilities in Adults. Twin Res. Hum. Genet. 2020, 23, 23–32. [Google Scholar] [CrossRef]
- Luciano, M.; Gow, A.J.; Pattie, A.; Bates, T.C.; Deary, I.J. The Influence of Dyslexia Candidate Genes on Reading Skill in Old Age. Behav. Genet. 2018, 48, 351–360. [Google Scholar] [CrossRef]
- Doust, C.; Fontanillas, P.; Eising, E.; Gordon, S.D.; Wang, Z.; Alagoz, G.; Molz, B.; andMe Research, T.; Quantitative Trait Working Group of the GenLang, C.; Pourcain, B.S.; et al. Discovery of 42 genome-wide significant loci associated with dyslexia. Nat. Genet. 2022, 54, 1621–1629. [Google Scholar] [CrossRef] [PubMed]
- Tsigos, C.; Chrousos, G.P. Hypothalamic-pituitary-adrenal axis, neuroendocrine factors and stress. J. Psychosom. Res. 2002, 53, 865–871. [Google Scholar] [CrossRef] [PubMed]
- Espin, L.; Garcia, I.; Del Pino Sanchez, M.; Roman, F.; Salvador, A. Effects of psychosocial stress on the hormonal and affective response in children with dyslexia. Trends Neurosci. Educ. 2019, 15, 1–9. [Google Scholar] [CrossRef]
- Kuracki, K.; Dluzniewska, A. Exam stress and the metacognitive strategies of reading in students with dyslexia: The role of motivational mechanisms and educational support. PLoS ONE 2023, 18, e0294255. [Google Scholar] [CrossRef] [PubMed]
- Francis, D.; Hudson, J.L.; Kohnen, S.; Mobach, L.; McArthur, G.M. The effect of an integrated reading and anxiety intervention for poor readers with anxiety. PeerJ 2021, 9, e10987. [Google Scholar] [CrossRef]
- Wilmot, A.; Hasking, P.; Leitao, S.; Hill, E.; Boyes, M. Understanding Mental Health in Developmental Dyslexia: A Scoping Review. Int. J. Environ. Res. Public. Health 2023, 20, 1653. [Google Scholar] [CrossRef]
- Zakopoulou, V.; Mavreas, V.; Christodoulides, P.; Lavidas, A.; Fili, E.; Georgiou, G.; Dimakopoulos, G.; Vergou, M. Specific learning difficulties: A retrospective study of their co morbidity and continuity as early indicators of mental disorders. Res. Dev. Disabil. 2014, 35, 3496–3507. [Google Scholar] [CrossRef]
- Allsopp, K.; Read, J.; Corcoran, R.; Kinderman, P. Heterogeneity in psychiatric diagnostic classification. Psychiatry Res. 2019, 279, 15–22. [Google Scholar] [CrossRef]
- Zankert, S.; Bellingrath, S.; Wust, S.; Kudielka, B.M. HPA axis responses to psychological challenge linking stress and disease: What do we know on sources of intra- and interindividual variability? Psychoneuroendocrinology 2019, 105, 86–97. [Google Scholar] [CrossRef] [PubMed]
- Hunter, A.L.; Minnis, H.; Wilson, P. Altered stress responses in children exposed to early adversity: A systematic review of salivary cortisol studies. Stress 2011, 14, 614–626. [Google Scholar] [CrossRef] [PubMed]
- Bunea, I.M.; Szentagotai-Tatar, A.; Miu, A.C. Early-life adversity and cortisol response to social stress: A meta-analysis. Transl. Psychiatry 2017, 7, 1274. [Google Scholar] [CrossRef] [PubMed]
- Roy, B.; Shelton, R.C.; Dwivedi, Y. DNA methylation and expression of stress related genes in PBMC of MDD patients with and without serious suicidal ideation. J. Psychiatr. Res. 2017, 89, 115–124. [Google Scholar] [CrossRef]
- Palma-Gudiel, H.; Cordova-Palomera, A.; Leza, J.C.; Fananas, L. Glucocorticoid receptor gene (NR3C1) methylation processes as mediators of early adversity in stress-related disorders causality: A critical review. Neurosci. Biobehav. Rev. 2015, 55, 520–535. [Google Scholar] [CrossRef]
- McGowan, P.O.; Sasaki, A.; D’Alessio, A.C.; Dymov, S.; Labonte, B.; Szyf, M.; Turecki, G.; Meaney, M.J. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat. Neurosci. 2009, 12, 342–348. [Google Scholar] [CrossRef]
- Wang, W.; Feng, J.; Ji, C.; Mu, X.; Ma, Q.; Fan, Y.; Chen, C.; Gao, C.; Ma, X.C.; Zhu, F. Increased methylation of glucocorticoid receptor gene promoter 1(F) in peripheral blood of patients with generalized anxiety disorder. J. Psychiatr. Res. 2017, 91, 18–25. [Google Scholar] [CrossRef]
- Labonte, B.; Yerko, V.; Gross, J.; Mechawar, N.; Meaney, M.J.; Szyf, M.; Turecki, G. Differential glucocorticoid receptor exon 1(B), 1(C), and 1(H) expression and methylation in suicide completers with a history of childhood abuse. Biol. Psychiatry 2012, 72, 41–48. [Google Scholar] [CrossRef]
- Turecki, G.; Meaney, M.J. Effects of the Social Environment and Stress on Glucocorticoid Receptor Gene Methylation: A Systematic Review. Biol. Psychiatry 2016, 79, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Hompes, T.; Izzi, B.; Gellens, E.; Morreels, M.; Fieuws, S.; Pexsters, A.; Schops, G.; Dom, M.; Van Bree, R.; Freson, K.; et al. Investigating the influence of maternal cortisol and emotional state during pregnancy on the DNA methylation status of the glucocorticoid receptor gene (NR3C1) promoter region in cord blood. J. Psychiatr. Res. 2013, 47, 880–891. [Google Scholar] [CrossRef] [PubMed]
- Schur, R.R.; Boks, M.P.; Rutten, B.P.F.; Daskalakis, N.P.; de Nijs, L.; van Zuiden, M.; Kavelaars, A.; Heijnen, C.J.; Joels, M.; Kahn, R.S.; et al. Longitudinal changes in glucocorticoid receptor exon 1(F) methylation and psychopathology after military deployment. Transl. Psychiatry 2017, 7, e1181. [Google Scholar] [CrossRef]
- Romens, S.E.; McDonald, J.; Svaren, J.; Pollak, S.D. Associations between early life stress and gene methylation in children. Child. Dev. 2015, 86, 303–309. [Google Scholar] [CrossRef] [PubMed]
- van der Knaap, L.J.; Riese, H.; Hudziak, J.J.; Verbiest, M.M.; Verhulst, F.C.; Oldehinkel, A.J.; van Oort, F.V. Glucocorticoid receptor gene (NR3C1) methylation following stressful events between birth and adolescence. The TRAILS study. Transl. Psychiatry 2014, 4, e381. [Google Scholar] [CrossRef] [PubMed]
- Parade, S.H.; Ridout, K.K.; Seifer, R.; Armstrong, D.A.; Marsit, C.J.; McWilliams, M.A.; Tyrka, A.R. Methylation of the Glucocorticoid Receptor Gene Promoter in Preschoolers: Links With Internalizing Behavior Problems. Child. Dev. 2016, 87, 86–97. [Google Scholar] [CrossRef] [PubMed]
- Daskalakis, N.P.; Yehuda, R. Site-specific methylation changes in the glucocorticoid receptor exon 1F promoter in relation to life adversity: Systematic review of contributing factors. Front. Neurosci. 2014, 8, 369. [Google Scholar] [CrossRef] [PubMed]
- Dee, G.; Ryznar, R.; Dee, C. Epigenetic Changes Associated with Different Types of Stressors and Suicide. Cells 2023, 12, 1258. [Google Scholar] [CrossRef] [PubMed]
- Carmi, L.; Zohar, J.; Juven-Wetzler, A.; Desarnaud, F.; Makotkine, L.; Bierer, L.M.; Cohen, H.; Yehuda, R. Promoter methylation of the glucocorticoid receptor following trauma may be associated with subsequent development of PTSD. World J. Biol. Psychiatry 2023, 24, 578–586. [Google Scholar] [CrossRef] [PubMed]
- Tyrka, A.R.; Parade, S.H.; Welch, E.S.; Ridout, K.K.; Price, L.H.; Marsit, C.; Philip, N.S.; Carpenter, L.L. Methylation of the leukocyte glucocorticoid receptor gene promoter in adults: Associations with early adversity and depressive, anxiety and substance-use disorders. Transl. Psychiatry 2016, 6, e848. [Google Scholar] [CrossRef] [PubMed]
- Yehuda, R.; Flory, J.D.; Bierer, L.M.; Henn-Haase, C.; Lehrner, A.; Desarnaud, F.; Makotkine, I.; Daskalakis, N.P.; Marmar, C.R.; Meaney, M.J. Lower methylation of glucocorticoid receptor gene promoter 1F in peripheral blood of veterans with posttraumatic stress disorder. Biol. Psychiatry 2015, 77, 356–364. [Google Scholar] [CrossRef]
- Brenet, F.; Moh, M.; Funk, P.; Feierstein, E.; Viale, A.J.; Socci, N.D.; Scandura, J.M. DNA methylation of the first exon is tightly linked to transcriptional silencing. PLoS ONE 2011, 6, e14524. [Google Scholar] [CrossRef]
- Capp, J.P. Interplay between genetic, epigenetic, and gene expression variability: Considering complexity in evolvability. Evol. Appl. 2021, 14, 893–901. [Google Scholar] [CrossRef] [PubMed]
- Turner, J.D.; Muller, C.P. Structure of the glucocorticoid receptor (NR3C1) gene 5′ untranslated region: Identification, and tissue distribution of multiple new human exon 1. J. Mol. Endocrinol. 2005, 35, 283–292. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tyrka, A.R.; Parade, S.H.; Eslinger, N.M.; Marsit, C.J.; Lesseur, C.; Armstrong, D.A.; Philip, N.S.; Josefson, B.; Seifer, R. Methylation of exons 1D, 1F, and 1H of the glucocorticoid receptor gene promoter and exposure to adversity in preschool-aged children. Dev. Psychopathol. 2015, 27, 577–585. [Google Scholar] [CrossRef] [PubMed]
- Giarraputo, J.; DeLoach, J.; Padbury, J.; Uzun, A.; Marsit, C.; Hawes, K.; Lester, B. Medical morbidities and DNA methylation of NR3C1 in preterm infants. Pediatr. Res. 2017, 81, 68–74. [Google Scholar] [CrossRef] [PubMed]
- Tseng, C.C.; Wong, M.C.; Liao, W.T.; Chen, C.J.; Lee, S.C.; Yen, J.H.; Chang, S.J. Genetic Variants in Transcription Factor Binding Sites in Humans: Triggered by Natural Selection and Triggers of Diseases. Int. J. Mol. Sci. 2021, 22, 4187. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Wang, G.; Qian, J. Transcription factors as readers and effectors of DNA methylation. Nat. Rev. Genet. 2016, 17, 551–565. [Google Scholar] [CrossRef] [PubMed]
- Hop, P.J.; Luijk, R.; Daxinger, L.; van Iterson, M.; Dekkers, K.F.; Jansen, R.; Consortium, B.; van Meurs, J.B.J.; t Hoen, P.A.C.; Ikram, M.A.; et al. Genome-wide identification of genes regulating DNA methylation using genetic anchors for causal inference. Genome Biol. 2020, 21, 220. [Google Scholar] [CrossRef]
- Spainhour, J.C.; Lim, H.S.; Yi, S.V.; Qiu, P. Correlation Patterns Between DNA Methylation and Gene Expression in The Cancer Genome Atlas. Cancer Inform. 2019, 18, 1176935119828776. [Google Scholar] [CrossRef]
- Sklar, B.; Hanley, J.; Simmons, W.W. An EEG experiment aimed toward identifying dyslexic children. Nature 1972, 240, 414–416. [Google Scholar] [CrossRef]
- Ulett, G.A.; Gleser, G. The Effect of Experimental Stress upon the Photically Activated EEG. Science 1952, 115, 678–682. [Google Scholar] [CrossRef]
- Newson, J.J.; Thiagarajan, T.C. EEG Frequency Bands in Psychiatric Disorders: A Review of Resting State Studies. Front. Hum. Neurosci. 2018, 12, 521. [Google Scholar] [CrossRef] [PubMed]
- Cainelli, E.; Vedovelli, L.; Carretti, B.; Bisiacchi, P. EEG correlates of developmental dyslexia: A systematic review. Ann. Dyslexia 2023, 73, 184–213. [Google Scholar] [CrossRef] [PubMed]
- Klimesch, W.; Doppelmayr, M.; Wimmer, H.; Gruber, W.; Rohm, D.; Schwaiger, J.; Hutzler, F. Alpha and beta band power changes in normal and dyslexic children. Clin. Neurophysiol. 2001, 112, 1186–1195. [Google Scholar] [CrossRef]
- João, A.; Benjamin, D.S.; Varghese, P.; Kanad, M.; Marina, K.; Annabel, M.; Fiona, G.; Angela, W.; Giovanni, M.D.L.; Denis, B.; et al. Atypical cortical encoding of speech identifies children with Dyslexia versus Developmental Language Disorder. bioRxiv 2022. [Google Scholar] [CrossRef]
- Knyazev, G.G.; Schutter, D.J.; van Honk, J. Anxious apprehension increases coupling of delta and beta oscillations. Int. J. Psychophysiol. 2006, 61, 283–287. [Google Scholar] [CrossRef]
- Ismail, L.E.; Karwowski, W. Applications of EEG indices for the quantification of human cognitive performance: A systematic review and bibliometric analysis. PLoS ONE 2020, 15, e0242857. [Google Scholar] [CrossRef]
- Turker, S.; Kuhnke, P.; Jiang, Z.; Hartwigsen, G. Disrupted network interactions serve as a neural marker of dyslexia. Commun. Biol. 2023, 6, 1114. [Google Scholar] [CrossRef]

| Frequency Range | Frequency Band |
|---|---|
| 0.5–4 Hz | Delta |
| 4–8 Hz | Theta |
| 8–12 Hz | Alpha |
| 12–20 Hz | Beta1 |
| 20–30 Hz | Beta2 |
| 30–45 Hz | Gamma |
| Demographic Information | Dyslexia (n = 10) | Control (n = 10) | Total (n = 20) | p-Value | |
|---|---|---|---|---|---|
| Age (years), mean ± SD | 21.3 ± 2.1 | 20.7 ± 1.2 | 21 ± 1.7 | 0.6168 | |
| Sex (M/F) | 4/6 | 2/8 | 6/14 | - | |
| DASS21 Scores | Dyslexia (n = 10) | Control (n = 10) | Total (n = 20) | p-Value | |
| Depression, mean ± SD | 17 ± 10.7 | 13.8 ± 11.8 | 15.4 ± 11.1 | 0.4445 | |
| Anxiety, mean ± SD | 15.4 ± 12.4 | 8.8 ± 5.4 | 12.1 ± 9.9 | 0.2212 | |
| Stress, mean ± SD | 19.4 ± 9.6 | 19.6 ± 12.1 | 19.5 ± 10.6 | 0.8975 | |
| Allelic Frequencies | Dyslexia (n = 10) | Control (n = 8) | Total (n = 18) | ||
| FKBP5 (rs1360780) | C | 0.7 | 0.625 | 0.7 | |
| T | 0.3 | 0.375 | 0.3 | ||
| SLC6A4 (5-HTTLRP) | L | 0.4 | 0.625 | 0.5 | |
| S | 0.6 | 0.375 | 0.5 | ||
| SLC6A4 (rs25531) | A | 1 | 1 | 1 | |
| G | 0 | 0 | 0 | ||
| Gene | Brain Area | Rhythm | Correlation | Spearman’s r | p-Value |
|---|---|---|---|---|---|
| Control | |||||
| NR3C1 | T7 | Beta 1 | Negative | −0.783 | <0.05 * |
| T7 | Beta 2 | Negative | −0.700 | <0.05 * | |
| P7 | Alpha 2 | Negative | -0.750 | <0.05 * | |
| O2 | Delta | Positive | 0.821 | <0.05 * | |
| GILZ | P7 | Alpha 2 | Negative | −0.917 | <0.01 ** |
| P8 | Theta | Positive | 0.786 | <0.05 * | |
| O1 | Beta 2 | Negative | −0.827 | <0.05 * | |
| FKBP5 | P7 | Beta 2 | Negative | −0.883 | <0.01 ** |
| P8 | Alpha 1 | Negative | −0.929 | <0.01 ** | |
| DD | |||||
| NR3C1 | P8 | Theta | Positive | 0.883 | <0.01 ** |
| P8 | Beta 1 | Negative | −0.717 | <0.05 * | |
| GILZ | O1 | Theta | Positive | 0.881 | <0.01 ** |
| O1 | Delta | Positive | 0.786 | <0.05 * | |
| FKBP5 | T8 | Delta | Positive | 0.717 | <0.05 * |
| Gene | Interacting Gene | Networks |
|---|---|---|
| NR3C1 | NR3C2 | Physical Interactions, Shared Protein Domains |
| CNTNAP2 | Genetic Interactions | |
| PCNT | Genetic Interactions | |
| CYP19A1 | Physical Interactions, Genetic Interactions | |
| DIP2A | Genetic Interactions | |
| CMIP | Genetic Interactions | |
| FKBP5 | Physical Interactions | |
| S100B | Genetic Interactions | |
| PAXBP1 | Genetic Interactions | |
| AKAP9 | Co-Expression | |
| EPB41L3 | Genetic Interactions | |
| CNTN2 | Genetic Interactions | |
| CPE | Genetic Interactions | |
| NR3C2 | NR3C1 | Physical Interactions, Shared Protein Domains |
| FKBP5 | Physical Interactions | |
| SLC6A4 | Co-Expression | |
| MIB1 | Genetic Interactions | |
| CNTN2 | Co-Expression | |
| CPE | Co-Expression | |
| FKBP5 | NR3C1 | Physical Interactions |
| NR3C2 | Physical Interactions | |
| ROBO1 | Co-Expression | |
| GILZ | Co-Expression | |
| TSC22D1 | Co-Expression, Genetic Interactions | |
| CNTNAP2 | Genetic Interactions | |
| PRMT2 | Genetic Interactions | |
| P2RY1 | Genetic Interactions | |
| SLC6A4 | NR3C2 | Co-Expression |
| PRMT2 | Co-Expression | |
| CNTNAP2 | Genetic Interactions | |
| MRFAP1 | Genetic Interactions | |
| CNTN2 | Genetic Interactions | |
| GILZ | FKBP5 | Co-Expression |
| AKR1B1 | Co-Expression | |
| EPB41L3 | Co-Expression | |
| ROBO1 | Genetic Interactions | |
| TSC22D1 | Genetic Interactions, Shared Protein Domains | |
| TSC22D2 | Physical Interactions, Shared Protein Domains | |
| TSC22D4 | Physical Interactions, Shared Protein Domains |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Theodoridou, D.; Tsiantis, C.-O.; Vlaikou, A.-M.; Chondrou, V.; Zakopoulou, V.; Christodoulides, P.; Oikonomou, E.D.; Tzimourta, K.D.; Kostoulas, C.; Tzallas, A.T.; et al. Developmental Dyslexia: Insights from EEG-Based Findings and Molecular Signatures—A Pilot Study. Brain Sci. 2024, 14, 139. https://doi.org/10.3390/brainsci14020139
Theodoridou D, Tsiantis C-O, Vlaikou A-M, Chondrou V, Zakopoulou V, Christodoulides P, Oikonomou ED, Tzimourta KD, Kostoulas C, Tzallas AT, et al. Developmental Dyslexia: Insights from EEG-Based Findings and Molecular Signatures—A Pilot Study. Brain Sciences. 2024; 14(2):139. https://doi.org/10.3390/brainsci14020139
Chicago/Turabian StyleTheodoridou, Daniela, Christos-Orestis Tsiantis, Angeliki-Maria Vlaikou, Vasiliki Chondrou, Victoria Zakopoulou, Pavlos Christodoulides, Emmanouil D. Oikonomou, Katerina D. Tzimourta, Charilaos Kostoulas, Alexandros T. Tzallas, and et al. 2024. "Developmental Dyslexia: Insights from EEG-Based Findings and Molecular Signatures—A Pilot Study" Brain Sciences 14, no. 2: 139. https://doi.org/10.3390/brainsci14020139
APA StyleTheodoridou, D., Tsiantis, C.-O., Vlaikou, A.-M., Chondrou, V., Zakopoulou, V., Christodoulides, P., Oikonomou, E. D., Tzimourta, K. D., Kostoulas, C., Tzallas, A. T., Tsamis, K. I., Peschos, D., Sgourou, A., Filiou, M. D., & Syrrou, M. (2024). Developmental Dyslexia: Insights from EEG-Based Findings and Molecular Signatures—A Pilot Study. Brain Sciences, 14(2), 139. https://doi.org/10.3390/brainsci14020139












