Transcriptomic Profile of Mouse Brain Ageing in Early Developmental Stages
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Microarray-Based Transcriptomic Profile Measurement
2.3. Bioinformatics Analysis
2.4. Data Description
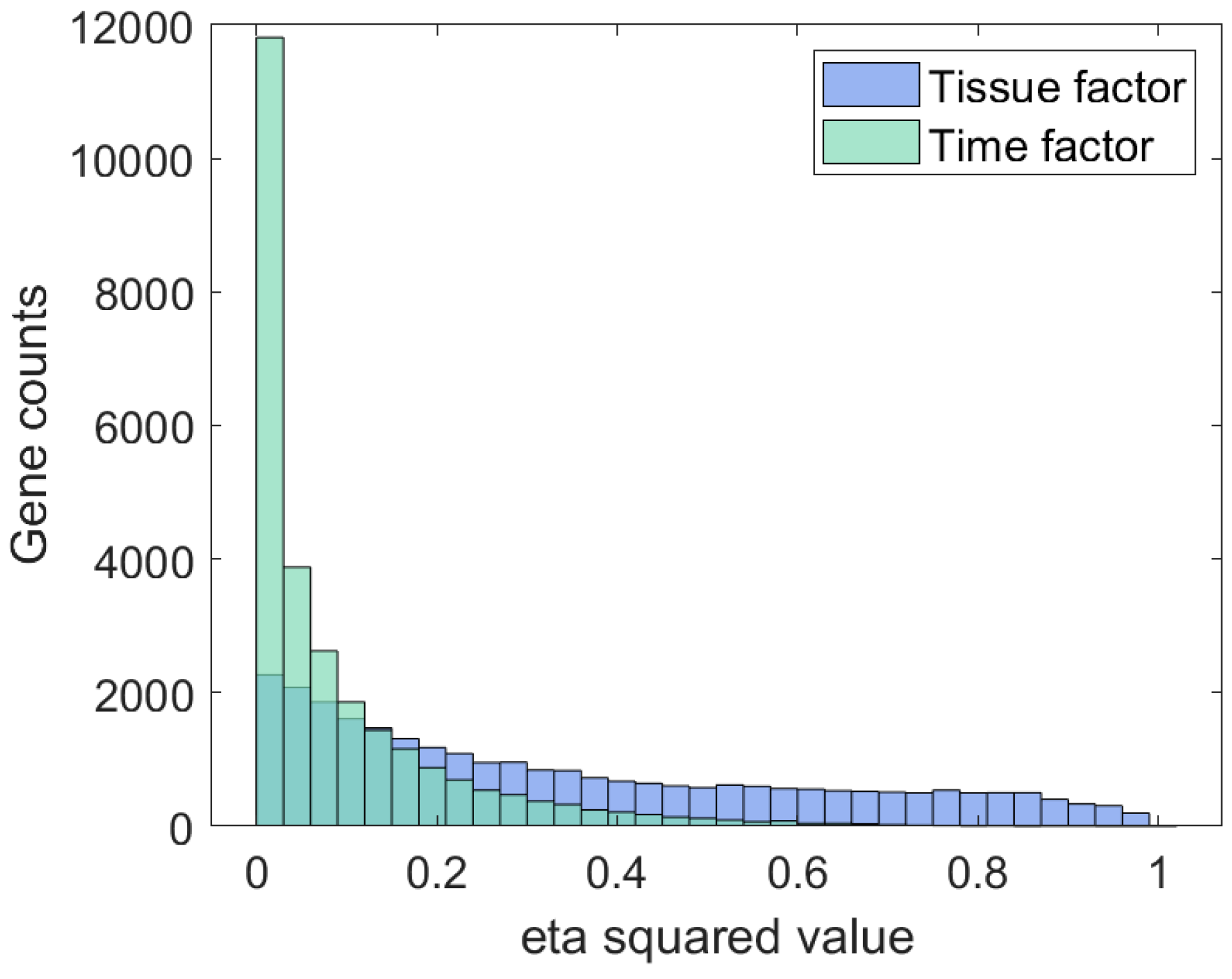
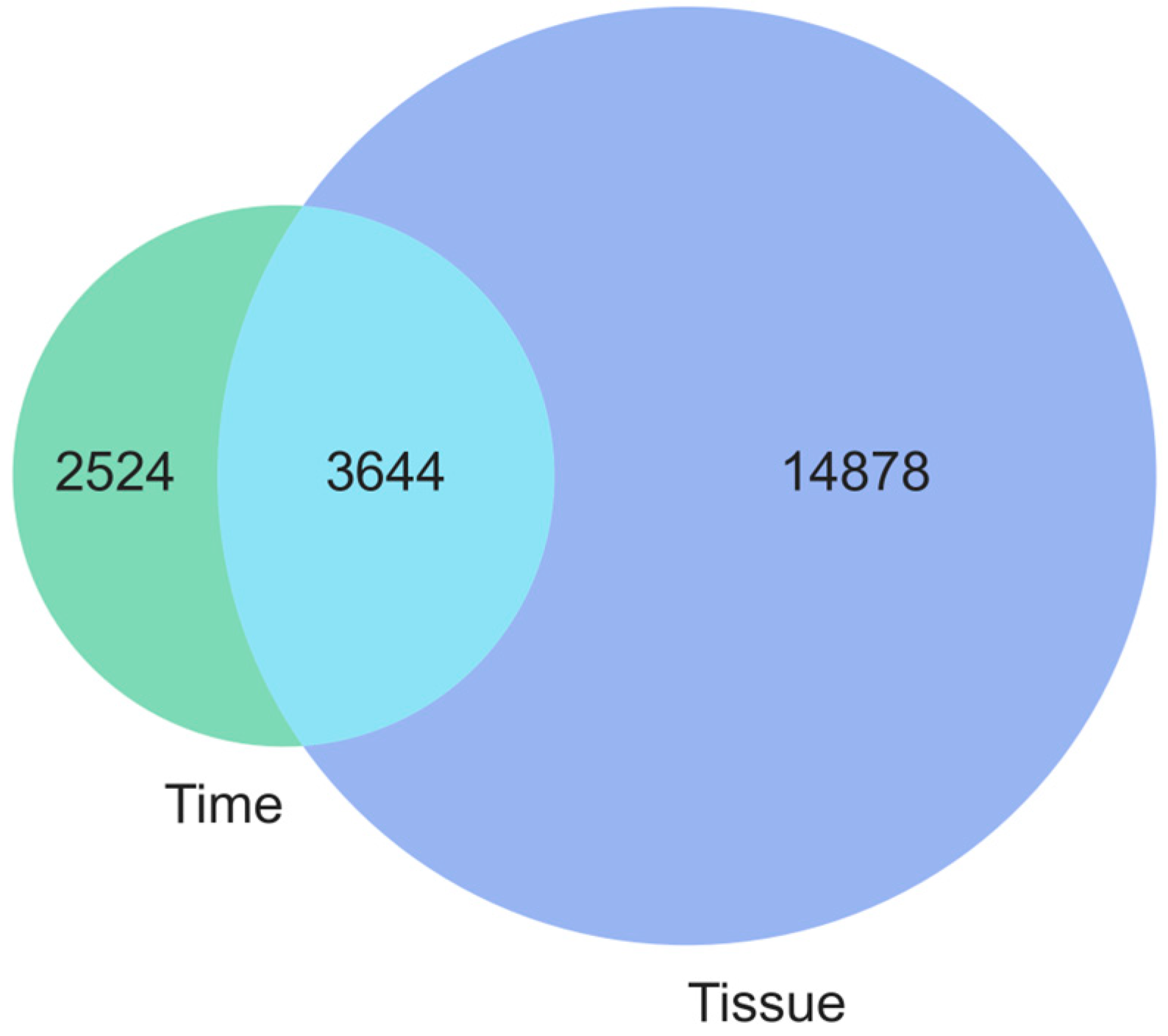
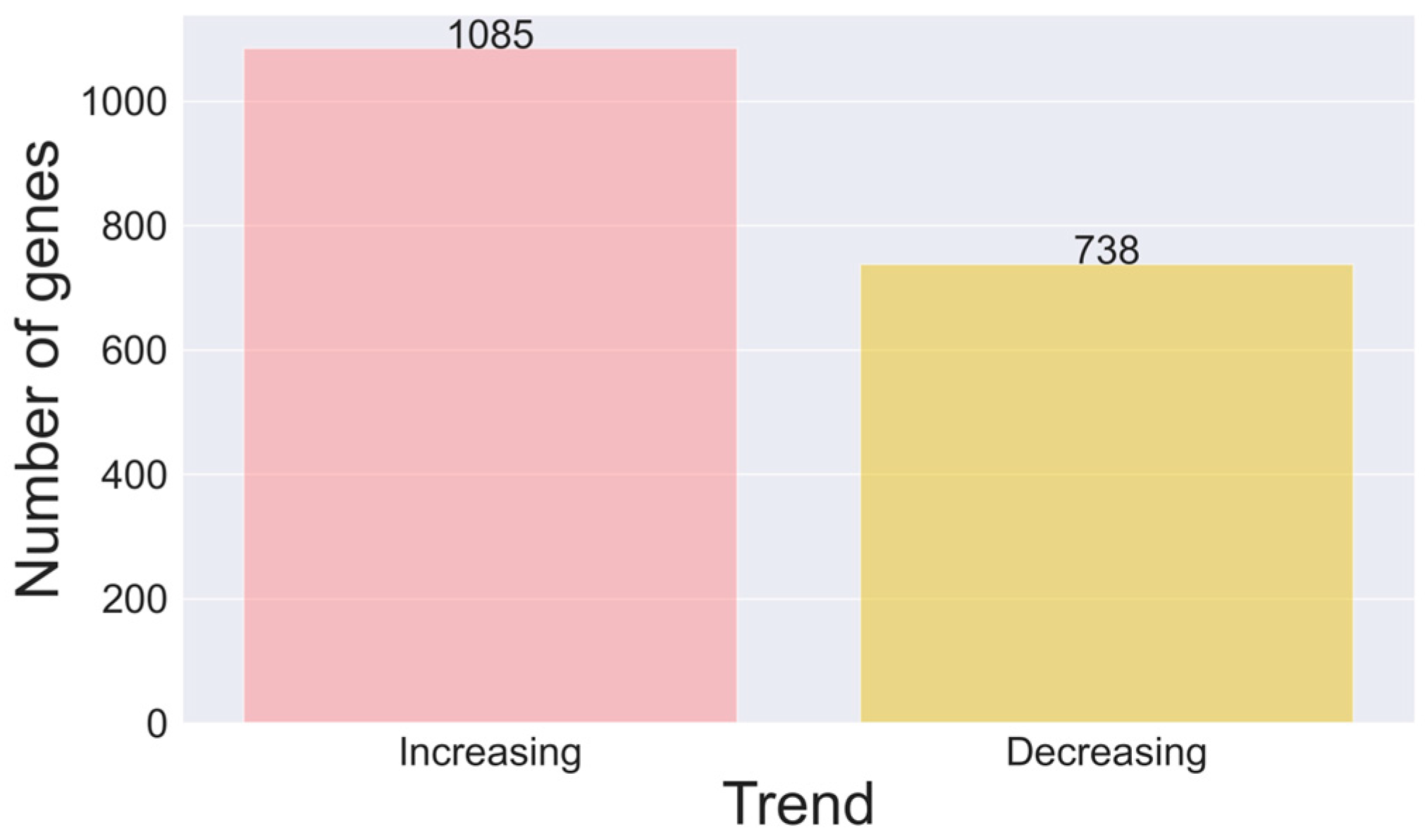
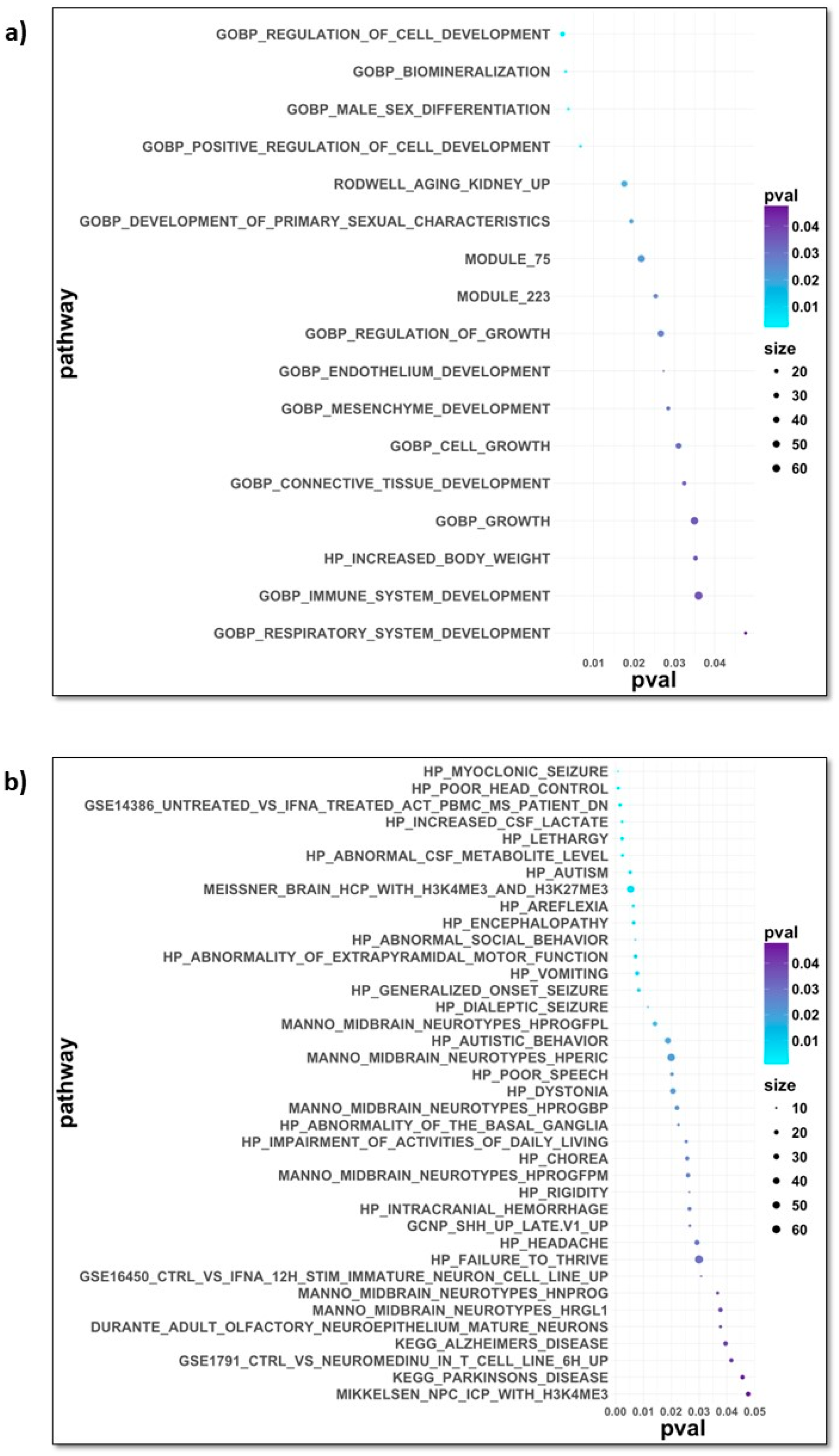
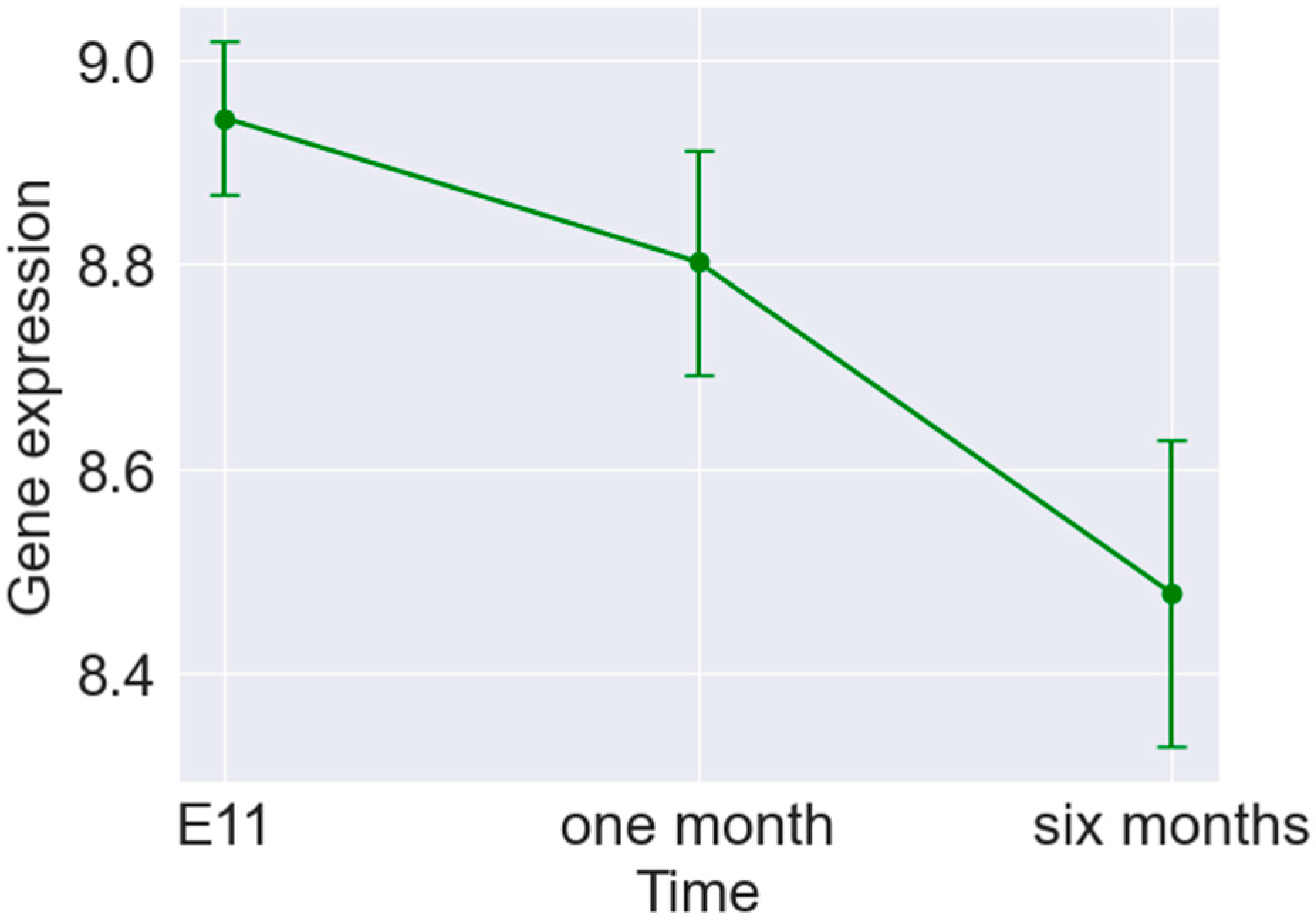
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Neutzner, M.; Kohler, C.; Frank, S.; Killer, H.E.; Neutzner, A. Impact of Aging on Meningeal Gene Expression. Fluids Barriers CNS 2023, 20, 12. [Google Scholar] [CrossRef]
- Srivastava, A.; Barth, E.; Ermolaeva, M.A.; Guenther, M.; Frahm, C.; Marz, M.; Witte, O.W. Tissue-Specific Gene Expression Changes Are Associated with Aging in Mice. Genom. Proteom. Bioinform. 2020, 18, 430–442. [Google Scholar] [CrossRef]
- Zhang, M.J.; Pisco, A.O.; Darmanis, S.; Zou, J. Mouse Aging Cell Atlas Analysis Reveals Global and Cell Type-Specific Aging Signatures. Elife 2021, 10, e62293. [Google Scholar] [CrossRef]
- Hahn, O.; Foltz, A.G.; Atkins, M.; Kedir, B.; Moran-Losada, P.; Guldner, I.H.; Munson, C.; Kern, F.; Palovics, R.; Lu, N.; et al. Atlas of the Aging Mouse Brain Reveals White Matter as Vulnerable Foci. Cell 2023, 186, 4117–4133.e22. [Google Scholar] [CrossRef]
- Martínez-Cué, C.; Rueda, N. Cellular Senescence in Neurodegenerative Diseases. Front. Cell. Neurosci. 2020, 14, 16. [Google Scholar] [CrossRef]
- Ibragimova, M.; Kussainova, A.; Aripova, A.; Bersimbaev, R.; Bulgakova, O. The Molecular Mechanisms in Senescent Cells Induced by Natural Aging and Ionizing Radiation. Cells 2024, 13, 550. [Google Scholar] [CrossRef]
- Guo, J.; Huang, X.; Dou, L.; Yan, M.; Shen, T.; Tang, W.; Li, J. Aging and Aging-Related Diseases: From Molecular Mechanisms to Interventions and Treatments. Signal Transduct. Target. Ther. 2022, 7, 391. [Google Scholar] [CrossRef]
- Flurkey, K.; Joanne, M.; Currer, D.E.H. Mouse Models in Aging Research. In The Mouse in Biomedical Research, 2nd ed.; American College of Laboratory Animal Medicine: Chester, NH, USA, 2007; Volume III, pp. 637–672. [Google Scholar]
- Radulescu, C.I.; Cerar, V.; Haslehurst, P.; Kopanitsa, M.; Barnes, S.J. The Aging Mouse Brain: Cognition, Connectivity and Calcium. Cell Calcium 2021, 94, 102358. [Google Scholar] [CrossRef]
- Castellano, J.M.; Mosher, K.I.; Abbey, R.J.; McBride, A.A.; James, M.L.; Berdnik, D.; Shen, J.C.; Zou, B.; Xie, X.S.; Tingle, M.; et al. Human Umbilical Cord Plasma Proteins Revitalize Hippocampal Function in Aged Mice. Nature 2017, 544, 488–492. [Google Scholar] [CrossRef]
- Wyss-Coray, T. Ageing, Neurodegeneration and Brain Rejuvenation. Nature 2016, 539, 180–186. [Google Scholar] [CrossRef]
- Dixon, W.J. Analysis of Extreme Values. Ann. Math. Stat. 1950, 21, 488–506. [Google Scholar] [CrossRef]
- Cohen, J. Statistical Power Analysis for the Behavioral Sciences; Routledge: New York, NY, USA, 1988. [Google Scholar]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene Set Enrichment Analysis: A Knowledge-Based Approach for Interpreting Genome-Wide Expression Profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Mootha, V.K.; Lindgren, C.M.; Eriksson, K.-F.; Subramanian, A.; Sihag, S.; Lehar, J.; Puigserver, P.; Carlsson, E.; Ridderstråle, M.; Laurila, E.; et al. PGC-1α-Responsive Genes Involved in Oxidative Phosphorylation Are Coordinately Downregulated in Human Diabetes. Nat. Genet. 2003, 34, 267–273. [Google Scholar] [CrossRef]
- Milacic, M.; Beavers, D.; Conley, P.; Gong, C.; Gillespie, M.; Griss, J.; Haw, R.; Jassal, B.; Matthews, L.; May, B.; et al. The Reactome Pathway Knowledgebase 2024. Nucleic Acids Res. 2024, 52, D672–D678. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING Database in 2023: Protein-Protein Association Networks and Functional Enrichment Analyses for Any Sequenced Genome of Interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef]
- Snel, B.; Lehmann, G.; Bork, P.; Huynen, M.A. STRING: A Web-Server to Retrieve and Display the Repeatedly Occurring Neighbourhood of a Gene. Nucleic Acids Res. 2000, 28, 3442–3444. [Google Scholar] [CrossRef]
- Zhou, Z.; Sun, B.; Yu, D.; Bian, M. Roles of TRNA Metabolism in Aging and Lifespan. Cell Death Dis. 2021, 12, 548. [Google Scholar] [CrossRef]
- Johann Holzmann, W.R. TRNA Recognition, Processing, and Disease: Hypotheses around an Unorthodox Type of RNase P in Human Mitochondria. Mitochondrion 2009, 9, 284–288. [Google Scholar] [CrossRef]
- Singh, P.K.; Badimon, A.; Chen, Z.L.; Strickland, S.; Norris, E.H. The Contact Activation System and Vascular Factors as Alternative Targets for Alzheimer’s Disease Therapy. Res. Pract. Thromb. Haemost. 2021, 5, e12504. [Google Scholar] [CrossRef]
- Nokkari, A.; Abou-El-Hassan, H.; Mechref, Y.; Mondello, S.; Kindy, M.S.; Jaffa, A.A.; Kobeissy, F. Implication of the Kallikrein-Kinin System in Neurological Disorders: Quest for Potential Biomarkers and Mechanisms. Prog. Neurobiol. 2018, 165–167, 26–50. [Google Scholar] [CrossRef]
- Li, Y.; Tian, X.; Luo, J.; Bao, T.; Wang, S.; Wu, X. Molecular Mechanisms of Aging and Anti-Aging Strategies. Cell Commun. Signal. 2024, 22, 285. [Google Scholar] [CrossRef]
- Romay, M.C.; Knutsen, R.H.; Ma, F.; Mompeón, A.; Hernandez, G.E.; Salvador, J.; Mirkov, S.; Batra, A.; Sullivan, D.P.; Procissi, D.; et al. Age-Related Loss of Notch3 Underlies Brain Vascular Contractility Deficiencies, Glymphatic Dysfunction, and Neurodegeneration in Mice. J. Clin. Investig. 2024, 134, e166134. [Google Scholar] [CrossRef]
- Stoka, V.; Turkb, V.; Bredesen, D.E. Differential Regulation of the Intrinsic Pathway of Apoptosis in Brain and Liver during Ageing. Bone 2008, 23, 3739–3745. [Google Scholar] [CrossRef]
- Schneider, A.; Chatterjee, S.; Bousiges, O.; Selvi, B.R.; Swaminathan, A.; Cassel, R.; Blanc, F.; Kundu, T.K.; Boutillier, A.L. Acetyltransferases (HATs) as Targets for Neurological Therapeutics. Neurotherapeutics 2013, 10, 568–588. [Google Scholar] [CrossRef]
- Confettura, A.D.; Cuboni, E.; Ammar, M.R.; Jia, S.; Gomes, G.M.; Yuanxiang, P.A.; Raman, R.; Li, T.; Grochowska, K.M.; Ahrends, R.; et al. Neddylation-Dependent Protein Degradation Is a Nexus between Synaptic Insulin Resistance, Neuroinflammation and Alzheimer’s Disease. Transl. Neurodegener. 2022, 11, 2. [Google Scholar] [CrossRef]
- Stroo, E.; Koopman, M.; Nollen, E.A.A.; Mata-Cabana, A. Cellular Regulation of Amyloid Formation in Aging and Disease. Front. Neurosci. 2017, 11, 64. [Google Scholar] [CrossRef]
- Grantham, J. The Molecular Chaperone CCT/TRiC: An Essential Component of Proteostasis and a Potential Modulator of Protein Aggregation. Front. Genet. 2020, 11, 172. [Google Scholar] [CrossRef]
- Sladitschek-Martens, H.L.; Guarnieri, A.; Brumana, G.; Zanconato, F.; Battilana, G.; Xiccato, R.L.; Panciera, T.; Forcato, M.; Bicciato, S.; Guzzardo, V.; et al. YAP/TAZ Activity in Stromal Cells Prevents Ageing by Controlling CGAS–STING. Nature 2022, 607, 790–798. [Google Scholar] [CrossRef]
- Westphal, D.; Kluck, R.M.; Dewson, G. Building Blocks of the Apoptotic Pore: How Bax and Bak Are Activated and Oligomerize during Apoptosis. Cell Death Differ. 2014, 21, 196–205. [Google Scholar] [CrossRef]
- Shi, M.M.; Shi, C.H.; Xu, Y.M. Rab GTPases: The Key Players in the Molecular Pathway of Parkinson’s Disease. Front. Cell. Neurosci. 2017, 11, 81. [Google Scholar] [CrossRef]
- Loerch, P.M.; Lu, T.; Dakin, K.A.; Vann, J.M.; Isaacs, A.; Geula, C.; Wang, J.; Pan, Y.; Gabuzda, D.H.; Li, C.; et al. Evolution of the Aging Brain Transcriptome and Synaptic Regulation. PLoS ONE 2008, 3, e3329. [Google Scholar] [CrossRef]
- GeneCards—The Human Gene Database. Available online: https://www.genecards.org (accessed on 6 February 2024).
- Brioschi, S.; Wang, W.L.; Peng, V.; Wang, M.; Shchukina, I.; Greenberg, Z.J.; Bando, J.K.; Jaeger, N.; Czepielewski, R.S.; Swain, A.; et al. Heterogeneity of Meningeal B Cells Reveals a Lymphopoietic Niche at the CNS Borders. Science 2021, 373, eabf9277. [Google Scholar] [CrossRef]
- Liu, H.; Yang, Y.; Xia, Y.; Zhu, W.; Leak, R.K.; Wei, Z.; Wang, J.; Hu, X. Aging of Cerebral White Matter. Ageing Res. Rev. 2017, 34, 64–76. [Google Scholar] [CrossRef]
- Schroer, A.B.; Ventura, P.B.; Sucharov, J.; Misra, R.; Chui, M.K.K.; Bieri, G.; Horowitz, A.M.; Smith, L.K.; Encabo, K.; Tenggara, I.; et al. Platelet Factors Attenuate Inflammation and Rescue Cognition in Ageing. Nature 2023, 620, 1071–1079. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kulis, K.; Tabury, K.; Benotmane, M.A.; Polanska, J. Transcriptomic Profile of Mouse Brain Ageing in Early Developmental Stages. Brain Sci. 2024, 14, 581. https://doi.org/10.3390/brainsci14060581
Kulis K, Tabury K, Benotmane MA, Polanska J. Transcriptomic Profile of Mouse Brain Ageing in Early Developmental Stages. Brain Sciences. 2024; 14(6):581. https://doi.org/10.3390/brainsci14060581
Chicago/Turabian StyleKulis, Karolina, Kevin Tabury, Mohammed Abderrafi Benotmane, and Joanna Polanska. 2024. "Transcriptomic Profile of Mouse Brain Ageing in Early Developmental Stages" Brain Sciences 14, no. 6: 581. https://doi.org/10.3390/brainsci14060581




